Summary
The RNA exosome has a key role in RNA decays and RNA quality control. In 2012, two human Mendelian diseases: syndromic diarrhea/tricho-hepato-enteric syndrome (SD/THE) and Ponto-cerebellar hypoplasia type 1(PCH1) were linked to the RNA exosome or its cofactor's defect. SD/THE's main features are an intractable diarrhea of infancy associated with hair abnormalities, facial dysmorphism, intra uterine growth restriction and immune deficiency. SD/THE is caused by a defect of the SKI complex (TTC37 and SKIV2L), the cytoplasmic co-factor of the RNA exosome for mRNA degradation. PCH1's main features are atrophy of the pons and of the cerebellum, a progressive microcephaly with developmental delay and muscle atrophy secondary to spinal anterior horn cell loss. In 30–40% of patients, PCH1 is caused by a defect in EXOSC3 which encodes RRP40, a protein of the cap of the RNA exosome. Thanks to knowledge about other forms of PCH it could be assumed that the altered substrates are probably transfer RNA However, as there exists no patient with two null mutations, residual RNA exosome functionality is probably required to preserve viability. Thus, to date two very different human Mendelian diseases have been related to the dysfunctioning of the RNA exosome. It illustrates the versatility of the RNA exosome function and substrate.
Keywords: Syndromic diarrhea, tricho-hepato-enteric syndrome, RNA exosome, Ponto-cerebellar hypoplasia type 1, EXOSC3, TTC37, SKIV2L
1. Introduction
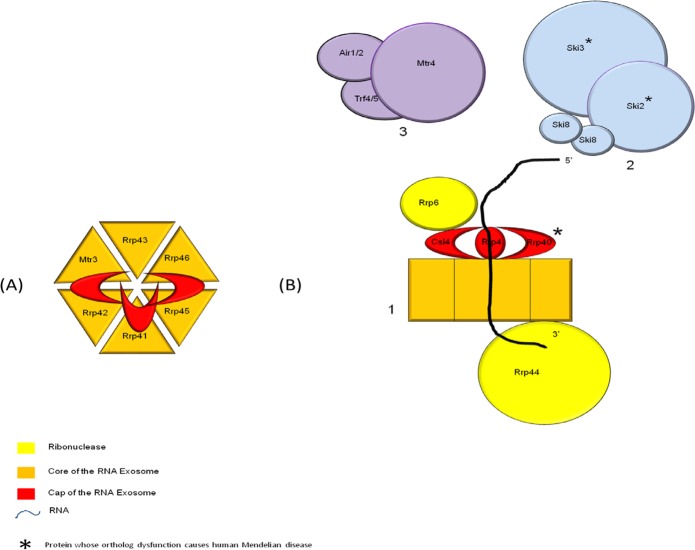
In 2012, two human Mendelian diseases were linked to the RNA exosome or its cofactor. The RNA exosome, discovered in 1997, has a key role in the RNA decays and RNA quality control (1,2). Mostly studied in yeast, it is conserved among eukaryotes and there exist homologs in archea. In yeast, the RNA exosome is a multiprotein complex (Figure 1). The core of the RNA exosome consists of 9 proteins which form a barrel-like structure. Six proteins form the ring of the exosome (Rrp41, Rrp42, Rrp43, Rrp45, Rrp46, Mtr3) on top of which stands the cap of exosome, formed by three proteins (Csl4, Rrp40, Rrp4). Most of the enzymatic activity is brought by associated proteins RRP44 and RRP6 which also have other functions and subcellular localization. Finally two cofactors are needed for RNA exosome function: Mtr4 either alone or with the TRAMP complex (Air1/2, Trf4/5 and Mtr4) and the SKI complex (Ski2, Ski3 and Ski8). The RNA substrates are varied and include messenger RNA, small nuclear RNA, ribosomal RNA, transfer RNA, cryptic unstable transcripts. The RNA decay starts at the 3′ end. The human exosome structure is globally the same except for the existence of three paralogs of Rrp44 called DIS3, DIS3L1 and DIS3L2. (1,2) (Table 1, Figure 1). Until 2012, only auto immune diseases were linked to the RNA exosome (3). Since, two human Mendelian diseases: syndromic diarrhea/tricho-hepato-enteric syndrome (SD/THE) and Pontocerebellar hypoplasia type 1 (PCH1) were linked to the RNA exosome or its cofactor's defect. These diseases emphasize the importance of the RNA exosome in humans but their distinctive phenotypes with few overlaps of clinical signs show that an alteration of the function of the RNA exosome can be quite segmentary, probably reflecting the various exosome substrates and functions.
Figure 1.
Model of yeast RNA exosome and cofactor. (A), Transversal view of RNA exosome. (B), Front view of RNA exosome (1), SKI complex (2), and TRAMP complex (3).
