Summary
Exposure of cells to reactive oxygen species (ROS) causes a rapid and significant drop in intracellular ATP-levels. This energy depletion negatively affects ATP-dependent chaperone systems, making ROS-mediated protein unfolding and aggregation a potentially very challenging problem. Here we show that Get3, a protein involved in ATP-dependent targeting of tail-anchored (TA) proteins under non-stress conditions, turns into an effective ATP-in dependent chaperone when oxidized. Activation of Get3’s chaperone function, which is a fully reversible process, involves disulfide bond formation, metal release and its conversion into distinct, higher oligomeric structures. Mutational studies demonstrate that the chaperone activity of Get3 is functionally distinct from and likely mutually exclusive with its targeting function, and responsible for the oxidative stress sensitive phenotype that has long been noted for yeast cells lacking functional Get3. These results provide convincing evidence that Get3 functions as a redox regulated chaperone, effectively protecting eukaryotic cells against oxidative protein damage.
Introduction
The cytosolic ATPase Get3 in yeast (mammalian TRC40) is a central player of the Guided Entry of Tail-anchored proteins (GET) pathway, which is responsible for the posttranslational integration of tail-anchored (TA) proteins into the membrane of the endoplasmic reticulum (Favaloro et al., 2008; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). In this role, Get3 shuttles between a cytosolic multi-protein complex that receives the TA precursor proteins from the ribosome, and a Get1/Get2 receptor complex at the ER membrane, where the TA-protein precursors are released and integrated into the lipid bilayer (Mariappan et al., 2010; Mariappan et al., 2011; Stefer et al., 2011). This cycle is associated with significant conformational changes in Get3, induced by substrate binding and ATP hydrolysis. Importantly, while mutations in components of the GET system are lethal in higher eukaryotes (Mukhopadhyay et al., 2006), yeast cells survive their absence (Metz et al., 2006; Shen et al., 2003). Nevertheless, deletion of the GET3 gene in yeast leads to several, seemingly unrelated phenotypes, including hygromycin sensitivity, copper and H2O2 sensitivity, heat sensitivity and the inability to grow on iron-limiting media (Schuldiner et al., 2008; Shen et al., 2003). It still remains to be elucidated whether these different phenotypes are all due to the reduced integration of specific TA proteins into the ER membrane, or are caused by the absence of a potentially second role of Get3 in maintaining metal homeostasis and/or mediating oxidative stress resistance.
Get3 is a zinc-binding protein with four highly conserved cysteines, two of which (C285/C288) are essential for complementing the growth defect of a get3 deletion strain under various stress conditions (Metz et al., 2006). Arranged in a C-X-C –X43-C X-X-C motif, this cysteine arrangement is highly reminiscent of the oxidation sensitive zinc-binding motif found in Hsp33, a redox-regulated ATP-independent chaperone in bacteria. Once activated by disulfide bond formation and zinc release, Hsp33 prevents oxidative stress mediated protein aggregation, thereby increasing bacterial oxidative stress resistance (Jakob et al., 1999). Like oxidized Hsp33, aerobically purified Get3 was recently found to also function as ATP-independent general chaperone in vitro, highly effective in protecting unfolding proteins against aggregation (Powis et al., 2013). In vivo studies agreed with these results by demonstrating that Get3 co-localizes with unfolding proteins and chaperones in distinct foci during ATP-depleting stress conditions in yeast (Powis et al., 2013).
The intriguing similarities between Get3 and the structurally unrelated Hsp33 prompted our investigations into the regulation of Get3’s ATP-independent chaperone function. We discovered that incubation of Get3 with the hydroxyl-radical producing combination of H2O2 and Cu2+ rapidly and reversibly switches the protein from an ATPase-driven TA-binding protein into a general, chaperone holdase. Triggered by disulfide bond formation and zinc release, Get3 undergoes massive conformational rearrangements, which result in the formation of highly active Get3 tetramers and higher oligomers, capable of binding unfolding proteins and preventing their nonspecific aggregation. In vitro and in vivo studies using established TA-binding or ATPase deficient Get3 variants confirmed our discovery that Get3 has two distinct functions, and moonlights as general chaperone under ATP-depleted oxidative stress conditions.
Results
Get3 – A redox-regulated chaperone in eukaryotes
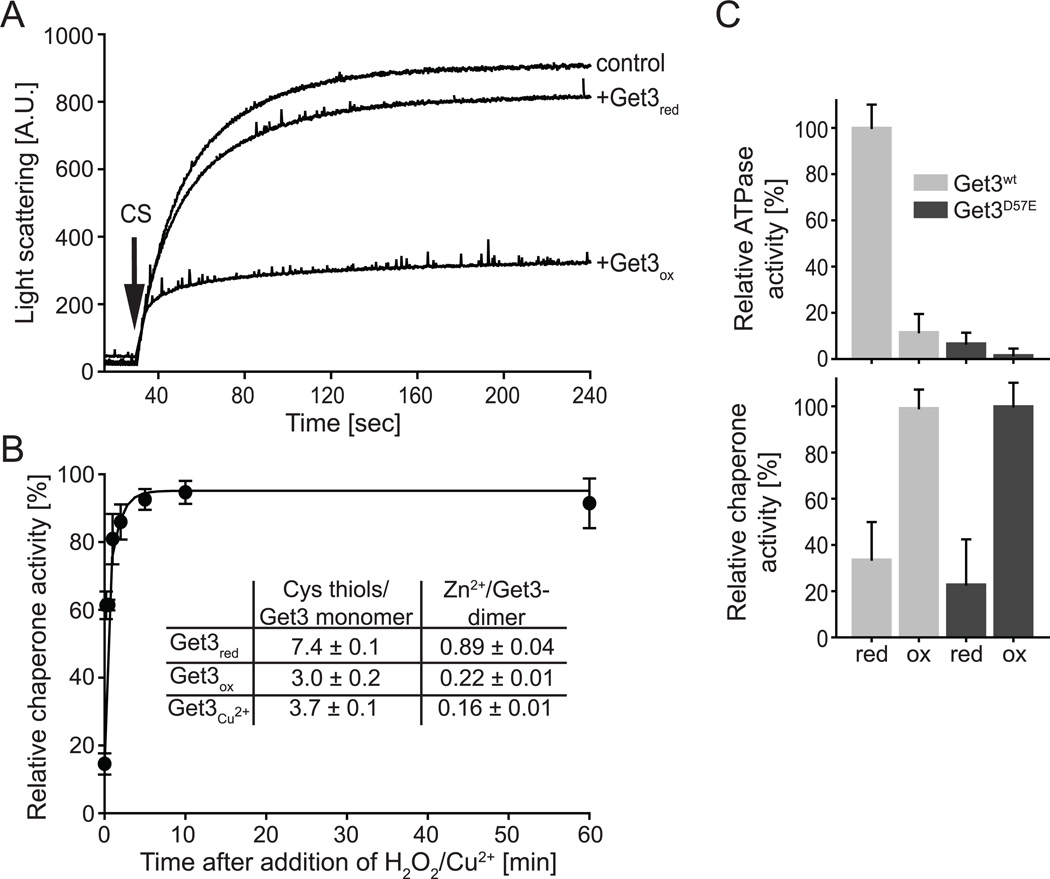
Get3’s copper sensitive phenotype, its ability to chaperone soluble proteins in vitro and the presence of disulfide-linked Get3 dimers in Get3 crystals (Suloway et al., 2009) raised the intriguing possibility that Get3 might serve as redox-controlled chaperone that protects proteins against oxidative stress-induced protein aggregation. ATP-independent chaperone function is particularly crucial under these stress conditions, since exposure of cells to ROS is known to cause a severe and rapid decrease in cellular ATP-levels, incapacitating ATP-dependent chaperones (Winter et al., 2005). To test if Get3’s general chaperone activity is indeed redox-regulated, we purified Get3 under reducing conditions (Get3red), and tested its ability to prevent the aggregation of chemically unfolding citrate synthase (CS). An eight-fold molar excess of Get3red over CS had only a minor effect on the aggregation behavior of CS (Figure 1A). However, incubation of Get3red with the hydroxyl-radical producing mixture of hydrogen peroxide (H2O2) and Cu2+ (Get3ox) led to the very rapid activation of Get3’s chaperone function (T1/2 < 1 min) (Figure 1B, black circles) and an almost complete inhibition of CS aggregation at an 8:1 molar ratio of Getox to CS (Figure 1A). Very similar results were obtained when we tested the influence of Get3red and Get3ox on chemically or thermally unfolding luciferase (Figure S1A). These results suggest that incubation with hydroxyl-producing oxidants converts Get3 into a general molecular chaperone for unfolding soluble proteins. Incubation of Get3 with 50 µM Cu2+ in the absence of H2O2 also activated the chaperone function of Get3 but displayed biphasic kinetics with an extremely fast initial activation followed by a slower activation phase (Figure S1B, triangles). At 200 µM copper, however, full activation was achieved within the mixing time of the experiment (Figure S1C). Removal of Cu2+ using the strong chelator tetrakis(2pyridylmethyl) ethylene-diamine (TPEN) immediately before testing the chaperone function of Get3 revealed no difference in the chaperone activity of Get3 (Figure S1C, insert), indicating that the activation of Get3’s chaperone function is not triggered by copper binding but is likely due to oxidative modification of Get3. Consistent with this conclusion we found that presence of 2 mM H2O2, a kinetically slow oxidant also converted Get3 into a chaperone but with slower activation kinetics (T1/2 ~ 5 min) (Figure S1B, squares). To test how oxidation affects the ATPase activity of Get3, we directly compared the ability of oxidized and reduced Get3 to hydrolyze ATP. As shown in Figure 1C upper panel, chaperone-active Get3ox showed less than 20% ATPase activity compared to Get3red, suggesting that oxidation leads to conformational changes that affect ATP hydrolysis. These results are fully consistent with previous findings revealing that Get3’s chaperone function is not affected by ATP, and that in vivo co-localization of Get3 with protein aggregates occurs under ATP-depleting stress conditions (Powis et al., 2013). Analysis of the chaperone function of the ATPase-deficient Get3 D57E mutant variant revealed that this mutant protein is fully chaperone-active when oxidized further agreeing with our conclusions (Figure 1C, lower panel). These results indicate that ATP-hydrolysis is neither required for the oxidative activation of Get3 nor for its general chaperone function. From these data we concluded that Get3 converts into a potent ATP-independent chaperone when exposed to oxidative stress conditions in vitro.
