Abstract
The t(12;21) translocation, which generates the TEL-AML1 (ETV6-RUNX1) fusion gene, is the most common structural chromosome change in childhood cancer and is exclusively associated with the common B cell precursor subset of acute lymphoblastic leukemia (ALL). Evidence suggests that the translocation usually occurs in utero during fetal hemopoiesis and most probably constitutes an initiating or first-hit mutation that is necessary but insufficient for the development of overt, clinical leukemia. The mechanism by which TEL-AML1 contributes to this early stage of leukemogenesis is unknown. To address this question we have analyzed hemopoiesis in mice syngeneically transplanted with TEL-AML1-transduced bone marrow stem cells. TEL-AML1 expression was associated with an accumulation/expansion of primitive c-kit-positive multipotent progenitors and a modest increase in myeloid colony-forming cells. TEL-AML1 expression was, however, permissive for myeloid differentiation. Analysis of B lymphopoiesis revealed an increase in early, pro-B cells but a differentiation deficit beyond that stage, resulting in reduced B cell production in the marrow. TEL-AML1-positive B cell progenitors exhibited reduced expression of the surrogate light-chain component λ5 and the IL-7 receptor, both of which may contribute to impedance of differentiation in vivo and account for their reduced in vitro clonogenicity in IL-7. A selective differentiation deficit of B lineage progenitors (i) is consistent with the phenotype of TEL-AML1-associated leukemia in children and (ii) provides a potential mechanism for the protracted preleukemic state that often precedes ALL. These results provide mechanistic insight into the role of the t(12;21) translocation in the initiation of common B cell precursor ALL.
TEL (ETV6)-AML1 (RUNX1), generated by the chromosomal translocation t(12;21)(p13;q22) (1, 2), is the most prevalent fusion gene in pediatric cancer, occurring in some 25% of childhood acute lymphoblastic leukemias (ALL) (3). The chimeric gene arises predominantly during fetal hemopoiesis (4) and at a rate that considerably exceeds that of overt clinical ALL (5), indicating the requirement for additional secondary and postnatal genetic events.
Twin studies and retrospective analysis of archived neonatal blood spots of patients with ALL indicates that TEL-AML1-expressing fetal clones expand and can persist in a clinically covert state for more than a decade (4, 6). These insights into the natural history of childhood leukemia raise fundamental issues on the nature of the transformed, preleukemic state.
In accord with these observations on clinical samples, TEL-AML1 by itself is insufficient for leukemogenesis in murine in vivo models (7) but, in common with AML1-ETO fusions (8, 9), may result in leukemia in the presence of complementary mutations (10).
The fetal cell initially transformed by TEL-AML1 is not precisely defined, but studies on the cells that were enriched and individually identified with a TEL-AML1 fusion in ALL itself (11) and in cord blood with preleukemic clones (5) suggest that it is probably an early B lineage progenitor or stem cell. In ALL with TEL-AML1, the predominant phenotype is that of B cell precursors (3). TEL-AML1 protein, in common with several other fusion proteins, e.g., AML1-ETO (12), appears to function as a transcriptional repressor recruiting corepressor molecules such as NCoR and Sin3A and histone deacetylases (13, 14), perhaps inhibiting normal AML1 target genes. The phenotypic impact of such activity has not been demonstrated, although, by analogy with AML1-ETO, one might anticipate an impedance to differentiation (12). This result would then be compatible with the view that the requisite additional genetic changes required for conversion to full malignancy might be those that uncouple the cell cycle (15).
The challenge, therefore, is to identify how TEL-AML1 fusions produce sustained premalignant clones of cells. We have attempted to model this crucial first step in leukemogenesis by retrovirally transducing TEL-AML1 into murine hemopoietic stem cells, transferring these cells in vivo and analyzing recipients for alterations in progenitor cell populations of different lineages. We report a selective impact on early B cell differentiation that is compatible with the phenotype observed clinically.
Materials and Methods
Generation of Retrovirus. cDNA for TEL-AML1 tagged with the myc-epitope at the carboxyl terminus was cloned upstream of the internal ribosome entry site element into the EcoRI site of the murine stem cell virus-internal ribosome entry site-GFP vector (16). Retrovirus vector plasmids were transiently transfected into Phoenix-Eco packaging cells [obtained from American Type Culture Collection (ATCC)] by a standard calcium phosphate precipitation method. Virus supernatants were then concentrated by centrifugation before infecting cells.
Retroviral Infection of Bone Marrow (BM) Cells and Transplantation. BM cells were harvested from tibiae and femora of BALB/c mice 4 days after i.v. administration of 5-fluorouracil (150 mg/kg) and treated with ACK (0.15 M NH4Cl/1.0 mM KHCO3/0.1 mM EDTA) to lyse red blood cells. BM cells were then prestimulated with stem cell factor (100 ng/ml) plus IL-3 (10 ng/ml) plus IL-6 (6 ng/ml) (PeproTech, London) for 24 h and spin-infected with concentrated virus in the presence of cytokines and Polybrene (4 μg/ml) (Sigma). Cells (1 × 106) were transplanted via the retroorbital vein into lethally irradiated (7.5 Gy) recipient mice maintained on acidified water throughout the experimental period. This study is based on the analysis of 14 independently transplanted TEL-AML1 animals; any single analysis used no fewer than 3 independently transplanted animals.