Table 1. Proteins and genes of the RNA exosome and cofactor in human and yeast.
| Items | Yeast protein (S. cerevisiae) | Human protein | Human gene | Size of the protein (Human, AA) |
 | ||||
*The role of the TRAMP complex seems to differ between yeast and human.
2. Human disease related to the Ski complex
The Syndromic diarrhea/ Tricho hepato enteric (SD/THE) (OMIM : #222466, #614602 ): SD/THE has been linked to the alteration of TTC37 and of SKIV2L, the orthologs of the yeast's Ski2 and Ski3, two components of the SKI complex (4–6). The SKI complex is an obligatory co-factor of the RNA exosome in the cytoplasm in yeast. To date, about 50 patients have been described (7). The main clinical features are an intractable diarrhea of infancy requiring parenteral nutrition, facial dysmorphism with a prominent forehead, a broad nose and hair abnormalities (wooly, easily removable), an intra uterine growth restriction, an immune deficiency (lack of immunoglobulin or absence of vaccine response). In half of the cases, liver abnormalities and skin abnormalities (mostly café au lait spots) are observed. Rarely, a congenital heart defect and platelet abnormalities are associated. The mortality is high with one third of the children deceased before 10 years. About half of the children can be weaned off parenteral nutrition and nearly all have a small size below the 3rd percentile (8). The phenotypes resulting from an alteration of either SKIV2L or TTC37 cannot be differentiated, suggesting that it is the absence of the SKI complex which is responsible for the disease. As the SKI complex is linked to a degradation of mRNA in yeast it could be assumed that SD/THE is caused either by the presence of aberrant mRNA or the stabilization of normal transcripts which should be degraded. The clinical management is essentially supportive with nutritional support (mainly parenteral nutrition) and immunoglobulin supplementation.
3. Core RNA Exosome linked human disease
Pontocerebellar hypoplasia type 1 (PCH1) (OMIM #614676): PCH1 has been linked to mutation of EXOSC3 encoding human RRP40, a protein of the cap of the RNA exosome (9). PCH is a group of inherited progressive neurodegenerative disorders with seven subtypes identified to date. All PCH associates atrophy of the pons and of the cerebellum and progressive microcephaly with developmental delay. PCH1 presents the distinctive feature of muscle atrophy secondary to spinal anterior horn cell loss which can lead to hypotonia and respiratory deficiency (10). Some patients develop growth retardation (9). EXOSC3 mutations are found in 37% of PCH1 patients (11). Patients harbouring mutation in EXOSC3 differ from other PCH1 patients by a longer lifespan, abnormal oculomotor function, and with time could appear a respiratory deficiency. Nearly 50 patients with a mutation in EXOSC3 have been described since 2012 (9,11–13). Most of the patients present two missense mutations or, more rarely, a compound heterozygosity associating a missense and a null mutation. To date, no patient with homozygous null mutation has been reported. As some level of hypomorphy of the missense mutations exists, it seems that a total lack of RRP40 is not viable. There is some degree of genotype/phenotype correlation: indeed patients with homozygous missense mutation have a milder course, contrary to patients with null mutation and missense mutation. A clinical form with a milder severity has also been described: it associated spasticity, mild intellectual retardation and cerebellar atrophy, and is caused by compound missense mutations (14). The clinical management of the patient is symptomatic with nutritional support, and management of dyskinesia, dystonia and seizure (10).
4. Discussion
To date, two very different diseases have been related to a defect of the RNA exosome, one concerning the core exosome and the other, one of its co-factors. Except for the short stature found both in SD/THE and in PCH1, there are no shared clinical signs (Table 2). This could be related to an alteration of different types of RNA degradation. Indeed, the RNA exosome is implicated in different RNA degradation pathways: mRNA, Sn RNA, ribosomal RNA, transfer RNA, cryptic unstable transcripts. The ski complex is only necessary for the mRNA degradation (15) whereas RRP40 is part of the core of the RNA exosome and is involved in all the RNA exosome functions. Most of the genes causing the other subtypes of PCH are related to the Transfer RNA pathway (10). In PCH1, there are no cases of homozygous null mutation and, at least for one mutation, the RNA accumulation typical of exosome knock-out has not been observed (9), suggesting that some level of the exosome activity is conserved in PCH1 and that RRP40 is mainly necessary for the transfer RNA degradation, at least in humans. In 2012, the Perlman syndrome, an association of a congenital overgrowth syndrome with tumour susceptibility, was related to DIS3L2 defect (16). Dis3L2 is an exoribonuclease degrading cytoplasmic mRNA specially uridylated in an exosome-independent pathway (17,18). Thus these findings confirm the importance of a controlled homeostasis of RNA product and the potential consequences of imbalance.