Figure 1. ROS-mediated activation of Get3 as ATP-independent chaperone.
A. Effect of an 8-fold molar excess of Get3red or Get3ox on the light scattering of 75 nM chemically denatured citrate synthase (CS). Light scattering of CS in the absence of added chaperones is shown as control. B. The chaperone function of 0.3 µM Get3red before and at defined time points after incubation in 2 mM H2O2/50 µM Cu2+ at 37°C was determined by analyzing the influence of Get3 on the aggregation of 75 nM chemically denatured CS. The light scattering signal of CS in the absence of added chaperones was set to 0% chaperone activity, whereas the light scattering signal in the presence of fully oxidized Get3 was set to 100%. Insert: The number of cysteine thiols in reduced and oxidized Get3 was determined under denaturing conditions using Ellman’s assay. The precise amount of zinc associated with reduced and oxidized Get3 was determined by ICP analysis. C. Relative ATPase activity (upper panel) and chaperone activity (lower panel) of reduced and oxidized wild-type Get3 and the ATPase deficient Get3 D57E mutant. A four-fold excess of Get3 to CS was used for the chaperone assays. At least 3–6 replicates were performed and the standard error is shown.
Mechanism of Get3’s activation process
Whereas mixtures of peroxide and copper produce highly reactive hydroxyl radicals that rapidly react with cysteine thiols, Cu2+ is known to catalyze the auto-oxidation of cysteine thiols displaying biphasic oxidation kinetics (Kachur et al., 1999). To investigate whether incubation of Get3 with H2O2/Cu2+ or Cu2+ alone leads indeed to the oxidation of Get3’s cysteines, we compared the thiol oxidation status of Get3red with that of the two chaperone-active species, Get3 treated with 2 mM H2O2 / 50 µM Cu2+ for 10 min (Get3ox) or Get3 treated with 50 µM Cu2+ for 240 min (Get3Cu2+)(Figure 1B, insert). The reaction revealed about seven cysteines for Get3red, representing the four absolutely conserved cysteines as well as three non-conserved cysteines. In contrast, we detected only about three cysteines in the chaperone-active species. These results suggest the formation of two disulfide bonds in Get3ox, whose formation either precedes or parallels the activation of Get3 as chaperone holdase. Since the cysteine thiols of the C-X-Y-C motif contribute to dimer formation via zinc coordination (Mateja et al., 2009), we next tested whether the observed oxidation of Get3 causes zinc release. We therefore performed metal analysis using inductively coupled plasma mass spectrometry (ICP) of both oxidized and reduced species. Indeed, ICP analysis revealed the presence of about 0.9 zinc equivalents per Get3 dimer in the reduced form (Figure 1B, insert), and only about 0.2 equivalents of zinc in the oxidized species. According to the 4-(2-pyridylazo)resorcinol/parachloromercuri-benzoic acid (PAR/PCMB) assay, which allows discrimination between high-affinity cysteinecoordinated and low-affinity surface metal binding (Ilbert et al., 2007), only reduced Get3 had metals bound via cysteine thiols (Figure S1D). From these experiments we concluded that Get3’s activation as ATP-independent chaperone involves thiol oxidation and concomitant release of the cysteine-coordinated zinc.
Activation of Get3’s chaperone function is a fully reversible process in vitro
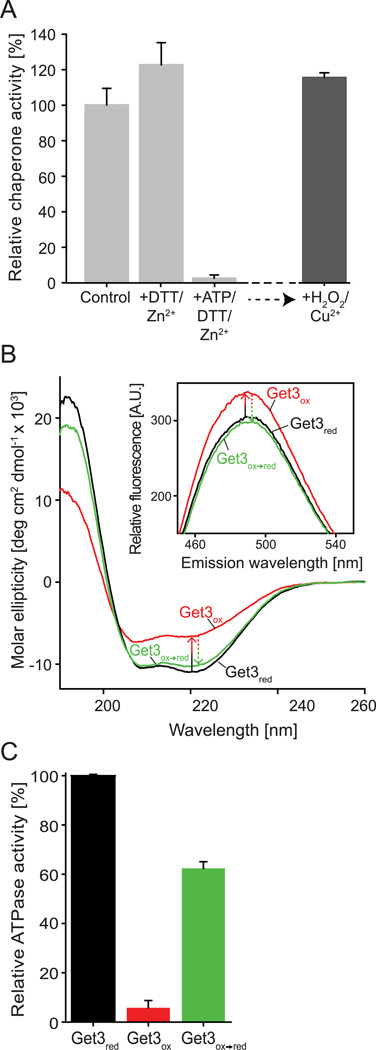
Reversibility is a major aspect of every posttranslational regulation event. To test whether oxidative activation of Get3’s chaperone function and, correspondingly, the inactivation of Get3’s ATPase activity are reversible processes, we incubated Get3ox with DTT and Zn to reduce any reversible thiol modifications and reconstitute the zinc site. We found that treatment with DTT rapidly re-reduced the cysteines, with six cysteines detectable after 5 min of DTT treatment and all seven cysteines accessible within 30 min of incubation. However, even after 6-hour treatment with DTT and zinc, the chaperone activity of Get3 was not significantly different from untreated Get3ox incubated over the same amount of time (Figure 2A). These results were highly reminiscent of oxidized Hsp33, whose inactivation was found to be very slow despite the very rapid reduction of its disulfide bonds (Hoffmann et al., 2004). We therefore reasoned that like Hsp33, Get3 might undergo conformational rearrangements, whose re-organization upon cysteine reduction and zinc-coordination might become rate-limiting in the inactivation process. Indeed, comparison of the secondary structure of Get3red and Get3ox by far-UV-CD revealed extensive conformational changes upon oxidation, consistent with a substantial loss in α-helices and the accumulation of random-coil structure (Figure 2B, compare black and red traces). Binding studies with bis(4-anilino-5-napththalenesulfonic acid) (bis-ANS), a fluorescent sensor molecule of hydrophobic surfaces revealed that this partial unfolding of Get3ox was accompanied by the exposure of hydrophobic surfaces, a hallmark of active chaperones (Figure 2B, insert).
