Abstract
Nuclear receptors and their coactivators have been shown to function as key regulators of adipose tissue biology. Here we show that a ligand-dependent transcriptional repressor for nuclear receptors plays a crucial role in regulating the balance between energy storage and energy expenditure. Mice devoid of the corepressor protein RIP140 are lean, show resistance to high-fat diet-induced obesity and hepatic steatosis, and have increased oxygen consumption. Although the process of adipogenesis is unaffected, expression of certain lipogenic enzymes is reduced. In contrast, genes involved in energy dissipation and mitochondrial uncoupling, including uncoupling protein 1, are markedly increased. Therefore, the maintenance of energy homeostasis requires the action of a transcriptional repressor in white adipose tissue, and ligand-dependent recruitment of RIP140 to nuclear receptors may provide a therapeutic target in the treatment of obesity and related disorders.
Energy homeostasis is a highly regulated process that requires precise control of food intake and energy expenditure (1). The major and most efficient storage of energy occurs in the form of triglycerides in white adipose tissue (WAT), and it is now clear that the adipocyte itself may act as an endocrine cell such that altered adipocyte function would cause changes in systemic energy balance (2, 3). From adipogenic stores, fatty acids are easily mobilized during periods of energy restriction or increased physical activity to provide enough fuel for energy synthesis in the form of ATP. In addition, energy is dissipated by generating heat in brown adipose tissue (BAT) and skeletal muscles by regulating the uncoupling of ATP production from respiration. Many of these metabolic processes are controlled in part by nuclear receptors (4, 5), including peroxisome proliferator-activated receptors (PPARs) (6, 7), thyroid hormone receptor (8, 9), estrogen receptor α (ERα) (10, 11), and ER-related receptor α (ERRα) (12). The best characterized of these are the PPARs, with PPARγ and PPARα playing an essential role in adipogenesis (13-16) and in thermogenesis and fatty acid oxidation (17-19), respectively, whereas recent studies have implicated a role for PPARδ in lipid homeostasis (20).
Nuclear receptors stimulate target gene transcription by recruiting coactivators that are required to remodel chromatin and facilitate the assembly of the basal transcription machinery (21). The key coactivator in metabolic processes is the PPARγ coactivator 1α (PGC-1α) (22-25), which was initially shown to promote adaptive thermogenesis in BAT and which has emerged as a target for integrating signals in the regulation of specific metabolic programs in other tissues, including muscle and liver. More recently, the related coactivator PGC-1β (26, 27) has been implicated in the regulation of energy expenditure as a potential coactivator for ERRα (28). In addition, the p160 family of coactivators has also been found to control energy balance in adipose tissue. For instance, SRC1, in association with PGC-1α, enhances thermogenesis in BAT, whereas TIF2 promotes fat accumulation in WAT (29).
In contrast to nuclear receptor-dependent coactivator recruitment, relatively little is known about the role of nuclear receptor corepressors in adipogenesis and adipocyte function. In the absence of ligands, many nuclear receptors prevent gene transcription by recruitment of corepressors such as N-CoR (30) and SMRT (31), which are proposed to function by antagonizing the actions of coactivators and maintaining a more repressed state in chromatin structure. It has been shown that enhanced corepressor binding to mutant forms of PPARγ blocks the thiazolidinedione-induced differentiation of primary human adipocytes by acting as a dominant negative form of the receptor (32). Transcription by nuclear receptors may also be inhibited in the presence of ligand by the recruitment of corepressors such as RIP140 and L-CoR, which are reported to function by both histone deacetylase-dependent and -independent mechanisms (33-37). In the case of RIP140, this inhibition involves the action of four independent repression domains (38) as well as the recruitment of additional repressor proteins such as the C-terminal binding protein (CtBP) (39, 40).
In this study, we provide evidence that transcriptional repression plays a crucial role in controlling the balance between fat accumulation and energy dissipation. In particular, the absence of the RIP140 corepressor results in increased expression in WAT of key regulatory genes involved in energy metabolism, including uncoupling protein 1 (UCP1), which is typically expressed in BAT and plays a major role in uncoupling ATP synthesis from respiration (4, 41). Thus, the genetic ablation of RIP140 in mice leads to a lean phenotype and resistance to diet-induced obesity.
Experimental Procedures
Animals. The generation of RIP140-null mice has previously been described (42). Mice used in this study were backcrossed eight generations to C57BL/6J background, except those used for a glucose tolerance test, which were on a 129/BL6 background. Mice were maintained under standard conditions, with controlled light and temperature, and fed a chow diet ad libitum, except for high-fat diet experiments wherein mice were fed a 35% (wt/wt) fat diet (Lillico Biotechnology, Betchnorth, U.K.). Leptin measurements were taken by using the Linco Research (St. Charles, MO) Mouse Leptin RIA kit. Triglycerides were measured by using Triglyceride GPO-Trinder reagent (Sigma), and free fatty acids were determined by using ADIFAB free fatty acid indicator (Molecular Probes). Oxygen consumption was measured in WT and RIP140-null mice fed a chow diet (seven mice per group) by using oxymax system version 4.66 (Columbus Instruments, Columbus, OH) with room air as a reference. Animals were placed individually in 0.3-liter chambers. Results are expressed as ml/kg per min. All assays were carried out according to the manufacturers' protocols. All experiments were performed according to guidelines of the Home Office of the United Kingdom.