Western Blot Analysis. Total cell lysates were fractionated by SDS/PAGE and blotted onto a nylon membrane. TEL-AML1 was detected with an anti-AML1 antibody (Ab-2, Oncogene Research Products, Boston) or an anti-myc antibody (A-14, Santa Cruz Biotechnology). Primary staining was visualized with a goat anti-rabbit Ig-horseradish peroxidase conjugate secondary antibody by using enhanced chemiluminescence (Amersham Bioscience).
Flow Cytometry. After red blood cell lysis, cells were incubated with the indicated antibodies, conjugated with phycoerythrin, peridinin chlorophyll protein, or biotin; biotinylated antibodies were then stained with streptavidin-APC. Nonspecific binding was blocked by preincubation with an anti-Fc receptor antibody. Antibodies used were as follows: anti-B220 (RA3-6B2), anti-CD19 (1D3), anti-CD3 (145-2C11), anti-CD5 (53-7.3), anti-CD4 (GK1.5), anti-CD8 (53-6.7), anti-TER-119 (TER-119), anti-Gr-1 (RB6-8C5), anti-Mac-1 (M1/70), anti-c-kit (2B8), anti-CD43 (S7), anti-IgM (1B4B1), anti-BP1 (FG35.4), anti-heat-stable antigen (HSA) (CD24) (30-F1), anti-IL7Rα (A7R34), anti-AA4.1 (AA4.1), and anti-λ 5 (LM34) (purchased from BD Bioscience, San Diego). Cells were washed with PBS containing 2% BSA and analyzed on a FACSCalibur (BD Bioscience) with cellquest software.
In Vitro Colony-Forming Assays. Cells were plated in triplicate in methylcellulose under myeloid [stem cell factor (50 ng/ml), IL-3 (10 ng/ml), IL-6 (10 ng/ml), and erythropoietin (3 units/ml)] or B cell [IL-7 (10 ng/ml)] differentiation conditions. Colonies were counted and typed at days 8-12. ST2 coculture was performed with sorted pro-B cells at limiting dilution in the presence of IL-7 as described (17).
Results
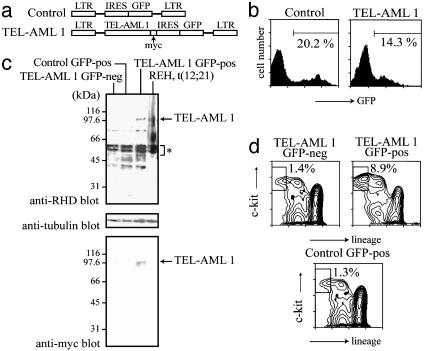
In Vivo Modeling of t(12;21). A full-length TEL-AML1 cDNA was myc-epitope tagged at its carboxyl terminus and inserted upstream of an internal ribosome entry site-GFP cassette in an murine stem cell virus-based retroviral vector (16) (Fig. 1a). BM cells, isolated from 5-fluorouracil-treated mice, were transduced with ecotropically packaged TEL-AML1-containing and control retroviruses and subsequently transplanted into lethally irradiated syngeneic hosts. Four months after transplantation, BM cells were harvested and analyzed. GFP-positive cells accounted for between 10% and 25% of BM cells in both experimental (TEL-AML1) and control (vector only) groups of animals (Fig. 1b). GFP-positive and -negative cells from both experimental and control animals were separated by flow cytometry and subsequently analyzed for TEL-AML1 expression by Western blotting by using an antibody directed against a peptide within the runt homology domain of AML1 that is conserved between mice and humans (Fig. 1c). Endogenous AML1 proteins were detected in all samples (asterisk), but TEL-AML1 expression was exclusively restricted to the GFP-positive fraction of BM cells isolated from the experimental group; the identity of the TEL-AML1 protein was additionally confirmed by Western blotting by using an antibody directed against the myc-epitope tag. Note that the level of TEL-AML1 expression is not dissimilar to that seen in the t(12;21) containing REH cell line derived from a common B-cell ALL patient (18, 19), but lower than that of endogenous murine AML1-proteins. These results confirm expression of an authentic TEL-AML1 protein within murine BM cells that is (i) concordant with GFP expression (thereby allowing use of GFP as a surrogate indicator of TEL-AML1 expression) and (ii) at a level consistent with that seen in TEL-AML1-associated human leukemia.
Fig. 1.