Figure 2. Activation of Get3’s chaperone function is a fully reversible process.
A. Chaperone-active wild-type Get3ox (0.3 µM) was incubated in the presence of either 5 mM DTT / 5 µM zinc (Get3ox→DTT/Zn) or 2 mM MgATP, 5 mM DTT / 5 µM zinc (Get3ox→red) for 6 h at 30°C. The reductants were removed and the various Get3 preparations were tested for their ability to prevent aggregation of chemically denatured CS as outlined in Figure legend 1. To investigate whether Get3 can undergo multiple rounds of oxidation and reduction processes, we incubated the chaperone-inactive Get3ox→red with 2 mM H2O2 and 50 µM Cu2+ for 10 min and tested its chaperone function as described. B. Analysis of secondary structure and surface hydrophobicity (insert) of Get3red (black trace), Get3ox (red trace) and Get3ox→red (green trace) using far-UV circular dichroism and bis-ANS fluorescence spectroscopy (insert), respectively. C. ATPase activity of Get3ox and Get3ox→red relative to untreated Get3red.
Since the ATP-binding site in Get3 is located in close vicinity to two of the four highly conserved cysteines (C242/C244), we reasoned that Mg-ATP binding should stabilize the reduced form and hence accelerate refolding. Indeed, as shown in Figure 2A, in the presence of DTT, Zn2+ and MgATP, Get3ox was completely inactivated within the time frame of the experiment (Get3ox→red), and could be fully reactivated upon subsequent incubation with Cu2+/H2O2. These results suggest that presence of MgATP plays a critical role for the inactivation of Get3’s chaperone function. This makes physiological sense since restoration of cellular ATP levels serves to indicate a cell’s return to pre-stress conditions. Analysis of the secondary structure revealed almost complete reversal of the structural rearrangements and decrease in surface hydrophobicity in Get3 upon incubation with DTT, Zn2+ and MgATP (Figures 2B, compare red and green traces), paralleled by a substantial reactivation of Get3’s ATPase activity (Figure 2C). These results demonstrate that the oxidative activation of Get3 as an chaperone holdase is a reversible process, enabling Get3 to potentially undergo multiple rounds of oxidative activation as molecular chaperone.
The minimal unit of chaperone-active Get3 is the oxidized tetramer
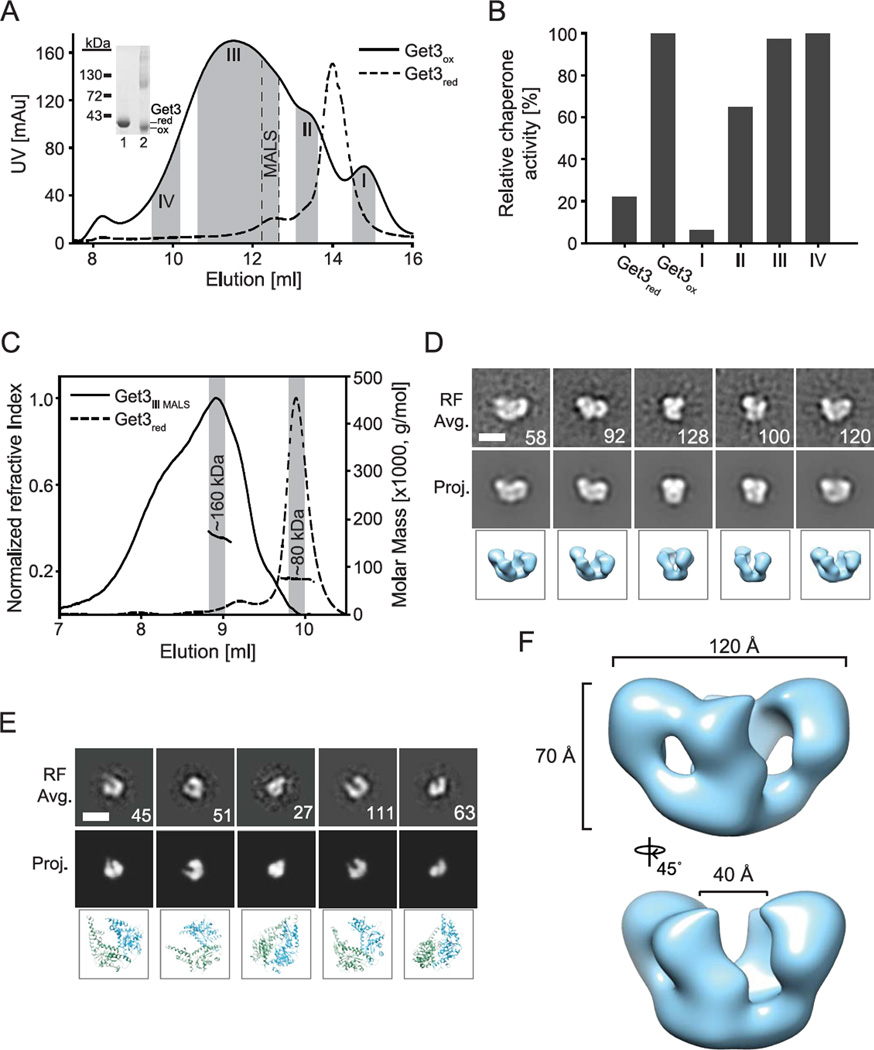
Analysis of reduced and oxidized Get3 on non-reducing SDS-PAGE revealed the expected monomeric migration behavior of Get3red and a slightly faster migrating Get3ox monomer, indicative of intramolecular disulfide bonds (Figure 3A, insert). In addition, however, we also noted the presence of some dimers and higher oligomers in our Get3ox preparation. To determine if oligomerization correlates with the chaperone function of Get3ox, we separated oxidized Get3 on size exclusion chromatography, collected individual peak fractions (Figure 3A, shaded areas I–IV), and determined their respective chaperone activity (Figure 3B). Whereas the specific chaperone activity of Get3ox was negligible in fractions harboring species that eluted in a similar volume as the reduced Get3 dimer, it was high in fractions that contained higher oligomeric Get3-species. To better characterize the stoichiometry of the minimal oligomeric unit of Get3ox that confers chaperone function, we further separated fraction III, and re-chromatographed each of the sub-fractions using a second size exclusion column connected to a static light scattering instrument for molecular weight determination (SEC-MALS). The fraction corresponding to the smallest chaperone-active Get3ox complex (indicated as “MALS” in Figure 3A) eluted as a broad single peak at ~9.0 ml in the SEC-MALS (Figure 3C). The average molecular weight was determined to be about 160 kDa, indicating a tetramer complex. Notably, we also observed a large shoulder and an increasing molecular weight across the peak, indicating larger oligomers are present as well. By comparison, Get3red eluted at ~ 10 ml with a calculated molecular weight of 80 kDa, confirming that it is a dimer under these conditions (Figure 3C).
Figure 3. Chaperone-active Get3ox forms distinct oligomeric species.
A. Get3red and Get3ox were analyzed on size exclusion chromatography. Peak fractions (indicated by gray bars) containing various oligomeric states of Get3ox were collected (fractions I–IV) and B. subsequently analyzed for their chaperone activity as described in figure legend 1. Insert: Analysis of Get3red (lane 1) and Get3ox (lane 2) on non-reducing SDS-PAGE. C. Get3red and the Get3ox sub-fraction of fraction III (indicated as MALS in A) were analyzed by SEC-MALS and calculated to be approximately 160 and 80 kDa, respectively. Distinct fractions of the SEC-MALS elution were taken (gray bars in C.) and used to determine negative-stain EM 2D reference-free class averages of D. Get3ox and E. Get3red using SPIDER (Frank et al., 1996). Matching projections and corresponding images of the D. Get3ox 3D model and E. Get3red dimer crystal structure (PDB: 3H84) are shown (Hu et al., 2009). The number of single particles is shown for each average and the scale bars equal 100 Å. F. Final 3D model of the Get3ox tetramer complex, filtered to 19 Å.