MRI and Magnetic Resonance Spectroscopy (MRS). Mice were scanned by using a 4.7T Varian system. Whole-body images (between 40 and 45 slices, each 2 mm thick) were obtained for each mouse by using a spin-echo sequence [repetition time (TR)/echo time (TE) = 4,500:20]. Whole-body spectra were obtained by using a TR of 10 s. Localized proton spectra of liver and muscle were obtained from a 3 × 3 × 3-mm voxel by using a PRESS sequence (TR/TE = 10,000:14).
Morphological and Immunohistochemical Analysis. Tissues were fixed in neutral buffered formalin, embedded in paraffin, and sectioned at 5 μm onto poly-l-lysine-coated slides. For histology, sections were stained with hematoxylin/eosin. For immunohistochemistry, deparaffinized sections were incubated in 0.3% hydrogen peroxide (Sigma) in methanol for 30 min to inactivate endogenous peroxidase, rinsed in PBS, and incubated in 1:75 normal goat serum/PBS for 30 min to reduce nonspecific background staining. Sections were incubated overnight at 4°C with polyclonal rabbit anti-mouse primary antibody to UCP1 (AB3038, Chemicon) diluted 1:800 in PBS. Primary antibody was detected by using the Vectastain Elite ABC kit (Vector Laboratories), with enzymatic detection by using 0.25 mg/ml diaminobenzidine (Sigma) and 0.06% hydrogen peroxide in PBS. Sections were counterstained with hematoxylin. Frozen sections (10 μm) of formalin-fixed liver were mounted on poly-l-lysine-coated slides and stained with Oil red O (0.15% in 60% isopropanol) for 5 min. Sections were counterstained with hematoxylin and mounted in glycerol gelatin (Sigma).
Cell Culture of Mouse Embryonic Fibroblasts (MEFs) and 3T3-L1 Cells. MEFs were isolated from 12-day-old embryos and cultured in DMEM/F12 media supplemented with 10% fetal bovine serum (Invitrogen). For differentiation experiments, MEFs cultured for four to six passages were used. 3T3-L1 cells were cultured in DMEM/F12 media supplemented with 10% newborn bovine serum (Invitrogen). Differentiation of 2-day-postconfluent MEFs and 3T3-L1 cells was performed as previously described (43) with the modification that MEFs were also supplemented with 2.5 nM rosiglitazone. Differentiated cells were visualized with Oil red O staining. β-Galactosidase activity was analyzed as previously described (42).
Expression Analysis. Total RNA was isolated by using TRIzol (Invitrogen) according to the manufacturer's instructions. To obtain first-strand cDNA for further analysis, 1 μg of total RNA was treated with DNase, and cDNA was prepared by using the SuperScript First-Strand Synthesis System for RT-PCR according to the manufacturer's instructions (Invitrogen). Real-time PCR was performed with the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). RIP140 and L19 expression was determined by using specific primers and TaqMan probes. Expression of all other genes was determined with SYBR-green reagent and specific primers according to the manufacturer's instructions. Expression levels for all genes were correlated to the mean value of two independent internal controls, the ribosomal coding gene L19 and cyclophilin. Primer and probe sequences are available on request.
Results
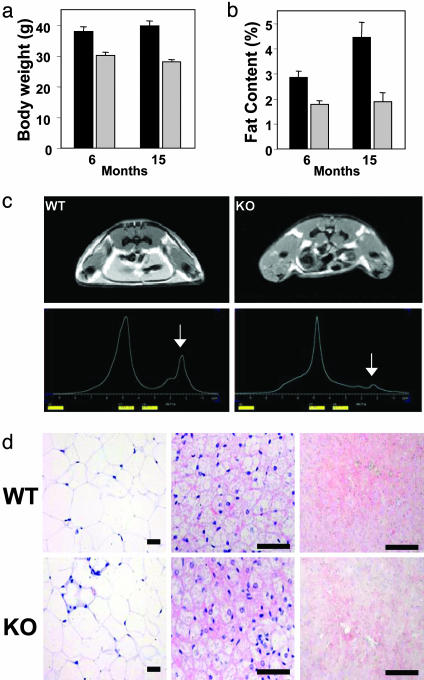
RIP140-Null Mice Exhibit a Lean Phenotype. RIP140-null mice are lean with a 20% reduction in body weight in both males and females compared with WT mice, both of which increase with age (Fig. 1a and data not shown). Total body fat content was reduced by ≈70% as analyzed by whole-body proton magnetic resonance spectroscopy (MRS), whereas the weight of inguinal fat was reduced by 40-60%, according to the age of the mice (Fig. 1 b and c). MRI and MRS of whole-body fat content revealed the almost complete absence of s.c. fat and a marked decrease in other fat depots (Fig. 1c). The reduction in body weight and fat content was not due to increased physical activity as judged by open field activity measurements (data not shown). In addition, food intake was 4.93 ± 0.37 and 4.70 ± 0.20 g per mouse per day in WT and RIP140-null mice respectively, corresponding to a small increase in the RIP140-null mice relative to total body weight. In conclusion, these observations suggest that the molecular mechanisms controlling energy expenditure may be altered in RIP140-null mice.