Modeling of t(12;21) in mice. (a) Diagram of recombinant viruses used (not to scale). (b) Fluorescence-activated cell sorter (FACS) analysis of GFP expression in BM cells of typical mice transplanted with control or TEL-AML1-containing viruses. (c) Western blot analysis of cell lysates by using anti-AML1, -myc, and -tubulin antibodies. (d) FACS analysis of the primitive c-kit+, lin- (B220, CD3, CD4, CD8, CD5, Gr-1, Mac-1, and TER-119) progenitor compartment. Results are shown from the GFP-positive and -negative fractions of a typical TEL-AML1 animal. GFP-positive cells from a typical mouse receiving vector-only-transduced cells provide a control.
Analysis of Primitive Progenitors. Morphological and immunophenotypic analysis of the distribution of different hematopoietic cell types within the GFP-positive and -negative fractions of BM cells from animals receiving TEL-AML1 or vector-only transduced grafts (Table 1) revealed a marked increase in blast-like, lineage-negative cells within the GFP-positive fraction of TEL-AML1 mice. This potential increase in the occurrence of immature progenitors is similar to that seen in AML1-ETO-transduced marrow (9, 20-25) and was confirmed by a two-color analysis with antibodies against c-kit and a mixture of lineage-specific antigens. The results show that within the TEL-AML1 animals, the c-kit-positive, lineage-negative compartment is increased 6-fold in the GFP-positive fraction relative to the GFP-negative fraction; this increase was not seen in the GFP-positive cells of control animals receiving vector-only-transduced BM (Fig. 1d). Similar results were obtained when phenotypic hematopoietic stem cells (Sca1-positive, c-kit-positive, lineage-negative) were examined (S.T. and T.E., unpublished observations). Preliminary data also indicate that the relative size of this fraction increases over time in the TEL-AML1 but not control virustransduced animals (unpublished observations), raising the possibility that this compartment may be responsible for maintenance and/or expansion of a preleukemic clone. We next asked whether this increase in the primitive progenitor compartment was reflected in altered colony-forming activity. GFP-positive and -negative BM cells were plated in methylcellulose under conditions that support the development of all myeloid lineages. Table 2 shows data from control and TEL-AML1 mice, which reveal increases in the frequency of mixed-lineage (CFU-GEMM) and bilineage (CFU-GM) colonies from the GFP-positive fraction of TEL-AML1 mice. The analysis of unilineagecommitted progenitors yielded mixed results. Whereas there was a decrease in CFU-G, we observed an increase in CFU-M. This increase predominantly reflects the novel appearance within the GFP-positive fraction of TEL-AML1 mice of small immature colonies with the appearance of CFU-M; these may, however, reflect more primitive progenitors of, for example, a blast colony type (26). These alterations in unilineage-committed and bilineage-committed myelomonocytic progenitors are not, however, reflected in any obvious alteration in the relative frequency of myelomonocytic cells in the marrow (Table 1). In contrast, the increase observed in CFU-E may in part underlie the relative increase in erythroid cells observed in the GFP-positive-marrow-fraction of mice transplanted with TEL-AML1-transduced cells.
Table 1. Immunophenotypic analysis: Distribution of lineage-affiliated cells in TEL-AML1-positive and -negative fractions.
| Distribution among lineage markers, %
|
|||||
|---|---|---|---|---|---|
| BM cells | B220 | CD3 | TER-119 | Gr-1/Mac-1 | Lineage-negative |
| TEL-AML1-GFP-negative | 29.4 ± 4.1 | 5.0 ± 1.1 | 4.2 ± 0.8 | 61.0 ± 4.1 | 1.8 ± 0.4 |
| TEL-AML1-GFP-positive | 9.3 ± 3.1 | 2.8 ± 1.0 | 19.4 ± 2.1 | 56.9 ± 6.4 | 12.1 ± 4.1 |
| Vector-only GFP-positive | 27.3 ± 5.6 | 5.8 ± 1.1 | 4.0 ± 1.2 | 60.2 ± 5.6 | 1.7 ± 0.4 |
BM cells from TEL-AML1 or GFP-only control mice were analyzed for lineage markers (B cell B220; T cell CD3; erythroid cells TER-119; granulomonocytic cells Gr-1/Mac-1). Data are presented as the mean ± SD of three mice analyzed.
Table 2. Colony-forming activities of sorted BM cells.
| Colony-forming units (CFU) per 1.5 × 104 cells
|
|||||
|---|---|---|---|---|---|
| BM cells | CFU-E | CFU-G | CFU-M | CFU-GM | CFU-GEMM |
| TEL-AML1-GFP-negative | 6.0 ± 2.1 | 7.7 ± 4 | 14.4 ± 3.6 | 15.7 ± 3.7 | 2.4 ± 1.8 |
| TEL-AML1-GFP-positive | 11.7 ± 3.6 | 4.8 ± 3.1 | 25.6 ± 3.4 | 32.1 ± 6.7 | 10.6 ± 1.7 |
| Vector-only GFP-positive | 5.4 ± 1.8 | 8.4 ± 3.1 | 13.3 ± 3.9 | 16.4 ± 3.4 | 3.2 ± 1.4 |
BM cells from TEL-AML1 or GFP-only control mice were sorted into GFP-positive and GFP-negative fractions and then assayed for colony-forming activities in multimyeloid conditions. Data are presented as the mean ± SD (triplicate).