Table 2. Clinical sign Frequency of Syndromic diarrhea/Tricho-hepato-enteric syndrome and Pontocerebellar hypoplasia type 1with mutation in Exosc3.
| Items | Syndromic diarrhea/Tricho-hepato-enteric syndrome | Pontocerebellar hypoplasia type 1with mutation in EXOSC3 |
| Nearly constant (> 90%) |
|
|
| Frequent (50–90%) |
|
|
| Inconstant (< 50%) |
|
|
In conclusion, to date, two very different human Mendelian diseases have been related to a dysfunction of the RNA exosome. They illustrate the versatility of the RNA exosome function and substrates. The question remains as to whether more diseases human Mendelian could be related to the dysfunction of the RNA exosome. Since non-functional exosome seems not to be viable, only moderate dysfunctions of the RNA exosome or cofactor alterations could be involved.
References
- 1. Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′–>5′ exoribonucleases. Cell. 1997; 91:457-466 [DOI] [PubMed] [Google Scholar]
- 2. Schneider C, Tollervey D. Threading the barrel of the RNA exosome. Trends Biochem Sci. 2013; 38:485-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Staals RH, Pruijn GJ. The human exosome and disease. Adv Exp Med Biol. 2010; 702:132-142 [PubMed] [Google Scholar]
- 4. Hartley JL, Zachos NC, Dawood B, et al. Mutations in TTC37 cause trichohepatoenteric syndrome (phenotypic diarrhea of infancy). Gastroenterology. 2010; 138:2388-2398, 2398.e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fabre A, Martinez-Vinson C, Roquelaure B, Missirian C, André N, Breton A, Lachaux A, Odul E, Colomb V, Lemale J, Cézard JP, Goulet O, Sarles J, Levy N, Badens C. Novel mutations in TTC37 associated with trichohepato-enteric syndrome. Hum Mutat. 2011; 32:277-281 [DOI] [PubMed] [Google Scholar]
- 6. Fabre A, Charroux B, Martinez-Vinson C, et al. SKIV2L mutations cause syndromic diarrhea, or trichohepatoenteric syndrome. Am J Hum Genet. 2012; 90:689-692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fabre A, Martinez-Vinson C, Goulet O, Badens C. Syndromicdiarrhea/Tricho-hepato-enteric syndrome. Orphanet J Rare Dis. 2013; 8:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fabre A, Breton A, Coste ME, et al. Syndromic (phenotypic) diarrhoea of infancy/tricho-hepato-enteric syndrome. Arch Dis Child. 2014; 99:35-38 [DOI] [PubMed] [Google Scholar]
- 9. Wan J, Yourshaw M, Mamsa H, et al. Mutations in the RNA exosome component gene EXOSC3 cause pontocerebellar hypoplasia and spinal motor neuron degeneration. Nat Genet. 2012; 44:704-708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Namavar Y, Barth PG, Poll-The BT, Baas F. Classification, diagnosis and potential mechanisms in pontocerebellar hypoplasia. Orphanet J Rare Dis. 2011; 6:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rudnik-Schöneborn S, Senderek J, Jen JC, et al. Pontocerebellar hypoplasia type 1: Clinical spectrum and relevance of EXOSC3 mutations. Neurology. 2013; 80:438-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biancheri R, Cassandrini D, Pinto F, Trovato R, Di Rocco M, Mirabelli-Badenier M, Pedemonte M, Panicucci C, Trucks H, Sander T, Zara F, Rossi A, Striano P, Minetti C, Santorelli FM. EXOSC3 mutations in isolated cerebellar hypoplasia and spinal anterior horn involvement. J Neurol. 2013; 260:1866-1870 [DOI] [PubMed] [Google Scholar]
- 13. Schwabova J, Brozkova DS, Petrak B, Mojzisova M, Pavlickova K, Haberlova J, Mrazkova L, Hedvicakova P, Hornofova L, Kaluzova M, Fencl F, Krutova M, Zamecnik J, Seeman P. Homozygous EXOSC3 mutation c.92G→C, p.G31A is a founder mutationcausing severe pontocerebellar hypoplasia type 1 among the Czech Roma. J Neurogenet. 2013; 27:163-169 [DOI] [PubMed] [Google Scholar]
- 14. Zanni G, Scotton C, Passarelli C, Fang M, Barresi S, Dallapiccola B, Wu B, Gualandi F, Ferlini A, Bertini E, Wei W. Exome sequencing in a family with intellectual disability, early onset spasticity, and cerebellar atrophy detects a novel mutation in EXOSC3. Neurogenetics. 2013; 14:247-250 [DOI] [PubMed] [Google Scholar]
- 15. Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006; 7:529-539 [DOI] [PubMed] [Google Scholar]
- 16. Astuti D, Morris MR, Cooper WN, et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012; 44:277-284 [DOI] [PubMed] [Google Scholar]
- 17. Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3′–5′ degradation pathway of human cytoplasmic mRNA. EMBO J. 2013; 13:1855-1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013; 19:1632-1638 [DOI] [PMC free article] [PubMed] [Google Scholar]