Fig. 1.
Metabolic phenotype of RIP140-null mice is shown. (a) Body weights of WT (black) and RIP140-null (gray) mice fed a regular chow diet at 6 months (n = 7) and 15 months (WT, n = 14; RIP140-null, n = 11) of age. (b) Inguinal WAT weights of WT (black) and RIP140-null (gray) mice at 6 months (n = 7 for both groups) and 15 months (WT, n = 14; RIP140-null, n = 11) of age. (c) MRI and magnetic resonance spectroscopy (MRS) of body fat content. Whole-body proton content is quantified, with the lipid peak indicated. Calculations from three representative animals revealed an ≈70% reduction of total fat content in RIP140-null (KO) mice. (d) Morphology of metabolic tissues from WT and RIP140-null mice. WAT (inguinal), BAT, and liver were isolated from WT and RIP140-null (KO) mice fed a control chow diet. WAT and BAT were stained with hematoxylin/eosin. Liver tissues were stained with Oil red O to demonstrate lipid accumulation and counterstained with hematoxylin. (Scale bars = 50 μm.)
Histological analysis of the inguinal WAT indicates that the diameter of adipocytes was reduced from 63.2 ± 3.7 μm in WT mice to 45.2 ± 3.5 μm in RIP140-null mice (Fig. 1d). This reduction is equivalent to a decrease in volume of ≈2-fold, indicating that the reduction in epididymal fat results from a decrease in cell size proportional to the decrease in fat content rather than to adipocyte number. The size and appearance of BAT cells was similar in both WT and RIP140-null mice. It is conceivable that decreased fat accumulation in WAT may have occurred in the context of a lipodystrophic phenotype; however, we observed no evidence that fat was being stored in alternate tissues, including liver and BAT, indicating that this was not the case (Fig. 1d). We also performed a glucose tolerance test in male mice after an overnight fast, but no statistically significant differences were observed (see Fig. 6, which is published as supporting information on the PNAS web site).
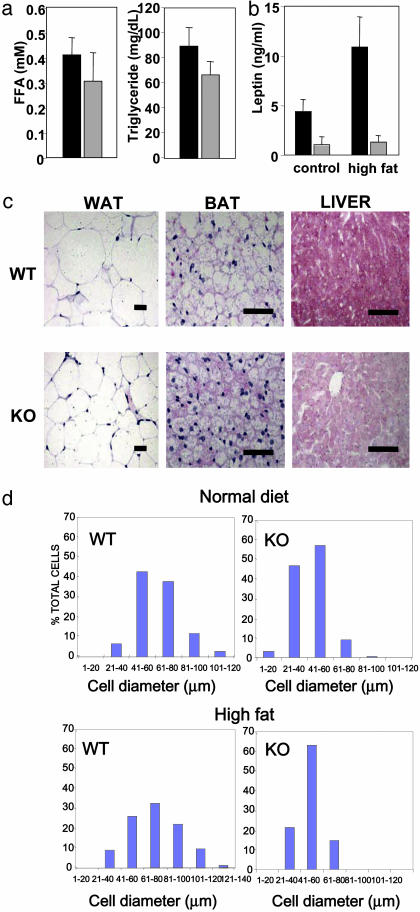
We next determined how the RIP140-null mice responded to the effects of being fed a high-fat diet for 10 days. The average weight gain in WT mice was 5.3 ± 0.29 g, corresponding to a 14.5% increase in body weight, whereas RIP140-null mice gained only 1.0 ± 0.15 g, corresponding to a 3.8% increase in body weight. Serum biochemistry did not reveal increased free fatty acids and hypertriglyceridemia (Fig. 2a). As expected, the reduced fat content in adipose tissue leads to a reduction in circulating leptin levels (Fig. 2b). Histology (Fig. 2c) and quantitative analysis of cell size in WAT (Fig. 2d) showed an increase in cell diameter to 69.9 ± 3.6 μm in WT mice and 50.7 ± 1.3 μm in RIP140-null mice. Thus, the 2-fold reduction in cell volume observed in RIP140-null mice was maintained as previously determined on a normal chow diet. An increase in cell size was also observed in BAT cells in WT mice fed the high-fat diet but not in RIP140-null mice. More importantly, Oil red O staining indicated a marked increase in triglyceride accumulation in the liver of WT mice fed a high-fat diet that was absent in RIP140-null mice (Fig. 2d), further supporting the nonlipodystrophic nature of the adipose tissue alterations. This difference was also evident in mice fed a normal diet as they aged, suggesting that RIP140-null mice were protected from hepatic steatosis. Comparison of oxygen consumption by WT and RIP140-null mice revealed an increase of 8.9% in RIP140-null mice (66.8 ± 5.1 ml/kg per min, n = 8) compared with WT animals (61.3 ± 4.3 ml/kg per min, n = 7) (P < 0.05). Thus, we conclude that RIP140-null mice are lean because they fail to store triglycerides and, strikingly, are resistant to high-fat diet-induced obesity, indicating that alternative mechanisms are involved in the dissipation of excess fuel.