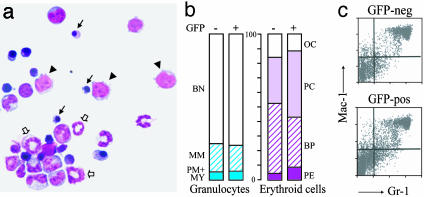
Permissiveness of TEL-AML1 for Myeloid Differentiation. Given these effects on myeloid progenitors, we next examined whether the expression of TEL-AML1 altered the differentiation of myeloid cells. TEL-AML1-expressing cells within the BM were sorted on the basis of GFP expression, centrifuged onto a glass slide, and morphologically examined alongside control samples. As shown in Fig. 2a, well differentiated granulocytic and erythroid cells were evident, and there was no apparent change in the relative distribution of cells between any of the maturation stages of normal granulocytic or erythroid differentiation (Fig. 2b). Flow-cytometric analysis of cells stained with myeloid-affiliated antigens, Gr-1 and Mac-1, also showed no marked difference between GFP-positive and negative cells from either TEL-AML1 or control animals (Fig. 2c). Taken together, these data indicate that expression of TEL-AML1 has no obvious deleterious effect on the production of normally differentiated granulocytes and erythroid cells and therefore suggests that the TEL-AML1 fusion gene is permissive for differentiation of the myeloid series in vivo.
Fig. 2.
Analysis of myeloid differentiation. (a) Cytospin of GFP-positive BM cells from a typical TEL-AML1 animal. Note the normal appearance of mature myeloid forms. Arrowheads indicate immature blasts. Small arrows indicate mature erythrocytes. Large arrows indicate mature granulocytes. (b) Proportion of cells representative of different erythroid and granulocytic maturation, within the marrow. BN, band neutrophil; MM, metamyelocyte; PM+My, promyelocyte + myelocyte; OC, orthrochromatic erythroblast; PC, polychromatic erythroblast; BP, basophilic erythroblast; PE, proerythrocyte. (c) Fluorescence-activated cell sorter analysis of myeloid marker expression in the GFP-positive and -negative fraction of BM cells of a typical TEL-AML1 animal.
TEL-AML1 Impedes B Cell Differentiation at Pro-B Cell Stage. Analysis of the BM from mice grafted with TEL-AML1-transduced cells revealed a marked reduction in B lymphocytes within the GFP-positive fraction (Table 1). We therefore examined the in vitro B lymphoid clonogenic potential of the GFP-positive and -negative fractions of TEL-AML1-BM by using a methylcellulose-colony formation assay for IL-7-responsive B cell progenitors (27). The results are summarized in Fig. 3a and show a dramatic decrease in the frequency of B lymphoid colony-forming cells in BM as a result of TEL-AML1 expression. A similar, although less marked, deleterious impact on B cell colony-forming activity was also observed when immunopurified pro-B cells were assayed in a stromal-dependent culture system (17). Typical data from one such experiment are shown in Fig. 3b. These in vitro assays suggest that B cell development may be deregulated at quite early stages of B cell differentiation.
Fig. 3.
Analysis of B lymphoid colony-forming cell activity. (a) Colonies formed per 5 × 104 total BM cells plated in semisolid medium in the presence of IL-7. (b) Analysis by limiting dilution of colony-forming potential of immunopurified pro-B cells on ST2 cell stromal layers. Logarithmic fractions of no cell growth after 7 days of culture are indicated along with input cell number per well.
We therefore next examined the extent to which TEL-AML1-expression impaired the differentiation potential of the B cell lineage in vivo. Mouse B cell development in BM is thought to be composed of at least five different stages, which may be identified on the basis of immunophenotyping according to the classification scheme developed by Hardy (28): pre-pro-B and pro-B cells are contained within Hardy's fractions A, B, and C, whereas fractions D, E, and F, respectively, contain pre-B, immature B, and mature B cells (Fig. 4a). B220-positive BM cells from TEL-AML1 mice were gated according to GFP expression and then analyzed for CD43 expression and IgM, HSA, or BP-1 (Fig. 4b). The B220+CD43+IgM- population (fractions A, B, and C) is markedly increased in GFP-positive (66.7%) relative to GFP-negative (12.1%) cells. Consistent with these data, analysis of B220+CD43+HSA- cells (fraction A) as well as B220+CD43+BP-1+ cells (fraction C) shows similar relative increases in the GFP-positive population. In contrast to this relative increase in the proportion of these pre-pro-B and pro-B cell-containing compartments, we observed a significant decrease in the proportion of pre-B cells (B220+IgM-CD43-/fraction D). The analysis of fractions E and F is shown in Fig. 4c and is consistent with a reduction in the frequency of immature and mature B cells in GFP-positive population from TEL-AML1 animals.