To further characterize the architecture of the oxidized Get3 oligomeric states, we conducted negative-stain electron microscopy (EM) on peak fractions following SEC-MALS. In micrograph images and single particles, Get3ox appeared larger and in a different arrangement compared to images of the Get3red dimer (Figure S2A). Single particle data sets were then collected and analyzed by generating 2D reference-free projection averages. For Get3ox, the 2D averages showed a globular arrangement with two defined lobes that form a ‘W’ shape in some views (Figure 3D). Larger, elongated complexes were also observed in the overall set of 2D averages, indicating the presence of multiple oligomeric states, however the smaller, compact form clearly predominated (Figure S2B). The arrangement was different than the crystallographic tetramer of the archaeal homologue, which forms an elongated ‘dumbbell’ shape (Suloway et al., 2012). Reference-free classification of the Get3red dimer was also performed and clearly showed a small ‘U’ shaped arrangement that matched well with 2D projections and corresponding 3D views of the crystal structure (Hu et al., 2009) (Figure 3E). Based on comparisons of these 2D projection images it is clear that Get3ox is larger then Get3red, and contains two lobes that are consistent with two dimers of Get3 connected in a tetrameric complex.
To further characterize the architecture of the Get3ox tetramer, we determined a 3D reconstruction. Rounds of refinement were performed using a sphere as an initial model with no imposed symmetry (Figure S2C). The final model, calculated to be 19 Å by the Fourier shell correlation procedure (Figure S2D), was achieved following additional rounds of refinement with 2-fold symmetry imposed. The reconstruction reveals a 70 by 120-Å complex with two oval-shaped domains that are connected along one side, resulting in a 40-Å cleft between the domains (Figure 3F). Importantly, the reference-free averages agree with the 2D projections of the model, supporting this tetramer arrangement (Figure 3D). The crystal structures of the dimer and tetramer forms of Get3 were docked into the EM map and found to be incompatible, however the dimensions of each oval domain in the 3D model are consistent with the size and shape of the dimer. Overall the SEC-MALS and EM structural work identifies that the minimal chaperone-active form of Get3ox is a distinct tetramer complex that is likely in a different conformation and arrangement than previously characterized structures of the reduced form.
Oxidative activation of Get3 causes massive structural rearrangements
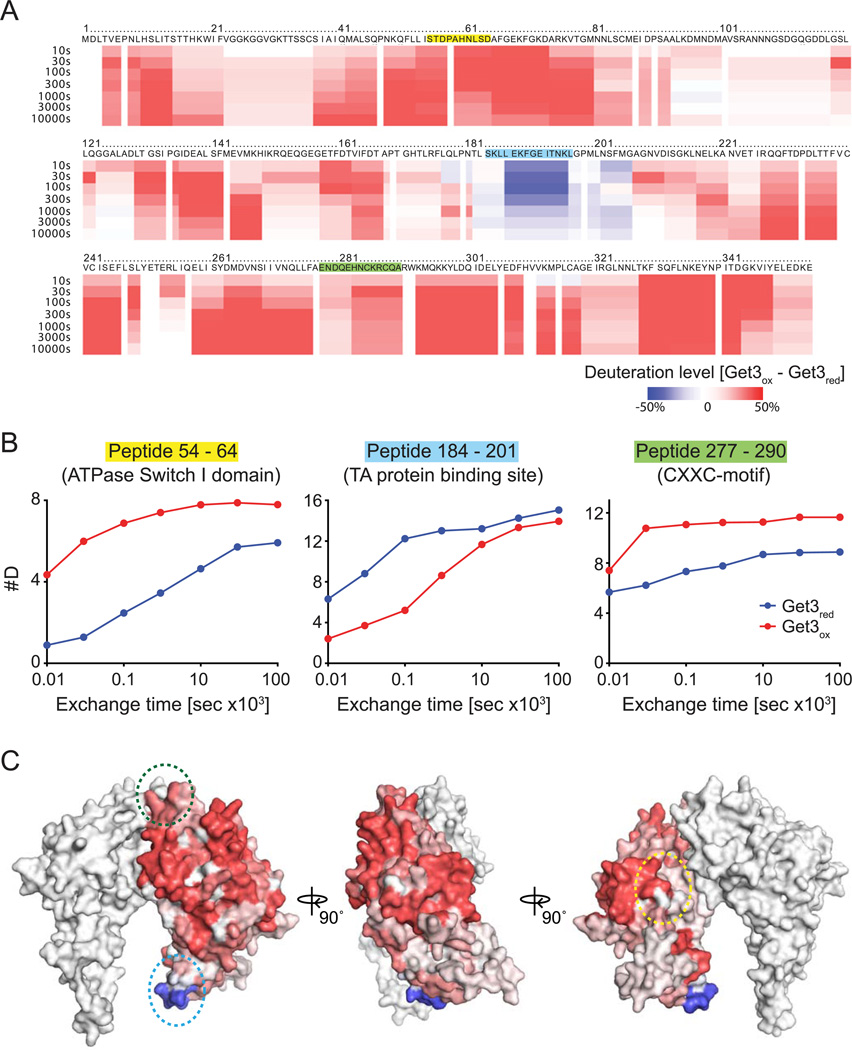
To obtain more detailed insights into the structural changes that Get3 undergoes during its oxidative activation process, we decided to conduct kinetic H/D exchange experiments in combination with mass spectrometry (Englander, 2006). Proteins of interest are incubated in deuterated buffer to allow amide protons to exchange with deuterium. After defined time points, the exchange is quenched by a sudden shift to pH 2.5. Then, the protein is digested with pepsin in the presence of 1 M Gdn-HCl, and the resulting peptides are analyzed by MS/MS. We first analyzed reduced Get3red and tested its proteolytic sensitivity towards pepsin, assessing the protein coverage. We found that a 30 sec treatment with pepsin was sufficient to digest Get3red into numerous, often overlapping fragments that covered ~99.8% of the entire Get3. When we compared the digestion pattern of oxidized, disulfide-bonded Get3ox in the absence and presence of the strong thiol reductant 100 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), however, we observed some striking differences; while the pepsin digest in the presence of TCEP looked indistinguishable from the pepsin digest of Get3red, the absence of TCEP led to a near-absence of proteolytic cleavage in the regions surrounding both pairs of conserved cysteines as well as the non-conserved Cys317 (Figure S3A, left panel). These results suggest that thiol oxidation prevents pepsin from accessing its target sites consistent with our conclusion that the conserved cysteines are involved in disulfide bonds. Manual inspection of the MS spectra allowed us to identify two additional peptides, which harbor the conserved cysteine pairs in their oxidized, disulfide-bonded state, indicating that the four conserved cysteines engage in intramolecular disulfide bonds, connecting the next-neighbor cysteines (Figure S3B). Since pepsin digestion of TCEP-reduced Get3ox was indistinguishable from the digestion pattern of Get3red that was never oxidized, we conducted all subsequent pH quench and pepsin digests in the presence of 100 mM TCEP. By using this method, which has been previously established to avoid differences in pepsin coverage when comparing oxidized and reduced proteins (Zhang et al., 2010), we were able to directly compare the mass differences in the individual Get3 peptides over time (Table S1). As shown in Figure S3C and Figure 4, analysis of the H/D exchange rates in Get3ox versus Get3red peptides revealed astounding differences. Nearly every region in Get3ox showed a faster exchange rate with deuterium than the corresponding region in Get3red as indicated by the red shading (Figure 4A), and shown for select peptides in Figure 4B (the complete set of H/D exchange rates for all identified peptides can be found in Table S1). These results are fully consistent with our CD data and indicative of an oxidation-induced partial unfolding of Get3’s structure and/or increase in dynamic properties of the protein. They are reminiscent of other recently identified stress-specific chaperones, including Hsp33 and HdeA, whose activation appears to be triggered by significant protein unfolding (Reichmann et al., 2012; Tapley et al., 2009). Astonishingly, the only region in Get3ox that showed a slower exchange with deuterium than the corresponding region in Get3red involved residues 184–201 of the alpha-helical subdomain in Get3 (Figure 4B middle panel; Figure 4C indicated by blue circle), which is proposed to form the composite hydrophobic binding site for TA-proteins (Mateja et al., 2009). Mutagenesis in this region revealed several Get3 variants that had lost their capacity to co-immunoprecipitate the TA-protein Sec61beta but retained high levels of ATPase activity indicative of a folded protein (Mateja et al., 2009). These results revealed that oxidative activation of Get3 leads to major conformational rearrangements, which cause the exposure of binding sites for unfolding proteins while potentially masking the binding sites for TA-proteins.