Fig. 2.
Histology of metabolic tissues and serum biochemistry is shown. (a) Serum free fatty acid (FFA) levels and triglyceride levels in WT (black) and RIP140-null (gray) mice fed a high-fat diet for 10 days (n = 6). (b) Serum levels of leptin in WT (black) or RIP140-null (gray) mice (n = 3-6) fed chow or a high-fat diet. (c) Morphology of inguinal WAT, BAT, and liver from WT and RIP140-null (KO) mice fed a high-fat diet (35% wt/wt) for 10 days. Tissues were stained as in Fig. 1d. (Scale bars = 50 μm.) (d) Comparison of cell size in WAT of WT and RIP140-null mice maintained on normal chow and high-fat (35%) diet.
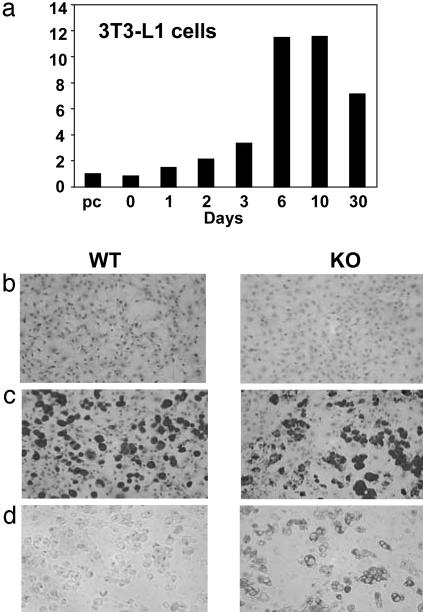
Impaired Adipogenesis Does Not Account for the Lean Phenotype of RIP140-Null Mice. A recent expression-profiling analysis of 3T3-L1 cells indicated that there was a marked, progressive increase in RIP140 expression during differentiation into adipocytes (43), which we confirmed by quantitative real-time PCR analysis and showed that maximum levels are reached and maintained 3-6 days after induction of differentiation (Fig. 3a). Given these observations and the lean phenotype exhibited by RIP140-null mice, we investigated the role of RIP140 in the differentiation and function of adipocytes.
Fig. 3.
RIP140 expression and adipocyte differentiation in vitro is shown. (a) Regulation of RIP140 mRNA levels during differentiation of 3T3-L1 adipocyte cells. Differentiation was induced in 2-day-confluent cells by using a standard hormone mixture of insulin, dexamethasone, and 3-isobutyl-1-methylxanthine preconfluent cells (pc). mRNA was quantitated by TaqMan real-time PCR analysis. (b and c) Embryonic fibroblasts at confluence (b) and after differentiation by using hormone mixture as above but with addition of a PPAR agonist, rosiglitazone (c), were stained with Oil red O. Differentiated cells were also subject to β-galactosidase activity analysis to demonstrate RIP140 promoter activity (d). Note the lack of β-galactosidase activity in WT cells and the correlation of fat droplet-containing cells staining with oil red O and β-galactosidase.
Initially we tested whether RIP140 was required for adipocyte differentiation by comparing the ability of fibroblasts from WT and RIP140-null embryos to differentiate in vitro. We found that confluent fibroblasts treated with a standard hormone differentiation mixture in the absence or presence of the PPARγ agonist rosiglitazone were fully capable of differentiating into adipocytes, as judged by Oil red O staining, even when cells were devoid of the RIP140 gene (Fig. 3 b and c and data not shown). Nevertheless, we were able to demonstrate a link between adipocyte differentiation and RIP140 gene transcription by analyzing the expression of the β-galactosidase gene, which has been inserted in place of the RIP140 coding sequence in the null mice (42). Fig. 3 b-d indicates that Oil red O-positive cells and β-galactosidase activity were detected only in differentiated cells, suggesting that the adipose phenotype is required for activation of expression from the RIP140 promoter. The ability of embryonic fibroblasts to differentiate in the absence of RIP140 and the maintenance of some WAT in the null animals demonstrates that adipogenesis does not depend on this nuclear receptor corepressor. We therefore analyzed the in vivo role of RIP140 in maintaining the function of adipose and other metabolic tissues.