Fig. 4.
Analysis of B cell differentiation. (a) Classification scheme of B cell differentiation according to Hardy. (b and c) Fluorescence-activated cell sorter analyses of progenitor and mature B cell compartments, respectively, of GFP-positive and GFP-negative fractions of BM cells from a typical animal grafted with TEL-AML1 transduced cells; the various antibodies used in the analyses are indicated. B220+CD43+ represents fractions A-C; B220+IgM+ represents fractions E and F; B220+CD43+HSA- represents fraction A; B220+CD43+BP-1+ represents fraction C; B220loIgM+ and B220hiIgM+ represent fraction E and F, respectively. (d) A detailed analysis of cell subpopulations contained within fraction A. The B220+CD19- fractions from GFP-positive and -negative populations (Top) were analyzed for expression of AA4.1 (Middle). CD43 and HSA expression by B220+CD19- cells is shown in Bottom. (E) Summary of the relative distribution of B cells with respect to the differentiation stages defined by Hardy. The results presented are derived from the analysis of five independent animals.
In addition to B lineage-restricted progenitors, fraction A is thought to contain progenitors with additional or alternative lineage potentials, including natural killer and myeloid progenitors (29, 30). We therefore examined the extent to which the frequency of B lineage-restricted progenitors contained within fraction A was altered in the GFP-positive BM population of TEL-AML1 animals (Fig. 4d). Bona fide B cell progenitors within fraction A do not express the characteristic B lineage marker CD19 but are known to express the AA4.1 antigen; approximately half of fraction A cells express AA4.1 (31). Consistent with the data described above, the B220+CD19- population was increased in the GFP-positive fraction of TEL-AML1 marrow. These cells were found to express the CD43+HSA- phenotype indicative of fraction A. Approximately half the B220+CD19- population expressed AA4.1, irrespective of GFP status. These results indicate that the proportion of very early CD19-negative, B lineage-restricted progenitors contained within fraction A is increased as a result of expression of the TEL-AML1 fusion gene.
The relative B cell distribution in respect of Hardy's classification is summarized in Fig. 4e for both GFP-positive and -negative cells sampled from the marrows of several TEL-AML1 animals. The results suggest that expression of TEL-AML1 impedes, or delays, B cell differentiation from the earliest stages of B cell development with a particularly marked impact at the transition from pro-B to pre-B cell stages.
Evidence for Impaired Signal-Responsiveness. The λ5 gene comprises one component of the surrogate B cell receptor and has been shown genetically to be crucial for the pro-B to pre-B cell transition (32). [λ5 has also been postulated as a transcriptional target of AML1 in the mouse (33).] We therefore examined expression of λ5 on the cell surface of large B cells as well as pro-B cells that normally express this protein; the cells used for this analysis are shown in Fig. 5a. We observed that λ5 expression was significantly reduced in the GFP-positive as compared to GFP-negative fractions of both these cell types in TEL-AML1 mice (Fig. 5b). In a similar vein, we examined expression of the IL-7 receptor; IL-7 receptor knockout mice exhibit a differentiation arrest at the fraction A stage of B cell development (34). The results presented in Fig. 5c indicate a reduction in IL-7 receptor expression in the GFP-positive fraction of B cells from TEL-AML1 mice. These results are consistent both with the TEL-AML1-associated differentiation delay that we have observed in vivo and with the reduced clonogenicity in vitro of TEL-AML1-expressing B cell progenitors.
Fig. 5.
Analysis of receptor expression. (a) Identification of large B and pro-B cell populations by flow cytometry. (b) Analysis of λ5 expression within the large B and pro-B cell fractions of GFP-positive and -negative BM cells from a TEL-AML1 animal. (c) A similar analysis for IL-7 receptor expression.
Discussion
Although several lines of evidence indicate that generation of t(12;21) translocation is usually a prenatal initiating mutation in the common B cell precursor ALL (4), the mechanism by which the TEL-AML1 fusion protein establishes a persistent preleukemic condition and thereby contributes to leukemogenesis has remained unclear. In the present study, we have provided an insight into how TEL-AML1 might function as a first-hit mutation. Our results show that the TEL-AML1 fusion has the capacity to inhibit the differentiation program of B lymphocytes producing a relative accumulation of early B cell progenitors at the expense of more differentiated forms. It has recently been proposed that development of leukemia requires the accumulation of two distinct classes of mutation (15). Class II mutations restrict differentiation and class I mutations dysregulate proliferation. In the context of this scheme our results identify TEL-AML1 as a class II mutation.