Figure 4. Get3 undergoes massive conformational rearrangements upon oxidation.
A. Incorporation of deuterium after select times into Get3ox versus Get3red. Quenching and pepsin digest was performed in the presence of 100 mM TCEP. The deuteration level was calculated as described in the experimental procedures. B. Direct comparison of the deuterium incorporation over time into select Get3 peptides prepared from either Get3red (blue trace) or Get3ox (red trace). C. Differences in deuterium incorporation between Get3ox and Get3red after 100 sec of H/D exchange, using the Get3red crystal structure (PDB: 3H84). Location of peptides shown in B is indicated using color-coded dotted circles.
Dissecting the two Get3 functions in vivo
Our structural analysis suggested that the binding of Get3 to TA-proteins under non-stress conditions and the interaction of Get3 with unfolding proteins under stress conditions might represent two independent and hence separable protein functions. We therefore decided to purify and test one of the mutant proteins (Get3 I193D), which has been previously shown to no longer bind TA-proteins (Mateja et al., 2009). We reasoned that if our hypothesis was correct, this mutant protein would still work as a redox-active chaperone and would complement specifically those get3 phenotypes that require the redox-regulated chaperone function but not the TA-protein targeting function of Get3.
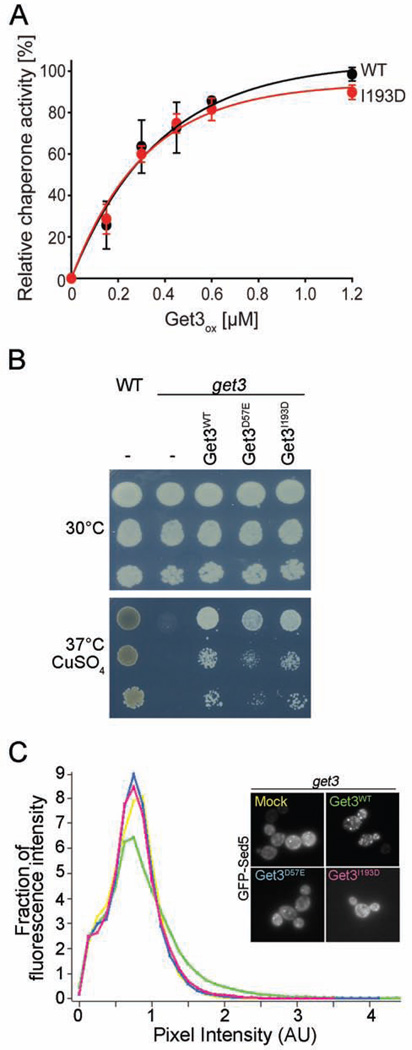
Purification of the Get3 I193D variant was indistinguishable from that of wild type Get3, excluding major structural alterations in the protein. Furthermore, analysis of its ability to prevent protein aggregation in vitro revealed wild-type like redox-regulated chaperone function with little chaperone activity in the reduced form (Figure S4A) and high chaperone function in the oxidized form (Figure 5A). When we tested the Get3 I193D mutant variant for its capacity to rescue the oxidative stress phenotype of a get3 strain, we found that the mutant protein rescued the growth defect of the deletion strain to the same extent as the wild-type protein, indicating that it is likely the chaperone function of Get3 I193D that is essential to cells under these stress conditions (Figure 5B). The phenotype was also partially complemented by expression of the ATPase-deficient Get3D57E. Although this result is consistent with the observation that the client binding function of Get3 is independent of ATP, we cannot exclude that the very low residual ATPase activity contributes to the growth rescue under oxidative stress conditions (Figure 5B). To further segregate the in vivo chaperone function from any residual activity in TA-protein targeting, we then assayed the mutant proteins for a potential dominant negative effect, which has been observed for Get3-expressing strains in the absence of the Get1/Get2 receptor complex. In this strain background, overexpression of wild-type Get3 is toxic (Figure S4B), thought to be due to Get3’s unproductive sequestration of TA protein precursors, which effectively prevents their membrane targeting by other GET-independent mechanisms (Schuldiner et al., 2008). Expression of the ATPase-deficient Get3 D57E mutant protein was similarly toxic, presumably because Get3 D57E is still able to bind and sequester TA-proteins (Powis et al., 2013). In contrast, we found no dominant negative effect for the Get3 I193D mutant variant (Figure S4B), whose expression levels were very similar to the levels of soluble wild-type Get3 and Get3 D57E (Figure S4C). These results are fully consistent with the conclusion that the Get3 I193D mutant variant is no longer capable of binding TA-proteins. We further corroborated this finding by directly testing our Get3 I193D mutant protein in a well-established in vivo TA-targeting assay (Jonikas et al., 2009). In this assay, we followed the fate of a GFP-tagged TA-protein that is known to be GET-dependent (i.e., GFP-Sed5) in a get3 deletion strain, that either expresses no Get3 (mock), wild-type Get3, the ATPase-deficient Get3 D57E variant or the Get3 I193D mutant (Figure 5C, insert). While the strain expressing wild-type Get3 showed the expected formation of multiple puncta against a dark cytosol, indicative of the correct targeting of GFP-Sed5 to the ER and subsequent trafficking to the Golgi, get3 cells expressing either no Get3 or any of the two mutants revealed a much higher cytosolic background and only very few puncta (Figure 5C, insert). These results are highly reminiscent of other reported conditions in which GFP-Sed5 was mistargeted and moved to deposition sites for aggregated proteins (Battle et al., 2010; Kohl et al., 2011; Vilardi et al., 2014). Quantitative analysis further confirmed that GFP-Sed5 (mis)targeting observed in cells expressing either Get3 D57E or I193D was comparable to the strain lacking Get3 altogether (Figure 5C). These results are in excellent agreement with structural studies that demonstrated that a Get3 protein with an altered hydrophobic groove is incapable of TA protein targeting or sequestration (Mateja et al., 2009). Moreover, our results provide strong evidence that TA protein targeting activity of Get3 can be segregated from its redox-dependent chaperone activity, which appears to be responsible for the fitness of yeast cells under copper-induced oxidative stress.
Figure 5. Get3I193D has chaperone activity independent of TA protein targeting.
A. Increasing concentrations of oxidized wild-type Get3 and the TA-binding deficient mutant Get3I193D were tested for their influence on the aggregation behavior of chemically denatured CS (see figure legend 1 for details). B. get3 cells were transformed with an empty construct or with constructs containing the coding sequence of wild-type or mutant variants of Get3. Serial dilutions were then spotted on control plates or plates containing 1 mM of CuSO4 at 37°C. WT (BY4741) cells served as control. C. get3 cells expressing GFP-tagged Sed5 were transformed with an empty construct or with constructs containing the coding sequence of Get3 variants. Subcellular localization of GFP-Sed5 was recorded by fluorescence microscopy (insert). At least 40 cells per strain were analyzed to determine the distribution of fluorescence across bins of different pixel fluorescence intensity.
