Summary
The purpose of our study was to screen preliminary differential expression bone-related microRNAs (miRNAs) in serum of patients with osteogenesis imperfacta and to clarify whether serum microRNA is a promising biomarker for osteogenesis imperfecta. geNorm and several other programes were performed to select suitable reference genes for quantitative detection of serum miRNAs from 6 candidate control genes. With geometric averaging of selected reference genes as a normalization factor, fluorescence-based quantitative real-time reverse transcriptase-polymerase chain reaction (qRT-PCR) was used to detect expression levels of more than 100 bone-related miRNAs obtained by means of miRanda, Targetscan and Pictar software calculations and reading the literature. Through analysis of expression stability and pairwise variations, all 6 candidate reference genes had a stable expression level in serum of 8 healthy controls and 8 patients with different characrteristics, and the optimal number of reference genes for normalization was 4 (snRNAU6, miR-92a, miR-16, and Let-7a). For further validation, the expression stability of 4 reference genes remained steady in serum of another 8 healthy controls and 16 patients with osteogenesis imperfecta (M < 1.5). When normalized using multiple control genes, 11 bone-related miRNAs showed differential expression in serum of 8 osteogenesis imperfecta patients compared with 8 healthy controls. In conclusion, we identified snRNAU6, miR-92a, miR-16, and Let-7a as an internal reference gene group for qRT-PCR normalization and screening results revealed that there existed many differential expression bone-related miRNAs in serum of patients with osteogenesis imperfecta compared with healthy controls, and that these miRNAs had potential to be biomarkers for serologic tests and diagnosis of osteogenesis imperfecta with analysis of bioinformation.
Keywords: Osteogenesis imperfecta, serum miRNAs, reference gene, biomarker
1. Introduction
Osteogenesis imperfecta (OI) is a rare hereditary disease characterized by abnormality of osteoblastic proliferation and differentiation and osteoclastic activity (1). Emerging evidence has suggested that microRNAs (miRNAs) may be important mediators of ossification through binding and repressing the expression of target mRNAs whose translation products are involved in osteoblastic proliferation and differentiation and osteoclastic activity (2).
miRNA is a class of small noncoding RNAs that principally function in the spatiotemporal regulation of protein translation by binding to the 3′-untranslated region (UTR) of target mRNAs, leading to mRNA degradation or translational repression (3). Since discovered in the early 1990s, miRNAs have been shown to play important regulatory roles in a wide range of biological and pathological processes (4–6). As an existing form, serum miRNAs are thought to be a greatly promising novel biomarker for cancers, nerve-development diseases, and some metabolic diseases with their high accuracy (sensitivity and specificity) and predictability (7). However, little attention has been devoted to the relationship between serum miRNAs expression and bone-related diseases.
To learn more about the potential relevance of serum miRNAs in bone-related diseases, we screened more than 100 bone-related miRNAs in serum samples from patients with OI and healthy controls to find OI-related miRNAs, and with bioinformation analysis, we examined whether these differential bone-related miRNAs have potential to be markers for OI diagnosis.
2. Materials and Methods
2.1. Serum collection and storage
The study protocol was approved by the Ethical Committee of Shandong Medicinal Biotechnology Center, Ji'nan, Shandong, China. Whole blood samples were collected from 22 osteogenesis imperfecta patients (age, 9.54 ± 3.65 year) and 10 healthy donors (age, 8.20 ± 0.42 year). Serum was isolated by centrifuging whole blood at 1,900 × g for 10 min at room temperature. To minimize degradation, samples were processed within 2 h. Twenty-two OI patients came from Tianjin Hospital and Shandong Provincial Hospital with patient's informed consent, and 10 healthy controls came from volunteers in Ji'nan.