Establishing the molecular mechanism by which TEL-AML1 impedes differentiation will clearly be an important area for future study. The simplest model consistent with available molecular evidence is that TEL-AML1, through recruitment of transcriptional corepressors, antagonizes the function of wildtype AML1 or AML1-related molecules (such as AML2 or -3) (12). Studies of AML1 have mainly focused on its activity in myeloid cells (12). Nevertheless AML1 is expressed throughout the B cell series, and several genes have been proposed as AML1 targets. Among these, the surrogate light-chain component of the pre-B cell receptor, λ5, is known to be a critical regulator of B cell development. Mice deficient in λ5 display an impairment in the transition of pro-B cells to pre-B cells (Hardy's fractions C and D). However, the block to differentiation is not complete, and a number of mature B cell forms are still produced (32). Aspects of this phenotype are very similar to those observed in the TEL-AML1 animals, and consistent with this we observed reduced expression of λ5 on TEL-AML1-expressing pro-B cells. Reduction of λ5 expression may thus provide one mechanism for the inhibition of B cell differentiation and colony formation resulting from TEL-AML1 expression. The reduced IL-7 responsiveness of TEL-AML1-expressing B cell progenitors prompted us to investigate expression of the IL-7 receptor, which we then found to be reduced. IL-7 receptor-deficient mice exhibit a block B cell differentiation from the very earliest stages of B cell development (Hardy's fraction A) (34), and consistent with this TEL-AML1 animals also exhibit an accumulation of cells at this stage.
Thus, the reduced expression of both λ5 and IL-7 receptor is consistent with, and may contribute to or be causal in, the B cell abnormalities observed as a result of TEL-AML1 expression. However, these may not constitute the primary or only mechanisms at play in this mouse model or indeed in the patient situation; the regulation of the pre-B cell receptor may well differ between humans and mice (35, 36), and differences in IL-7 responsiveness of B cells between the two species have been documented. Also IL-7 responsiveness may be different between fetal and adult cells. The likely target cell for transformation by TEL-AML1 in patients is a fetal cell, perhaps of BM or liver origin. Although our experiments have made use of young-adult BM stem cells, the possibility that TEL-AML1 may have a differential phenotypic impact depending on the precise developmental level of target suggests that similar experiments with fetal-derived hemopoietic cells would be informative.
These considerations aside, the preleukemic phenotype observed in this mouse model shares many of the features seen in the human disease. First, there is a selective differentiation block in the B cell pathway. Second, whereas differences in the staging of human and mouse B lymphopoiesis make precise comparisons between the two species difficult (37), in both cases the block is at an early progenitor cell level. Third, the block is incomplete, resulting in the presence of differentiated forms in both cases. Thus, mature B cells expressing the TEL-AML1 transgene are present in our mice, and equivalent κ- or λ-bearing B cells with TEL-AML1 are detectable within the candidate preleukemic population present at birth in human cord blood (5). Differentiation arrest in ALL itself appears to be more stringent.
The cell in which the t(12;21) translocation first arises in humans is unclear. The translocation has been documented in CD19-positive B cell progenitors (5, 38) but not in earlier CD34-positive, lineage-negative cells, although this may reflect a technical limitation of the methods used. To maximize the potential target cell range available for transformation by TEL-AML1, we elected to transduce multipotential stem cells in the current study; ipso facto, our experiments are not informative in regard to the nature of the target cell in which the TEL-AML1 translocation initially occurs. Importantly, however, they do reveal the target cells in which the TEL-AML1 molecule may have a biological impact that is functionally relevant to the preleukemic state/leukemogenesis.
The transduction of multipotent cells has allowed us to explore the effects of TEL-AML1 on different hemopoietic compartments and compare these with those reported for a related AML1 fusion, AML1-ETO generated by the t(8;21) translocation and commonly associated with myeloid leukemia. The accumulation of lineage-negative, c-kit-positive progenitors observed in our TEL-AML1 animals has been reported in mice reconstituted with AML1-ETO-transduced BM (22). These findings may suggest that recruitment of transcriptional corepressors (such as Sin3A and NCoR) to AML1 moieties by means of either fused TEL or ETO domains may have a similar effect in the early multipotential progenitor compartment and perturb a similar spectrum of transcriptional targets. Preliminary data further indicate that the size of the hematopoietic stem cell compartment increases significantly over time in TEL-AML1 animals (S.T. and T.E., unpublished observations). This increase in size is accompanied by a relative increase in GFP-positive cells in the marrow and raises the possibility that this very primitive compartment may be responsible for the maintenance and/or expansion of a preleukemic clone. This notion would also be in part consistent with the relative increases seen in some myeloid compartments.
In contrast to the observed blockade of B cell differentiation, the myelo-erythroid compartment of TEL-AML1 mice appeared grossly normal. Morphological examination of TEL-AML1-expressing BM cells showed an apparently normal distribution of cells of the different erythroid and granulocytic maturation stages; immunophenotypic analysis was also consistent with this normality. These observations contrast with those made when in a similarly derived model of AML1-ETO where granulopoiesis is arrested at the final stage of differentiation and B lymphopoiesis is unimpeded at early stages (20). Thus, whereas expression of both AML1-ETO and TEL-AML1 within the stem cell compartment appears to inhibit differentiation, the activities of the two molecules appear to be broadly selective for myeloid and B lymphoid lineages, respectively, and are consistent with the lineage selectivity seen in t(8;21)- and t(21;21)-associated leukemias. The data argue that this striking feature of fusion genes arises not necessarily because of a restrictive cell of origin of chromosome translocation itself but rather because of a cell context-dependent function of the encoded chimeric proteins (38).