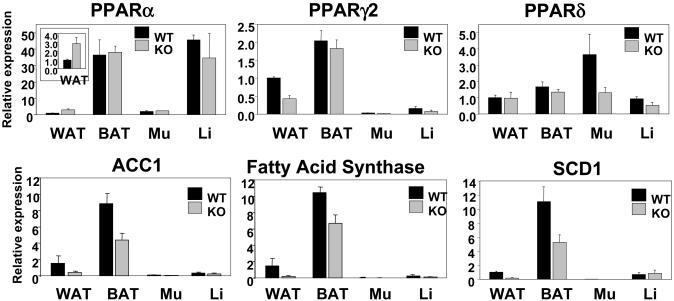
Expression Analysis of Regulators of Lipid Metabolism. To investigate potential molecular mechanisms leading to defects in WAT devoid of RIP140, we performed a detailed gene expression profile analysis in this tissue and in parallel studies also determined expression in BAT, muscle, and liver. Nuclear receptors, in particular the PPARs and thyroid receptors, play a central role in mediating metabolic signaling pathways, so we examined the expression of these transcriptional regulatory factors in metabolic tissues. In all cases, we found that the expression levels were similar in WT and RIP140-null mice, with the exception of WAT, wherein PPARα was increased ≈3-fold and PPARγ2 was reduced 2-fold (Fig. 4). Expression of thyroid hormone receptors α and β were unaltered or decreased by <2-fold in the tissues examined (Fig. 7, which is published as supporting information on the PNAS web site). Levels of expression of PGC-1α were not elevated in RIP140-null mice, and levels of SRC1 and TIF2, which have been found to control energy balance between WAT and BAT, were not altered (Fig. 7). Similarly, expression of the adipogenic factor C/EBPβ was unaffected, whereas C/EBPα and SREBP1c were reduced by 50-60% in WAT from RIP140-null mice but not in other tissues (Fig. 7). Consistent with these reductions, there was a decrease in the expression of corresponding target genes, including adiponectin, glucose transporter 4 (Glut4), and aP2 (Fig. 6). The expression of genes encoding the lipogenic enzymes acetyl CoA carboxylase 1 (ACC1), fatty acid synthase (FAS), and stearoyl CoA desaturase (SCD1) were reduced by up to 6-fold in WAT (Fig. 4). However, no differences were found in expression of ACC2, in acetyl CoA oxidase (ACOX1/AOX), in additional factors involved in the regulation of fat metabolism including leptin receptor, insulin receptor, and FOXC2, or in markers of mitochondrial biogenesis including ATP synthase subunit 8 and cytochrome c1 (Fig. 7).
Fig. 4.
Expression profile of adipogenic regulators and marker genes in metabolic tissues. TaqMan real-time PCR analysis of gene expression in metabolic tissues from WT (black) and RIP140-null (gray) mice (n = 3-7). Mu, skeletal muscle; Li, liver.
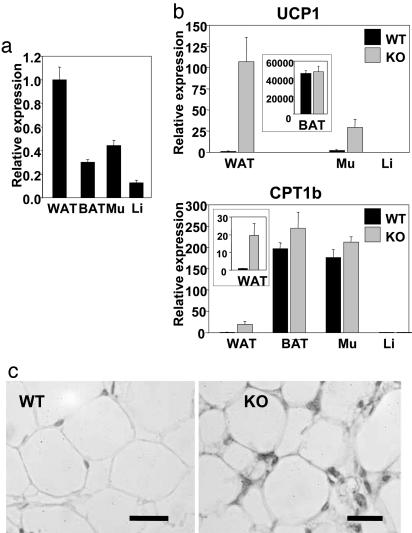
Up-Regulation of Genes Involved in Energy Dissipation in WAT. A survey of mouse tissues indicates that RIP140 mRNA is widely expressed with highest levels in WAT, followed by skeletal muscle, and with lower levels in BAT and liver (Fig. 5a). In contrast to the relatively small differences in RIP140-null animals in transcriptional regulators and many metabolic enzymes and markers in these tissues, we observed increased expression in WAT of carnitine palmitoyltransferase 1b (CPT1b; >20-fold) and the de novo expression of mitochondrial UCP1 (>100-fold); furthermore UCP1 was up-regulated by >10-fold in muscle (Fig. 5b). CPT1b is necessary for the transport of free fatty acids across the outer mitochondrial membrane (44, 45) and together with UCP1 is essential for thermogenesis in BAT. Immunohistochemical analysis shows the appearance of UCP1 protein in both unilocular and multilocular adipocytes in WAT from RIP140-null mice (Fig. 5c and data not shown). The staining for UCP1 is predominantly localized in the cytoplasmic area near the plasma membrane, similar to that described in mice expressing an aP2-Ucp1 transgene (46). Thus, up-regulation of CPT1b and UCP1 in RIP140-null mice may increase mitochondrial respiration and energy uncoupling resulting in reduced storage or depletion of fat in metabolic tissues.
Fig. 5.
Up-regulation of genes involved in energy dissipation in WAT. (a) TaqMan real-time PCR analysis of RIP140 mRNA levels in metabolic tissues. Tissue samples were from WAT, BAT, skeletal muscle (Mu), and liver (Li) from WT mice (n = 3-7). (b) TaqMan real-time PCR analysis of UCP1 and CPT1b gene expression in metabolic tissues from WT (black) and RIP140-null (gray) mice (n = 3-7). (c) Immunostaining of UCP1 expression in WAT from WT and RIP140-null (KO) mice. (Scale bars = 50 μm.)
Discussion
Previous work has clearly established the importance of transcriptional activation by nuclear receptors in both the control of adipogenesis and the regulation of energy balance by thermogenesis, with PPARs playing a central role (4-12). Spiegelman and coworkers (22, 23, 24) have provided substantial evidence to indicate that PGC-1α is a key transcriptional coactivator and metabolic regulator in BAT, muscle, and liver. This coactivator plays a crucial role in integrating cellular signals controlling energy balance and nutrient homeostasis by the coordination and induction of gene expression in a cell type- and tissue-specific manner. Recent studies also suggest a role for PGC-1β on the action of ERRα in adipose tissue and muscle (26-28, 47), whereas the balance among SRC1, TIF2, and PGC-1α appears to modulate energy homeostasis between WAT and BAT compartments (29).