2.2. Total RNA isolation
Total RNA from serum was isolated using TRIzol LS (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. In brief, 200 µL serum was mixed with 750 µL of TRIzol LS, and then 200 µL chloroform was added, the samples were shaken vigorously for 30 sec, and allowed to stand for 5 min at room temperature. After centrifugation at 12,000 × g for 15 min at 4°C, the upper aqueous phase was transferred to a fresh tube and 600 µL isopropanol was added followed by mild mixing and incubation for 5 min at room temperature. After centrifugation at 12,000 × g for 10 min at 4°C, the upper aqueous phase was discarded and the precipitate was washed twice with 750 µL of 75% ethanol. Finally, RNA was diluted in a final volume of 15 µL RNase-free water. The RNA integrity and concentration were determined using 2% agarose gel electrophoresis and a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.3. Quantification of serum miRNAs by real-time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis
The first-strand miRNA-cDNA PCR template was generated from ∼50 ng of total RNA using a miRNA first-strand cDNA synthesis kit (Tiangen, Beijing, China) according to the manufacturer's instructions. Then approximate 2.5 ng of cDNA was used to amplify using a miRcute miRNA qPCR detection kit (Tiangen) with a LightCycler 480 Sequence Detection System (Roche Applied Science, Mannheim, Germany). In the qRT-PCR, amplification was carried out as follows: 94°C for 2 min, 45 cycles of 94°C for 20 sec, 60°C for 20 sec, and 72°C for 20 sec. Data were generated from three independent qRT-PCR reactions and the relative quantity of miRNA expression was calculated using the comparative cycle threshold (Ct) method, in which the lowest expressed sample was used as a calibrator. Subsequently, the relative quantity was normalized using geometric averaging of selected reference genes.
2.4. Selection of reference genes
Six OI patients and 2 healthy controls with different characteristics (gender, age, and medication history) were selected as the objects for this study. In the selected 6 candidate reference genes (miR-16, miR-92a, miR-638, Let-7a, snRNAU6, and 18S rRNA), miR-16, miR-638, snRNAU6, and 18S rRNA have been regarded as common internal references (8), and miR-92a and Let-7a were two miRNAs which were stably expressed in our previous trials. Using qRT-PCR followed by geNorm software analysis, which was used to sort gene expression stability and find optimal reference gene(s) (9), the expression stability of 6 candidate reference genes was obtained and an optimal number of reference genes for normalization was determined. For verifying accuracy and universality of chose reference genes stable expression, validation was carried out by means of normFinder (10), Bestkeeper (11), and delta CT method analyses (12) and increasing sample size. Six candidate reference genes' primer sequences were as follows: for hsa-miR-16, 5′-TAG-CAG-CAC-GTA-AATATT-GGC-G-3′; for hsa-miR-92a, 5′-TAT-TGC-ACTTGT-CCC-GGC-CTG-T-3′; for hsa-miR-638, 5′-GGGTGG-CGG-CCT-AAA-AA-3′; for hsa-Let-7a, 5′-TTGAGG-TAG-TAG-GTT-GTA-TAG-TT-3′; for snRNAU6, 5′-TGC-GGG-TGC-TCG-CTT-CGG-CAG-C-3′; for 18S rRNA, 5′-CAG-CCA-CCC-GAG-ATT-GAG-CA-3′.
2.5. Screening of differential expression of bone-related miRNAs in serum of patients with OI
Through three miRNAs target genes analysis softwares, miRanda, Targetscan, and Pictar, we sorted out some miRNAs which could combine complementarily to mRNA 3’ UTR of genes functioning in osteogenesis. Together with miRNAs verified to regulate the course of bone formation, bone-related miRNAs have been chosen, and these miRNAs were quantified in serum of 8 OI patients without medication history and 8 age-matched healthy controls using qRT-PCR with multiple reference genes as endogenous controls. Finally, a matched t test was used to screen out the differential expression bone-related miRNAs.
3. Results
3.1. snRNAU6, miR-92a, miR-16, and Let-7a as internal references for qRT-PCR normalization
Based on that the expression ratio of two suitable reference genes should be identical in all samples regardless of the experimental conditions, we utilized geNorm software to assess the expression stability of 6 candidate control genes and found all 6 candidate reference genes had a stable expression level in serum of 8 healthy controls and 8 patients with different characrteristics (M < 1.5, the system default value is 1.5). The order of their expression stability is snRNAU6/miR-92a > miR-16 > Let-7a > 18S rRNA > miR-638 (Figure 1). With pairwise variation analysis, the optimal number of reference genes for normalization was determined to be 4 (V4/5 = 0.133 < 0.15, the system default value is 0.15) (Figure 2). Further validation by means of normFinder, Bestkeeper, and delta CT method analyses and increasing sample size revealed the same results (Figure 3), which revealed that snRNAU6, miR92a, miR-16, and Let-7a could be used as an internal reference group for qRT-PCR normalization.