Acknowledgments
This work was supported by Kay Kendall (to M.G.); Leukaemia Research Fund UK specialist programs (to T.E. and M.G.); the Medical Research Council (to T.E.); and grants-in-aid for (i) the Second Term Comprehensive 10-year Strategy for Cancer Control from the Ministry of Health and Welfare (to M.S.), (ii) Scientific Research from The Ministry of Education, Culture, Sports, Science, and Technology (to M.S.), (iii) Scientific Research from the Japanese Society for the Promotion of Science (to M.S.), and (iv) Scientific Research from the Japanese Society for the Promotion of Science (to S.T.).
Abbreviations: ALL, acute lymphoblastic leukemia; BM, bone marrow; HSA, heat-stable antigen; CFU, colony-forming units.
References
- 1.Romana, S. P., Mauchauffe, M., Le Coniat, M., Chumakov, I., Le Paslier, D., Berger, R. & Bernard, O. A. (1995) Blood 85, 3662-3670. [PubMed] [Google Scholar]
- 2.Golub, T. R., Barker, G. F., Bohlander, S. K., Hiebert, S. W., Ward, D. C., Bray-Ward, P., Morgan, E., Raimondi, S. C., Rowley, J. D. & Gilliland, D. G. (1995) Proc. Natl. Acad. Sci. USA 92, 4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shurtleff, S. A., Buijs, A., Behm, F. G., Rubnitz, J. E., Raimondi, S. C., Hancock, M. L., Chan, G. C., Pui, C. H., Grosveld, G. & Downing, J. R. (1995) Leukemia 9, 1985-1989. [PubMed] [Google Scholar]
- 4.Greaves, M. F. & Wiemels, J. L. (2003) Nat. Rev. Cancer 3, 639-649. [DOI] [PubMed] [Google Scholar]
- 5.Mori, H., Colman, S. M., Xiao, Z., Ford, A. M., Healy, L. E., Donaldson, C., Hows, J. M., Navarrete, C. & Greaves, M. (2002) Proc. Natl. Acad. Sci. USA 99, 8242-8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greaves, M. F., Maia, A. T., Wiemels, J. L. & Ford, A. M. (2003) Blood 102, 2321-2333. [DOI] [PubMed] [Google Scholar]
- 7.Andreasson, P., Schwaller, J., Anastasiadou, E., Aster, J. & Gilliland, D. G. (2001) Cancer Genet. Cytogenet. 130, 93-104. [DOI] [PubMed] [Google Scholar]
- 8.Yuan, Y., Zhou, L., Miyamoto, T., Iwasaki, H., Harakawa, N., Hetherington, C. J., Burel, S. A., Lagasse, E., Weissman, I. L., Akashi, K. & Zhang, D. E. (2001) Proc. Natl. Acad. Sci. USA 98, 10398-10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higuchi, M., O'Brien, D., Kumaravelu, P., Lenny, N., Yeoh, E. J. & Downing, J. R. (2002) Cancer Cell 1, 63-74. [DOI] [PubMed] [Google Scholar]
- 10.Bernardin, F., Yang, Y., Cleaves, R., Zahurak, M., Cheng, L., Civin, C. I. & Friedman, A. D. (2002) Cancer Res. 62, 3904-3908. [PubMed] [Google Scholar]
- 11.Hotfilder, M., Rottgers, S., Rosemann, A., Jurgens, H., Harbott, J. & Vormoor, J. (2002) Blood 100, 640-646. [DOI] [PubMed] [Google Scholar]
- 12.Tenen, D. G. (2003) Nat. Rev. Cancer 3, 89-101. [DOI] [PubMed] [Google Scholar]
- 13.Hiebert, S. W., Sun, W., Davis, J. N., Golub, T., Shurtleff, S., Buijs, A., Downing, J. R., Grosveld, G., Roussell, M. F., Gilliland, D. G., et al. (1996) Mol. Cell. Biol. 16, 1349-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guidez, F., Petrie, K., Ford, A. M., Lu, H., Bennett, C. A., MacGregor, A., Hannemann, J., Ito, Y., Ghysdael, J., Greaves, M., et al. (2000) Blood 96, 2557-2561. [PubMed] [Google Scholar]
- 15.Speck, N. A. & Gilliland, D. G. (2002) Nat. Rev. Cancer 2, 502-513. [DOI] [PubMed] [Google Scholar]
- 16.Persons, D. A., Allay, J. A., Allay, E. R., Ashmun, R. A., Orlic, D., Jane, S. M., Cunningham, J. M. & Nienhuis, A. W. (1999) Blood 93, 488-499. [PubMed] [Google Scholar]