Get3 co-localizes with unfolding proteins during oxidative stress in vivo
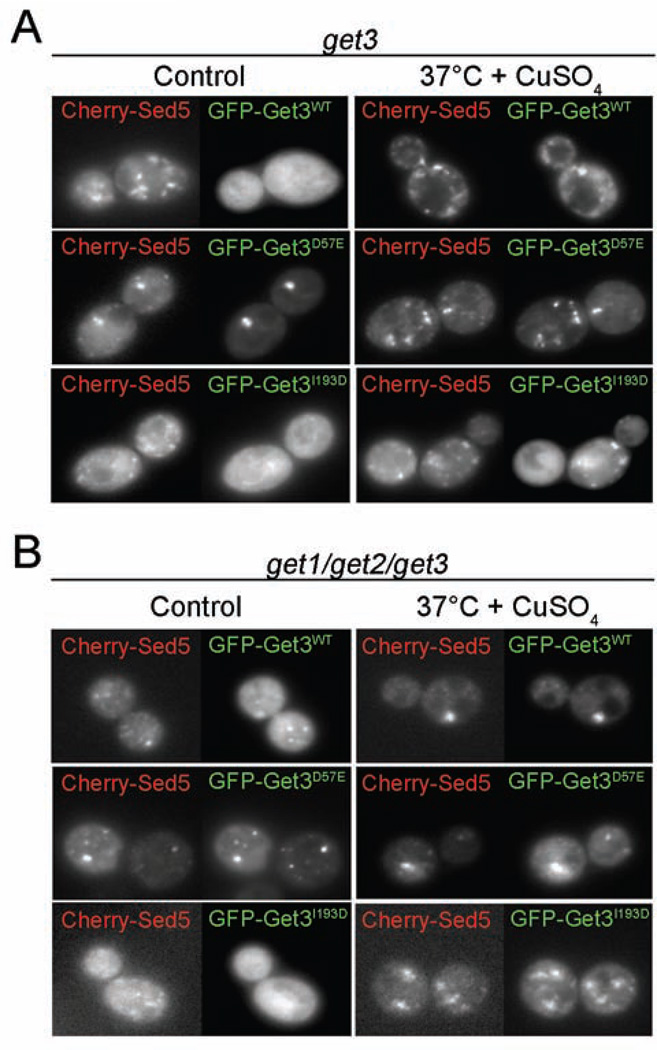
Co-localization studies using GFP-Get3 and Cherry-Sed5 revealed that under non-stress conditions, Get3 is diffusely distributed in the cytosol while the TA-protein is localized to its previously observed distinct Golgi foci (Figure 6A, control panel). Upon exposure of yeast cells to copper stress, however, Get3-GFP localizes to foci, which only partially overlap with the foci formed by Cherry-Sed5 (Figure 6A, right panel), and are reminiscent of previously observed co-deposition sites of Get3, aggregated proteins, and other chaperones (Powis et al., 2013). Intriguingly, we found a very similar oxidative stress-mediated redistribution of the TA-binding deficient Get3 I193D mutant, supporting the notion that the recruitment of Get3 to stress foci occurs through the oxidative activation of its chaperone function and not through TA-protein binding (Figure 6A, lower panel).
Figure 6. Get3 accumulates at deposition sites of mistargeted Sed5 under stress conditions.
A. get3 or B. get1/get2/get3 deletion cells expressing mCherry-tagged Sed5 in combination with GFP-tagged Get3 (wt or mutants) were grown under control conditions (30°C) or in presence of 1 mM CuSO4 at 37°C for 4 hours. Subcellular distribution of mCherry-Sed5 and GFP-Get3 was recorded by fluorescence microscopy.
Analysis of Get3-GFP localization in the GET receptor-deficient get1get2get3 deletion strain further confirmed these conclusions. In this strain background, wild-type Get3 is not freely diffusible but co-localizes with TA-proteins in distinct foci even under non-stress conditions (Figure 6B). This is presumably because TA-protein can no longer be released from Get3 and inserted into the membrane, which explains the previously shown dominant negative effect of wild-type Get3 expression in a strain that lacks the receptor complex (Schuldiner, 2008). As expected, the Get3 I193D mutant variant did not co-localize with Cherry-Sed5 in the strain lacking the ER receptor proteins (Figure 6B). However, once challenged with copper stress, we observed the re-localization the Get3 I193-GFP mutant variant into distinct stress foci. These results demonstrate that Get3 exerts a TA-binding-independent function under oxidative stress conditions in vivo, thereby playing an important role in protecting yeast cells during stress conditions that lead to protein unfolding and ATP-depletion.
Discussion
Reduced Get3 cycles between an open conformation with low affinity for TA-proteins in the absence of nucleotides and a closed, high affinity conformation in the presence of Mg-ATP or ADP•AIF4 (Mateja et al., 2009). Upon closing, a composite hydrophobic groove is formed by the alpha-helical subdomains of the two Get3 monomers, which accommodates the transmembrane segment of the TA-protein (Mateja et al., 2009). It has been noted that this alpha-helical subdomain differs substantially from that of the prokaryotic Get3 homologue ArsA as it is enriched in methionines like the hydrophobic binding region of the signal recognition particle (SRP) and features a 20-residue insert (TRC40-insert) unique to and conserved in eukaryotic homologues (Mateja et al., 2009). This observation strongly suggests different molecular functions for Get3 and ArsA, which has been shown to be involved in resistance to arsenite (Zhou et al., 2000). Analysis of yeast Get3, prior to the groundbreaking discovery that the homologous mammalian TRC40 cross-linked to TA-proteins during posttranslational membrane targeting (Favaloro et al., 2008; Stefanovic and Hegde, 2007) also clearly indicated a role for Get3 in metal resistance (Metz et al., 2006; Shen et al., 2003). However, due to the broad scope of TA-proteins in various cellular functions, and in particular the key role of SNARE TA-proteins in sustaining vesicular traffic and hence proper function of the endomembrane system in metal transport and sequestration, it became difficult to assess whether the copper stress phenotype of the get3 deletion strain was fully explained by the lack of TA-protein targeting via the GET pathway. Here we now demonstrate a second function of Get3 as oxidative stress-activated, ATP-independent chaperone holdase (Figure 7). By using a TA-protein targeting deficient yet fully chaperone-active variant of Get3, we discovered that the two Get3 functions can indeed be segregated in vivo, and that the copper sensitivity observed in a get3 strain is caused by the loss of the Get3 chaperone and not by impaired TA-protein targeting. Furthermore, our observation that both Get3 and the Get3 variant deficient in TA-protein targeting specifically accumulate in foci upon an oxidative challenge supports the idea that Get3ox serves as an integrated member of the cellular chaperone network, active at sites where unfolding and aggregation-prone proteins are sequestered. These results explain previous findings that the cellular abundance of Get3 (~17,300 molecules) is up to 10 times higher than that of other members of the GET pathway, such as Get1 (~2,250 molecules), and matches well with the abundance of other molecular chaperones, including Hsp104 (~32,800 molecules) (Ghaemmaghami et al., 2003). Our finding that Get3 appears to serve a dual purpose in vivo is also in excellent agreement with previous studies conducted in Caenorhabditis elegans, which revealed that deletion of the Get3 homologue TRC40 (ASNA1) causes two distinct phenotypes, a severe growth defect and increased sensitivity to cisplatin (Hemmingsson et al., 2010), an anti-cancer drug known to cause oxidative stress. Intriguingly, while both phenotypes were rescued by expression of wild-type TRC40, a TRC40-variant lacking the conserved, redox-sensitive cysteines rescued the growth defect but not the cisplatin-sensitivity (Hemmingsson et al., 2010). These results indicate that TRC40 works as a dual-function protein also in higher eukaryotes.