Figure 1.
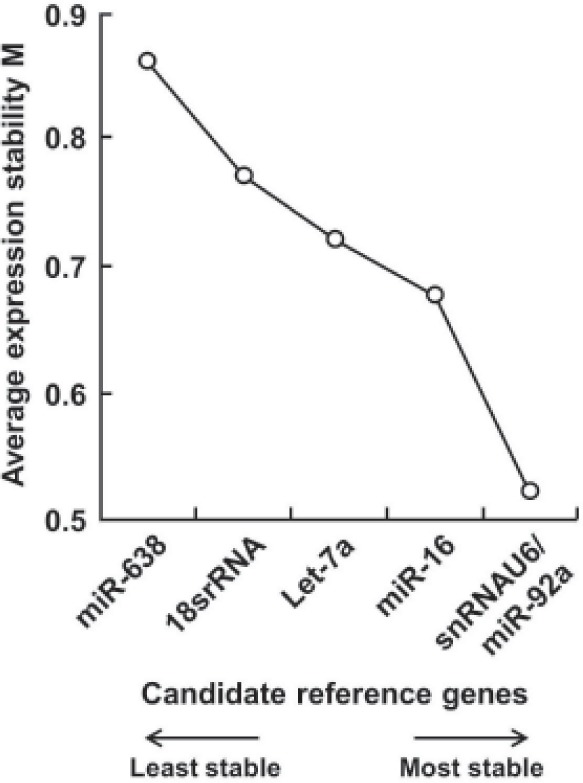
Expression stability values of 6 candidate reference genes. Data were obtained from analysis of gene expression stability using geNorm software.
Figure 2.
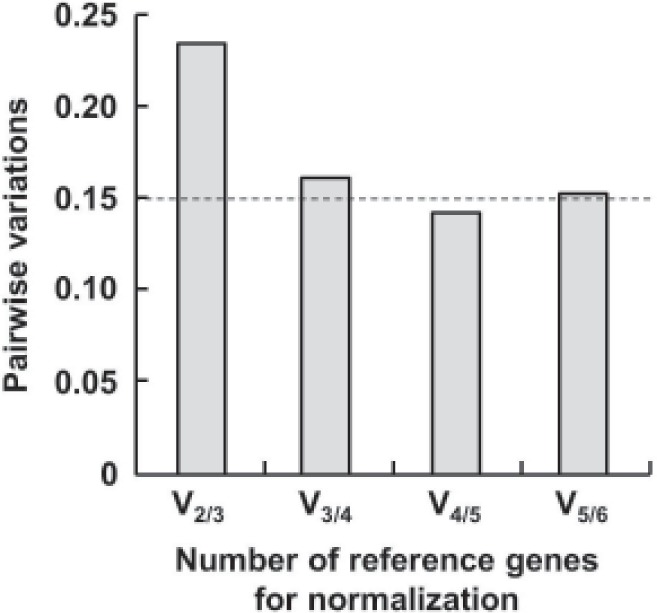
Determination of the optimal number of reference genes for normalization. Data were obtained from analysis ofpairwise variation using geNorm software. Dashed line indicatesa pairwise variation value of 0.15.
Figure 3.