In contrast, here we demonstrate an essential role for the corepressor RIP140 in the maintenance of energy homeostasis by regulating expression of lipogenic enzymes and preventing the expression of key metabolic genes in WAT. Transcriptional regulation of genes encoding enzymes such as SCD1, ACC1, and FAS is complex, and the direct targets for RIP140 action have yet to be elucidated. In BAT, transcription from the UCP1 gene may be regulated by PPARs, retinoids, and thyroid hormone, as well as through the activation of β-adrenergic receptor intracellular signaling pathways (19, 48-51). Expression of UCP1 has been shown to be induced in WAT in transgenic mice by introduction of an activated form of PPARδ (20) and also in human white adipocytes in culture as a result of expression of PGC-1α and activation by ligands for PPARγ (52). RIP140 is capable of interacting directly with all of these receptors in a ligand-dependent manner and of repressing their transcriptional activity (34, 35, 53). Although endogenous high-affinity ligands have not yet been identified for the PPARγ and PPARδ isoforms, it is apparent that the availability of PPARα ligands such as oleylethanolamide may influence the regulation of body weight (54). It is evident from numerous studies that the balance of levels of receptors, coactivators, and ligand availability is fundamental in controlling energy homeostasis. The results described in this study demonstrate the importance of ligand-dependent corepressor recruitment in this process. Recent studies have demonstrated that RIP140 has intrinsic repressive activity contained within four independent repression domains within the protein (37-40), although the role of these in adipose biology is yet to be determined. The temporal and cell type-specific control of RIP140 expression, however, provides an additional level of regulation of repressor function.
In primary MEFs induced to differentiate, both adipogenesis and the potential to store fat are reduced in the absence of the p160 coactivator TIF2 (29), and adipogenesis also depends on the action of the PPARγ cofactor TRAP220 (55). Thus, the delayed onset of RIP140 expression may be necessary to avoid interference with the action of PPARγ during the process of adipogenesis while facilitating a role in regulating the activity of the differentiated adipocyte. The up-regulation of genes involved in energy dissipation and mitochondrial uncoupling could clearly compromise the function of the adipocyte as a site of energy storage in the form of triglycerides.
The increasing prevalence of obesity may be considered to be a consequence of both increased energy availability and reduced energy expenditure superimposed on a genetic background evolved for survival in conditions of food deprivation (56). This study identifies a signal in the metabolic pathways regulating energy balance. In conclusion, our observations show that RIP140 plays a crucial role in energy homeostasis and demonstrate a role for corepressor action in addition to coactivator recruitment in determining adipocyte function. Because the interaction of RIP140 with nuclear receptors is a ligand-dependent process, specific recruitment of this corepressor may provide a therapeutic target for the treatment of obesity and related disorders.
Supplementary Material
Acknowledgments
We thank A. Soutar and members of the Molecular Endocrinology Laboratory for advice and comments on the manuscript; S. O'Rahilly for advice and discussion; and S. Macchiarulo and the staff of the CBS unit for technical assistance. This work was supported by Wellcome Trust U.K. Programme Grant 061930 (to M.G.P.), Cambridge Oxford Wellcome Trust Integrative Physiology Program Grant 065326 (to A.V.-P. and G.M.-G.), and Biotechnology and Biological Sciences Research Council Grant 28-S15893 (to S.M. and M.G.P.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: WAT, white adipose tissue; BAT, brown adipose tissue; PPAR, peroxisome proliferator-activated receptor; PGC, PPARγ coactivator; UCP1, uncoupling protein 1; MEF, mouse embryonic fibroblast.