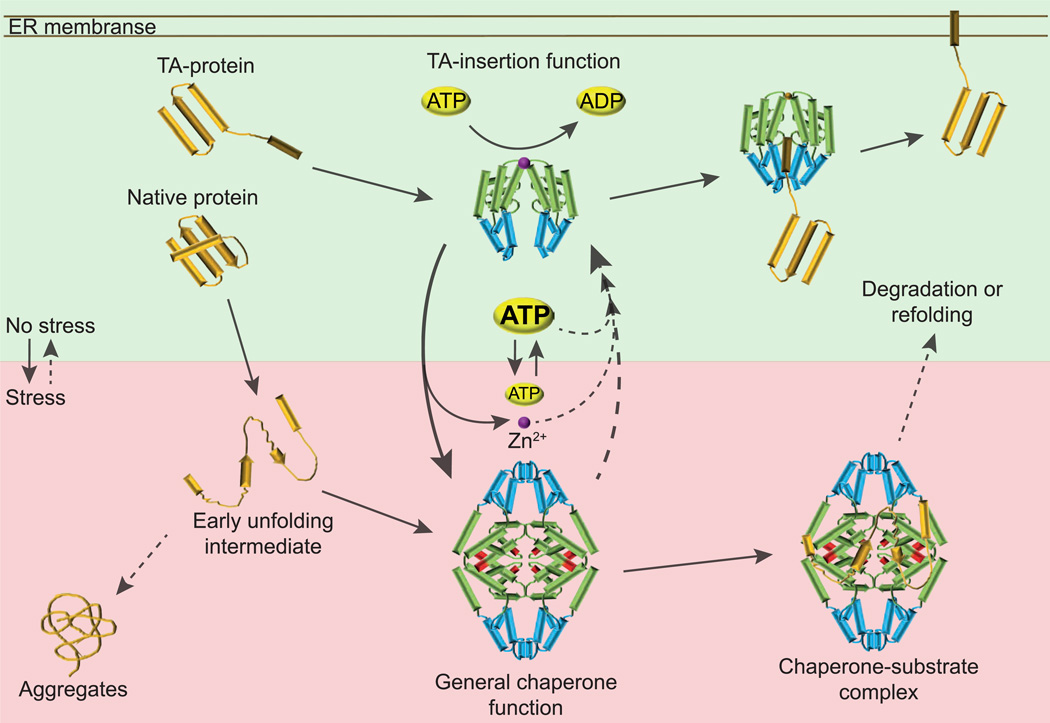
Figure 7. Get3 – A Dual-Function Protein.
Under non-stress conditions, Get3 functions as cytosolic component of the GET complex, which supports insertion of TA-proteins into the ER membrane. Reduced and homodimeric, Get3 is stabilized by zinc coordination (magenta sphere). Binding of TA-clients to the composite binding site in Get3 (indicated in blue), and subsequent release on the membrane is regulated by ATP-binding and hydrolysis (domain indicated in green), and involves several other cytosolic and membrane proteins, which are not shown for reasons of simplicity. Upon exposure to ATP-depleting oxidative stress conditions, Get3’s cysteines are oxidized (marked in red) and zinc is released. Oxidation causes major conformational rearrangements, which appear to bury the proposed TA-binding site on Get3 and turn Get3 into an ATP-independent, highly chaperone-active tetramer. Oxidized Get3 binds unfolding proteins and prevents their irreversible aggregation, protecting cells against oxidative stress. Upon return to non-stress condition and restoration of normal cellular ATP levels, Get3 returns into its initial dimer structure and presumably releases its substrate for refolding or degradation.
Hsp33 is a prokaryotic paradigm for an oxidation-activated chaperone and, although there is no sequence homology between Hsp33 and Get3, many analogies can be found between the two proteins: both proteins coordinate zinc via a highly conserved CXXC-motif, release zinc upon oxidative disulfide bond formation, and undergo partial unfolding that gives rise to an alternative protein structure, now acting as ATP-independent molecular chaperone. In addition to Hsp33, Get3 shares also several features with other ATP-independent chaperones, including small heat shock proteins (Richter et al., 2010). Both proteins form higher-order oligomers, contain intrinsically disordered regions, and show the propensity to partition into deposition sites for unfolded proteins in cells (Powis et al., 2013; Richter et al., 2010; Sudnitsyna et al., 2012). The chaperone function of Get3 protects cells against protein unfolding during oxidative stress conditions, either induced by exogenous application of ROS or endogenously elicited by ATP-depletion, which causes a loss in vacuolar ATPase activity, leading to chronic oxidative stress (Milgrom et al., 2007). Get3 then co-localizes with chaperones of the Hsp70 family and Hsp104 at deposition sites for aggregated proteins and can be chased out of this localization upon restoration of energy levels (Powis et al., 2013). These findings agree well with our in vitro inactivation studies, which showed that Mg-ATP massively accelerates the re-conversion of oxidized chaperone-active Get3 into its reduced ATPase-active form. These results imply that the availability of ATP rather than the end of the oxidative challenge may serve as the physiological cue involved in switching the Get3 chaperone off and returning the protein to the TA-protein targeting function.
In the context of the GET pathway, Get3 forms complexes with either the cytosolic proteins Get4 and Get5 further connecting it to Sgt2, which is thought to initially bind the TA-protein substrate and hand it over to Get3 (Wang et al., 2010), or with the ER membrane receptor formed by Get1 and Get2. It is currently unclear whether free Get3 exists in the cell and how interaction with Get4 and Get5 or the receptor would impact its capacity to be converted to a molecular chaperone by oxidation. Similarly, future experiments will have to test a potential role of Get3’s ATPase activity in its functional conversion and/or in linking its chaperone activity to other proteostatic mechanisms in vivo. Many genetic links exist between the GET pathway and major networks contributing to proteostasis (Schuldiner et al., 2008). So far these links have mainly been interpreted in the light of TA-protein precursors being an aggregation-prone protein species that may pose a threat to proteostasis when accumulating. However, in the context of our discovery that Get3 functions as a molecular chaperone specifically upon oxidative stress conditions, some of these links will now need to be reconsidered to reveal how the Get3 chaperone is integrated into the cellular network that safeguards proteostasis.
Experimental Procedures
Yeast strains, plasmids and growth conditions
Strains and plasmids used in this study are listed in Table S2. Construction of the plasmids not previously described is detailed in the supplemental Material. Cells were grown in Hartwell's Complete (HC) medium and experiments were performed at mid-log phase. Copper sensitivity was determined on HC agar plates in the absence or presence of 1 mM CuSO4 after 2–3 days at the indicated temperatures.
Get3 purification, reduction, oxidation and re-reduction
Wild-type Get3 and the mutant variants were purified as described in the Supplemental Material section. To prepare Get3red, ~100 µM Get3 in storage buffer (50 mM Tris, 50 mM NaCl, 5 mM MgAc, pH 7.5) was incubated with 5 mM dithiothreitol (DTT) for 1 h at 30°C. DTT was removed using a Zeba spin column (Thermo Scientific) or Micro Spin 30 (Bio-Rad) column equilibrated with 40 mM HEPES-KOH (pH 7.5). For oxidation, Get3red was incubated with either 2 mM H2O2/50 µM CuCl2 (Get3ox) for 4 or 10 min, various concentrations of CuCl2 (incubation time ranged between mixing time and 240 min) or 2 mM H2O2 for the indicated time points at 37°C. Oxidants were removed as described. To remove copper, the protein was incubated with 5 mM TPEN (10 min at 30°C) before loading it onto the spin column. To re-reduce Get3ox, 5 µM Get3ox (activated for 4 min) was incubated with 5 mM DTT/5 µM ZnCl2 for up to 6 h at 37°C in the absence or presence of 2 mM MgATP. The reductants were removed as described.
Chaperone and ATPase activity
To analyze chaperone activity, the influence of Get3 on the aggregation of chemically denatured citrate synthase (CS) was tested (Buchner et al., 1998). The ATPase activity of 2–4 mM Get3 was monitored using a NADH-coupled ATPase assay in a 96-well BMG FLUOstar Omega microplate reader (Kiianitsa et al., 2003).
Redox state and metal analysis
To determine the number of cysteine thiols in Get3, the proteins were denatured in 6 M Gdn-HCl, 40 mM KPi buffer, pH 7.5 followed by an Ellman’s assay (Riddles et al., 1983). To determine the amount of zinc bound to reduced and oxidized Get3, the cysteine-coordinated zinc was determined using the PAR/PCMB assay (Ilbert et al., 2007). For ICP analysis, samples were purified twice using Micro Spin 30 columns, equilibrated with metal-free 50 mM Tris, 50 mM NaCl, pH 7.5 buffer.
Monitoring structural changes in Get3
To determine changes in the secondary structure, far-UV CD spectra of Get3 (5 µM) in 20 mM KH2PO4, pH 7.5 were recorded (Jasco-J810) at 25°C. Changes in surface hydrophobicity were monitored in a Hitachi F4500 fluorescence spectrophotometer using 5 µM Get3 and 15 µM bis-ANS in 40 mM Hepes, pH 7.5.