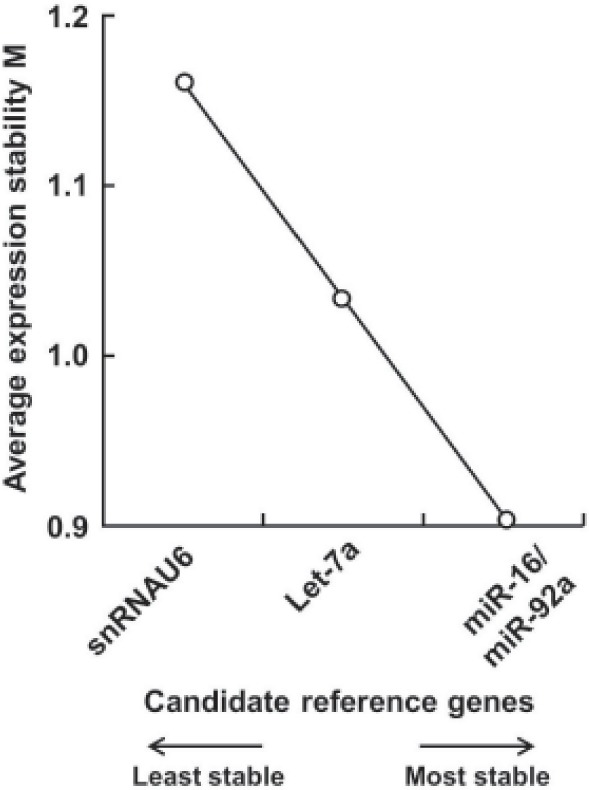
Expression stability values of 4 reference genes in serum of 8 healthy controls and 16 patients with OI. Data were obtained from analysis of gene expression stability by expanding the sample size.
3.2. Differential expression of bone-related miRNAs
In the more than 100 bone-related miRNAs we have chosen with the help of software forecasts and literature reports, miR-26a, miR-30e, and miR-21 were revealed to be up-expressed and miR-34c, miR-29a, miR-29b, miR-489, miR-133a, miR-145, miR-210, and miR-1297 down-expressed in serum of OI patients compared to healthy controls (p < 0.05) (Figure 4). In the differential expression, miRNAs, miR-29a, and miR-133a were significantly down-regulated, at 12.80-fold and 6.31-fold, respectively. Compared to the healthy control group, the quantity of miR-29a had a universal lower level in the patient group, and on the contrary, miR-26a had a universal upper level.
Figure 4.
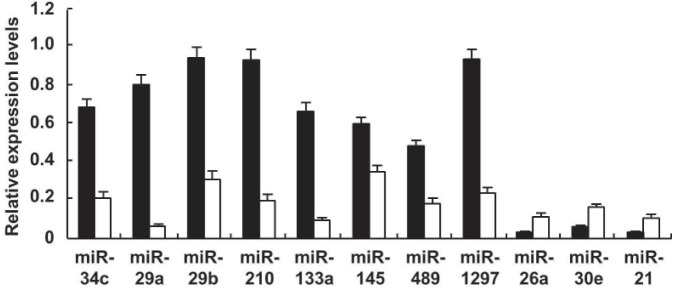
Differential expression levels of eleven serum miRNAs in healthy controls and patients with OI. Data were obtained from quantitative detection of bone-related miRNAs in serum of OI patients. Closed columns, healthy controls; open columns, patients.
4. Discussion
Although real-time reverse transcriptase PCR has become the technology of choice for high-throughput and accurate expression profiling of target genes, the selection of a proper internal reference was still a festering issue. Unconditional stable expression housekeeping genes are not known to exist in nature, and will lead to large errors with normalization based on a single reference gene (9). Therefore, we inputted geNorm software to seek out more stable genes as an internal reference group, and the qRT-PCR data was then normalized using a geometric mean of selected genes. In 4 selected reference genes, miR-16 was the most common reference for miRNAs detection in peripheral blood of various diseases, such as gastric cancer (8), prostate cancer (13), breast cancer (14), and liver cancer (15). Another gene, snRNAU6, a kind of intracellular stable expression ribosomal RNA, recently has been found to express stably in the peripheral circulation of diverse diseases, for instance, in the plasma of colorectal and oral squamous cell carcinoma, in the serum of breast cancer and in the urine of bladder cancer (15).