References
- 1.Spiegelman, B. M. & Flier, J. S. (2001) Cell 104, 531-543. [DOI] [PubMed] [Google Scholar]
- 2.Ahima, R. S. & Flier, J. S. (2000) Trends Endocrinol. Metab. 11, 327-332. [DOI] [PubMed] [Google Scholar]
- 3.Rajala, M. W. & Scherer, P. E. (2003) Endocrinology 144, 3765-3773. [DOI] [PubMed] [Google Scholar]
- 4.Lowell, B. B. & Spiegelman, B. M. (2000) Nature 404, 652-660. [DOI] [PubMed] [Google Scholar]
- 5.Francis, G. A., Fayard, E., Picard, F. & Auwerx, J. (2003) Annu. Rev. Physiol. 65, 261-311. [DOI] [PubMed] [Google Scholar]
- 6.Desvergne, B. & Wahli, W. (1999) Endocr. Rev. 20, 649-688. [DOI] [PubMed] [Google Scholar]
- 7.Lee, C. H., Olson, P. & Evans, R. M. (2003) Endocrinology 144, 2201-2207. [DOI] [PubMed] [Google Scholar]
- 8.Ribeiro, M. O., Carvalho, S. D., Schultz, J. J., Chiellini, G., Scanlan, T. S., Bianco, A. C. & Brent, G. A. (2001) J. Clin. Invest. 108, 97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viguerie, N., Millet, L., Avizou, S., Vidal, H., Larrouy, D. & Langin, D. (2002) J. Clin. Endocrinol. Metab. 87, 630-634. [DOI] [PubMed] [Google Scholar]
- 10.Heine, P. A., Taylor, J. A., Iwamoto, G. A., Lubahn, D. B. & Cooke, P. S. (2000) Proc. Natl. Acad. Sci. USA 97, 12729-12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohlsson, C., Hellberg, N., Parini, P., Vidal, O., Bohlooly, M., Rudling, M., Lindberg, M. K., Warner, M., Angelin, B. & Gustafsson, J. A. (2000) Biochem. Biophys. Res. Commun. 278, 640-645. [DOI] [PubMed] [Google Scholar]
- 12.Luo, J., Sladek, R., Carrier, J., Bader, J. A., Richard, D. & Giguere, V. (2003) Mol. Cell. Biol. 23, 7947-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazar, M. A. (2002) Genes Dev. 16, 1-5. [DOI] [PubMed] [Google Scholar]
- 14.Mueller, E., Drori, S., Aiyer, A., Yie, J., Sarraf, P., Chen, H., Hauser, S., Rosen, E. D., Ge, K., Roeder, R. G. & Spiegelman, B. M. (2002) J. Biol. Chem. 277, 41925-41930. [DOI] [PubMed] [Google Scholar]
- 15.Ren, D., Collingwood, T. N., Rebar, E. J., Wolffe, A. P. & Camp, H. S. (2002) Genes Dev. 16, 27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barak, Y., Nelson, M. C., Ong, E. S., Jones, Y. Z., Ruiz-Lozano, P., Chien, K. R., Koder, A. & Evans, R. M. (1999) Mol. Cell 4, 585-595. [DOI] [PubMed] [Google Scholar]
- 17.Kelly, D. P. (2003) Circ. Res. 92, 482-484. [DOI] [PubMed] [Google Scholar]
- 18.Peters, J. M., Hennuyer, N., Staels, B., Fruchart, J. C., Fievet, C., Gonzalez, F. J. & Auwerx, J. (1997) J. Biol. Chem. 272, 27307-27312. [DOI] [PubMed] [Google Scholar]
- 19.Barbera, M. J., Schluter, A., Pedraza, N., Iglesias, R., Villarroya, F. & Giralt, M. (2001) J. Biol. Chem. 276, 1486-1493. [DOI] [PubMed] [Google Scholar]
- 20.Wang, Y. X., Lee, C. H., Tiep, S., Yu, R. T., Ham, J., Kang, H. & Evans, R. M. (2003) Cell 113, 159-170. [DOI] [PubMed] [Google Scholar]
- 21.McKenna, N. J. & O'Malley, B. W. (2002) Cell 108, 465-474. [DOI] [PubMed] [Google Scholar]
- 22.Puigserver, P., Wu, Z., Park, C. W., Graves, R., Wright, M. & Spiegelman, B. M. (1998) Cell 92, 829-839. [DOI] [PubMed] [Google Scholar]
- 23.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C. & Spiegelman, B. M. (1999) Cell 98, 115-124. [DOI] [PubMed] [Google Scholar]
- 24.Puigserver, P. & Spiegelman, B. M. (2003) Endocr. Rev. 24, 78-90. [DOI] [PubMed] [Google Scholar]
- 25.Vega, R. B., Huss, J. M. & Kelly, D. P. (2000) Mol. Cell. Biol. 20, 1868-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, J., Puigserver, P., Donovan, J., Tarr, P. & Spiegelman, B. M. (2002) J. Biol. Chem. 277, 1645-1648. [DOI] [PubMed] [Google Scholar]
- 27.Meirhaeghe, A., Crowley, V., Lenaghan, C., Lelliott, C., Green, K., Stewart, A., Hart, K., Schinner, S., Sethi, J. K., Yeo, G., et al. (2003) Biochem. J. 373, 155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamei, Y., Ohizumi, H., Fujitani, Y., Nemoto, T., Tanaka, T., Takahashi, N., Kawada, T., Miyoshi, M., Ezaki, O. & Kakizuka, A. (2003) Proc. Natl. Acad. Sci. USA 100, 12378-12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picard, F., Gehin, M., Annicotte, J., Rocchi, S., Champy, M. F., O'Malley, B. W., Chambon, P. & Auwerx, J. (2002) Cell 111, 931-941. [DOI] [PubMed] [Google Scholar]