Analytical gel filtration, SEC-MALS and negative stain electron microscopy (EM)
Get3red and Get3ox was applied onto a Superdex 200 column (GE Healthcare), equilibrated with 40 mM HEPES, 140 mM NaCl, pH 7.5. Individual fractions of the gel filtration run were collected, pooled and tested for chaperone activity as described. The average Mw of was determined by separation using a WTC-050S5 SEC column (Wyatt Technology Corporation) with an Akta micro (GE Healthcare) and analysis using a DAWN HELEOS II MALS detector and Optilab rEX differential refractive index detector using ASTRA VI software (Wyatt Technology Corporation). The Mw was calculated from Raleigh ratio based on the static light scattering and corresponding protein concentration of a selected peak. For negative-stain EM, fractions were diluted and stained with 0.75% uranyl formate (pH 5.5–6.0) on thin carbon-layered 400 mesh copper grids (Pelco) (Ohi et al., 2004) and micrographs were taken at 52,000× magnification with 2.16 Å per pixel using a 4k × 4k CCD camera (Gatan). Sample imaging and analysis is described in detail in the supplemental material sections. The final model, at 19 Å resolution by the gold-standard FSC, was achieved after 9 rounds of refinement (Figure S2D).
Hydrogen/Deuterium exchange experiments combined with mass spectrometry
Prior to performing deuteration studies, enzymatic digestion conditions of Get3 were optimized as previously described (Marsh et al., 2013). A greater than 99.8% peptide coverage maps of Get3 was obtained using quench buffer containing 1 M Gdn-HCl, and 100 mM TCEP. The H/D exchange experiments and mass spectrometric analyses were conducted as described in detail in the supplemental material section. The deuteration level for each peptide was calculated according to equation 1:
Deuteration (%) = C(P)-C(N)) / (C(F)-C(N)) × C(N) × MaxD × 100
where C(P), C(N) and C(F) are the centroid values of partially deuterated peptide, non-deuterated peptide, and fully deuterated peptide at each time point. MaxD is the maximum deuterium incorporation for each peptide. It is calculated as the number of amino acids in the peptide minus 2 minus any proline that is present beyond position 2 of the peptide.
Live Cell Fluorescence microscopy
Images of yeast cells were acquired on a Delta Vision RT (Applied Precision) microscope using a 100×/0.35–1.5 Uplan Apo objective and specific band pass filter sets for GFP or mCherry. The images were acquired using a Coolsnap HQ (Photometrics) camera. Image processing was performed using ImageJ (http://rsbweb.nih.gov/ij/). Pixel fluorescence intensity of at least 40 cells per sample was quantified as described in (Jonikas et al., 2009; Vilardi et al., 2014) using Knime software (www.knime.org/knime).
Supplementary Material
Acknowledgements
We are grateful to Dr. Heather Tienson for initiating some of the experiments, Ted Houston for his help with metal analysis and Robert Keenan for the plasmid encoding Get I193D. This work was supported by the National Institute of Health awards [GM065318] to U.J., [R01AI081982, R01GM020501, R01AI1014360] to S.L. and by the DFG grant Schw823/3-1 to U.J. and B.S, which funded W.V. The Chemical Biology Interface training grant [GM008597] funded S.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Battle A, Jonikas MC, Walter P, Weissman JS, Koller D. Automated identification of pathways from quantitative genetic interaction data. Mol Syst Biol. 2010;6:379. doi: 10.1038/msb.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner J, Grallert H, Jakob U. Analysis of chaperone function using citrate synthase as nonnative substrate protein. Methods Enzymol. 1998;290:323–338. doi: 10.1016/s0076-6879(98)90029-5. [DOI] [PubMed] [Google Scholar]
- Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hemmingsson O, Kao G, Still M, Naredi P. ASNA-1 activity modulates sensitivity to cisplatin. Cancer Res. 2010;70:10321–10328. doi: 10.1158/0008-5472.CAN-10-1548. [DOI] [PubMed] [Google Scholar]
- Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U. Identification of a redox-regulated chaperone network. Embo J. 2004;23:160–168. doi: 10.1038/sj.emboj.7600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Li J, Qian X, Denic V, Sha B. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS One. 2009;4:e8061. doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007 doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachur AV, Koch CJ, Biaglow JE. Mechanism of copper-catalyzed autoxidation of cysteine. Free Radic Res. 1999;31:23–34. doi: 10.1080/10715769900300571. [DOI] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer WD. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal Biochem. 2003;321:266–271. doi: 10.1016/s0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- Kohl C, Tessarz P, von der Malsburg K, Zahn R, Bukau B, Mogk A. Cooperative and independent activities of Sgt2 and Get5 in the targeting of tail-anchored proteins. Biological Chemistry. 2011;392:601–608. doi: 10.1515/BC.2011.066. [DOI] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M, Mateja A, Dobosz M, Bove E, Hegde RS, Keenan RJ. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477:61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JJ, Guan HS, Li S, Chiles PG, Tran D, Morris TA. Structural insights into fibrinogen dynamics using amide hydrogen/deuterium exchange mass spectrometry. Biochemistry. 2013;52:5491–5502. doi: 10.1021/bi4007995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A, Szlachcic A, Downing ME, Dobosz M, Mariappan M, Hegde RS, Keenan RJ. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz J, Wachter A, Schmidt B, Bujnicki JM, Schwappach B. The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. J Biol Chem. 2006;281:410–417. doi: 10.1074/jbc.M507481200. [DOI] [PubMed] [Google Scholar]
- Milgrom E, Diab H, Middleton F, Kane PM. Loss of vacuolar proton-translocating ATPase activity in yeast results in chronic oxidative stress. J Biol Chem. 2007;282:7125–7136. doi: 10.1074/jbc.M608293200. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Ho YS, Swiatek PJ, Rosen BP, Bhattacharjee H. Targeted disruption of the mouse Asna1 gene results in embryonic lethality. FEBS Lett. 2006;580:3889–3894. doi: 10.1016/j.febslet.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis K, Schrul B, Tienson H, Gostimskaya I, Breker M, High S, Schuldiner M, Jakob U, Schwappach B. Get3 is a holdase chaperone and moves to deposition sites for aggregated proteins when membrane targeting is blocked. Journal of Cell Science. 2013;126:473–483. doi: 10.1242/jcs.112151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, Jakob U. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012;148:947–957. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Metz J, Schmid V, Denic V, Rakwalska M, Schmitt HD, Schwappach B, Weissman JS. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Hsu CM, Kang BK, Rosen BP, Bhattacharjee H. The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. Biometals. 2003;16:369–378. doi: 10.1023/a:1022504311669. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Stefer S, Reitz S, Wang F, Wild K, Pang YY, Schwarz D, Bomke J, Hein C, Lohr F, Bernhard F, et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 2011;333:758–762. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudnitsyna MV, Mymrikov EV, Seit-Nebi AS, Gusev NB. The role of intrinsically disordered regions in the structure and functioning of small heat shock proteins. Current protein & peptide science. 2012;13:76–85. doi: 10.2174/138920312799277875. [DOI] [PubMed] [Google Scholar]
- Suloway CJ, Chartron JW, Zaslaver M, Clemons WM., Jr Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci U S A. 2009;106:14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway CJ, Rome ME, Clemons WM., Jr Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue. The EMBO journal. 2012;31:707–719. doi: 10.1038/emboj.2011.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley TL, Korner JL, Barge MT, Hupfeld J, Schauerte JA, Gafni A, Jakob U, Bardwell JC. Structural plasticity of an acid-activated chaperone allows promiscuous substrate binding. Proc Natl Acad Sci U S A. 2009;106:5557–5562. doi: 10.1073/pnas.0811811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardi F, Stephan M, Clancy A, Janshoff A, Schwappach B. WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS One. 2014;9:e85033. doi: 10.1371/journal.pone.0085033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Molecular Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Zhang HM, McLoughlin SM, Frausto SD, Tang H, Emmett MR, Marshall AG. Simultaneous reduction and digestion of proteins with disulfide bonds for hydrogen/deuterium exchange monitored by mass spectrometry. Anal Chem. 2010;82:1450–1454. doi: 10.1021/ac902550n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Radaev S, Rosen BP, Gatti DL. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. The EMBO journal. 2000;19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.