- 17.Rolink, A., Kudo, A., Karasuyama, H., Kikuchi, Y. & Melchers, F. (1991) EMBO J. 10, 327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agape, P., Gerard, B., Cave, H., Devaux, I., Vilmer, E., Lecomte, M. C. & Grandchamp, B. (1997) Br. J. Haematol. 98, 234-239. [DOI] [PubMed] [Google Scholar]
- 19.Poirel, H., Lacronique, V., Mauchauffe, M., Le Coniat, M., Raffoux, E., Daniel, M. T., Erickson, P., Drabkin, H., MacLeod, R. A., Drexler, H. G., et al. (1998) Oncogene 16, 2895-2903. [DOI] [PubMed] [Google Scholar]
- 20.Schwieger, M., Lohler, J., Friel, J., Scheller, M., Horak, I. & Stocking, C. (2002) J. Exp. Med. 196, 1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tonks, A., Pearn, L., Tonks, A. J., Pearce, L., Hoy, T., Phillips, S., Fisher, J., Downing, J. R., Burnett, A. K. & Darley, R. L. (2003) Blood 101, 624-632. [DOI] [PubMed] [Google Scholar]
- 22.de Guzman, C. G., Warren, A. J., Zhang, Z., Gartland, L., Erickson, P., Drabkin, H., Hiebert, S. W. & Klug, C. A. (2002) Mol. Cell. Biol. 22, 5506-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hug, B. A., Lee, S. Y., Kinsler, E. L., Zhang, J. & Lazar, M. A. (2002) Cancer Res. 62, 2906-2912. [PubMed] [Google Scholar]
- 24.Mulloy, J. C., Cammenga, J., MacKenzie, K. L., Berguido, F. J., Moore, M. A. & Nimer, S. D. (2002) Blood 99, 15-23. [DOI] [PubMed] [Google Scholar]
- 25.Rhoades, K. L., Hetherington, C. J., Harakawa, N., Yergeau, D. A., Zhou, L., Liu, L. Q., Little, M. T., Tenen, D. G. & Zhang, D. E. (2000) Blood 96, 2108-2115. [PubMed] [Google Scholar]
- 26.Suda, T., Suda, J. & Ogawa, M. (1983) J. Cell. Physiol. 117, 308-318. [DOI] [PubMed] [Google Scholar]
- 27.Lee, G., Namen, A. E., Gillis, S., Ellingsworth, L. R. & Kincade, P. W. (1989) J. Immunol. 142, 3875-3883. [PubMed] [Google Scholar]
- 28.Hardy, R. R., Carmack, C. E., Shinton, S. A., Kemp, J. D. & Hayakawa, K. (1991) J. Exp. Med. 173, 1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tudor, K. S., Payne, K. J., Yamashita, Y. & Kincade, P. W. (2000) Immunity 12, 335-345. [DOI] [PubMed] [Google Scholar]
- 30.Rolink, A., ten Boekel, E., Melchers, F., Fearon, D. T., Krop, I. & Andersson, J. (1996) J. Exp. Med. 183, 187-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y. S., Wasserman, R., Hayakawa, K. & Hardy, R. R. (1996) Immunity 5, 527-535. [DOI] [PubMed] [Google Scholar]
- 32.Kitamura, D., Kudo, A., Schaal, S., Muller, W., Melchers, F. & Rajewsky, K. (1992) Cell 69, 823-831. [DOI] [PubMed] [Google Scholar]
- 33.Martensson, A., Xie, X. Q., Persson, C., Holm, M., Grundstrom, T. & Martensson, I. L. (2001) Eur. J. Immunol. 31, 3165-3174. [DOI] [PubMed] [Google Scholar]
- 34.Peschon, J. J., Morrissey, P. J., Grabstein, K. H., Ramsdell, F. J., Maraskovsky, E., Gliniak, B. C., Park, L. S., Ziegler, S. F., Williams, D. E., Ware, C. B., et al. (1994) J. Exp. Med. 180, 1955-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Y. H., Stephan, R. P., Scheffold, A., Kunkel, D., Karasuyama, H., Radbruch, A. & Cooper, M. D. (2002) Blood 99, 2459-2467. [DOI] [PubMed] [Google Scholar]
- 36.Lemmers, B., Gauthier, L., Guelpa-Fonlupt, V., Fougereau, M. & Schiff, C. (1999) Blood 93, 4336-4346. [PubMed] [Google Scholar]
- 37.Ghia, P., ten Boekel, E., Rolink, A. G. & Melchers, F. (1998) Immunol. Today 19, 480-485. [DOI] [PubMed] [Google Scholar]
- 38.Barr, F. G. (1998) Nat. Genet. 19, 121-124. [DOI] [PubMed] [Google Scholar]