- 30.Jepsen, K., Hermanson, O., Onami, T. M., Gleiberman, A. S., Lunyak, V., McEvilly, R. J., Kurokawa, R., Kumar, V., Liu, F., Seto, E., et al. (2000) Cell 102, 753-763. [DOI] [PubMed] [Google Scholar]
- 31.Nagy, L., Kao, H. Y., Chakravarti, D., Lin, R. J., Hassig, C. A., Ayer, D. E., Schreiber, S. L. & Evans, R. M. (1997) Cell 89, 373-380. [DOI] [PubMed] [Google Scholar]
- 32.Gurnell, M., Wentworth, J. M., Agostini, M., Adams, M., Collingwood, T. N., Provenzano, C., Browne, P. O., Rajanayagam, O., Burris, T. P., Schwabe, J. W., et al. (2000) J. Biol. Chem. 275, 5754-5759. [DOI] [PubMed] [Google Scholar]
- 33.Cavailles, V., Dauvois, S., L'Horset, F., Lopez, G., Hoare, S., Kushner, P. J. & Parker, M. G. (1995) EMBO J. 14, 3741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.L'Horset, F., Dauvois, S., Heery, D. M., Cavailles, V. & Parker, M. G. (1996) Mol. Cell. Biol. 16, 6029-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Treuter, E., Albrektsen, T., Johansson, L., Leers, J. & Gustafsson, J. A. (1998) Mol. Endocrinol. 12, 864-881. [DOI] [PubMed] [Google Scholar]
- 36.Fernandes, I., Bastien, Y., Wai, T., Nygard, K., Lin, R., Cormier, O., Lee, H. S., Eng, F., Bertos, N. R., Pelletier, N., et al. (2003) Mol. Cell 11, 139-150. [DOI] [PubMed] [Google Scholar]
- 37.Wei, L. N., Hu, X., Chandra, D., Seto, E. & Farooqui, M. (2000) J. Biol. Chem. 275, 40782-40787. [DOI] [PubMed] [Google Scholar]
- 38.Christian, M., Tullet, J. M. & Parker, M. G. (2004) J. Biol. Chem. 279, 15645-15651. [DOI] [PubMed] [Google Scholar]
- 39.Vo, N., Fjeld, C. & Goodman, R. H. (2001) Mol. Cell. Biol. 21, 6181-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar, V., Carlson, J. E., Ohgi, K. A., Edwards, T. A., Rose, D. W., Escalante, C. R., Rosenfeld, M. G. & Aggarwal, A. K. (2002) Mol. Cell 10, 857-869. [DOI] [PubMed] [Google Scholar]
- 41.Kozak, L. P. & Harper, M. E. (2000) Annu. Rev. Nutr. 20, 339-363. [DOI] [PubMed] [Google Scholar]
- 42.White, R., Leonardsson, G., Rosewell, I., Ann Jacobs, M., Milligan, S. & Parker, M. (2000) Nat. Med. 6, 1368-1374. [DOI] [PubMed] [Google Scholar]
- 43.Soukas, A., Socci, N. D., Saatkamp, B. D., Novelli, S. & Friedman, J. M. (2001) J. Biol. Chem. 276, 34167-34174. [DOI] [PubMed] [Google Scholar]
- 44.McGarry, J. D. & Brown, N. F. (1997) Eur. J. Biochem. 244, 1-14. [DOI] [PubMed] [Google Scholar]
- 45.Barrero, M. J., Camarero, N., Marrero, P. F. & Haro, D. (2003) Biochem. J. 369, 721-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossmeisl, M., Barbatelli, G., Flachs, P., Brauner, P., Zingaretti, M. C., Marelli, M., Janovska, P., Horakova, M., Syrovy, I., Cinti, S. & Kopecky, J. (2002) Eur. J. Biochem. 269, 19-28. [DOI] [PubMed] [Google Scholar]
- 47.Walczak, R. & Tontonoz, P. (2003) Nat. Med. 9, 1348-1349. [DOI] [PubMed] [Google Scholar]
- 48.del Mar Gonzalez-Barroso, M., Pecqueur, C., Gelly, C., Sanchis, D., Alves-Guerra, M. C., Bouillaud, F., Ricquier, D. & Cassard-Doulcier, A. M. (2000) J. Biol. Chem. 275, 31722-31732. [DOI] [PubMed] [Google Scholar]
- 49.Alvarez, R., de Andres, J., Yubero, P., Vinas, O., Mampel, T., Iglesias, R., Giralt, M. & Villarroya, F. (1995) J. Biol. Chem. 270, 5666-5673. [DOI] [PubMed] [Google Scholar]
- 50.Hernandez, A. & Obregon, M. J. (2000) Am. J. Physiol. 278, E769-E777. [DOI] [PubMed] [Google Scholar]
- 51.Sears, I. B., MacGinnitie, M. A., Kovacs, L. G. & Graves, R. A. (1996) Mol. Cell. Biol. 16, 3410-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiraby, C., Tavernier, G., Lefort, C., Larrouy, D., Bouillaud, F., Ricquier, D. & Langin, D. (2003) J. Biol. Chem. 278, 33370-33376. [DOI] [PubMed] [Google Scholar]
- 53.Tazawa, H., Osman, W., Shoji, Y., Treuter, E., Gustafsson, J. A. & Zilliacus, J. (2003) Mol. Cell. Biol. 23, 4187-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosen, E. D. (2003) Curr. Biol. 13, R961-R963. [DOI] [PubMed] [Google Scholar]
- 55.Ge, K., Guermah, M., Yuan, C. X., Ito, M., Wallberg, A. E., Spiegelman, B. M. & Roeder, R. G. (2002) Nature 417, 563-567. [DOI] [PubMed] [Google Scholar]
- 56.Flier, J. S. (2004) Cell 116, 337-350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.