Most alternatively expressed bone-related miRNAs we observed have been regarded as potential biomarkers for various cancers; for instance, miR-21 was called “oncogenic miRNA", precisely because it had abnormal expression in many cancers (16–21). Although no report directly indicated that there is a close relationship between differential expression of miRNAs and bone-related diseases, these miRNAs have been verified to be involved in the course of ossification (2). While investigating the relationship between bone morphogenetic proteins (BMPs) and miRNAs in phenotypic induction in osteoprogenitors, Li Z et al. found that miR-133 functionally inhibits osteoprogenitor differentiation by attenuating Runx2 and Smad5 pathways, which synergistically contribute to bone formation (22). miR-29a has been reported to suppress expression of Dkk1, Kremen2, and sFRP2, all of which are negative regulators of Wnt signaling, which promotes osteoblast differentiation (23). In a study to determine miR29b function, transforming growth factor beta 3 (TGFβ3) was found to inhibit osteoblast differentiation, while miR-29b was observed to promote osteoblastogenesis by directly targeting the 3′ UTR sequence of TGFβ3 mRNA (24). In a similar report, miR-210 was found to enhance osteoblastogenesis by inhibiting the TGFβ/activin signaling pathway through targeting and repressing translational products of the AcvR1b gene (25). miR-21, regulated by the RANK ligand (RANKL)-induced c-Fos, a critical transcription factor for osteoclastogenesis (26), conversely promotes c-Fos expression by downregulation of programmed cell death 4 (PDCD4) protein levels, due to repression removal from c-Fos (27). Therefore, a positive feedback is formed between miR-21 and c-Fos, which significantly enhances the regulation capability of miR-21 for osteoclast differentiation.
Although our findings demonstrate the potential utility of altered expression of serum miRNAs as OI biomarkers, it is necessary to acknowledge that the serum sample size of OI patients observed in this study was relatively small. Nonetheless, these data suggest that serum miRNAs could be a promising biomarker for diverse diseases.
In conclusion, the discovery of altered expression of bone-related miRNAs in serum of OI patents has opened up new possibilities for bone-related diseasediagnosis and expressed serum miRNAs are a promising biomarker for osteogenesis imperfecta.
Acknowlegements
This work was supported by a grant from National Key Technology R&D Program (No. SQ2011SF12C03081).
References
- 1. Iwamoto J, Takeda T, Ichimura S. Increased bone resorption with decreased activity and increased recruitment of osteoblasts inosteogenesis imperfect type I. J Bone Miner Metab. 2002; 20:174-179 [DOI] [PubMed] [Google Scholar]
- 2. Liu CJ. MicroRNAs in skeletogenesis. Front Biosci. 2009; 14:2757-2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lai EC. Micro RNAs are complementary to 3’ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002; 30:363-364 [DOI] [PubMed] [Google Scholar]
- 4. Takahashi RU, Makiko O, Ochiya T. Role of microRNA in cancer development: Biology and clinical applications. Nihon Geka Gakkai Zasshi. 2012; 113:197-203 (in Japanese) [PubMed] [Google Scholar]
- 5. Patel M, Hu BH. MicroRNAs in inner ear biology and pathogenesis. Hear Res. 2012; 287:6-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heinrich EM, Dimmeler S. MicroRNAs and stem cells: Control of pluripotency, reprogramming, and lineage commitment. Circ Res. 2012; 110:1014-1022 [DOI] [PubMed] [Google Scholar]
- 7. Ma R, Jiang T, Kang X. Circulating microRNAs in cancer: Origin, function and application. J Exp Clin Cancer Res. 2012; 31:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids — the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011; 8:467-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multipleinternal control genes. Genome Biol. 2002; 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004; 64:5245-5250 [DOI] [PubMed] [Google Scholar]
- 11. Lardizábal MN, Nocito AL, Daniele SM, Ornella LA, Palatnik JF, Veggi LM. Reference genes for real-time PCR quantification of microRNAs and messenger RNAs in rat models of hepatotoxicity. PLoS One. 2012; 7:e36323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012; 57:897-904 [DOI] [PubMed] [Google Scholar]
- 13. Mahn R, Heukamp LC, Rogenhofer S, von Ruecker A, Müller SC, Ellinger J. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology. 2011; 77:1265.e9-16 [DOI] [PubMed] [Google Scholar]
- 14. Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009; 2:89-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bihrer V, Waidmann O, Friedrich-Rust M, et al. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and withouthepatocellular carcinoma. PLoS One. 2011; 6:e26971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, Yuan Y, Li Z, Zen K, Ba Y, Zhang CY. Serum MicroRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012; 58:610-618 [DOI] [PubMed] [Google Scholar]
- 17. Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, Huang L, Li H, Tan W, Wang C, Lin D. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011; 50:136-142 [DOI] [PubMed] [Google Scholar]
- 18. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008; 141:672-675 [DOI] [PubMed] [Google Scholar]
- 19. Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011; 57:84-91 [DOI] [PubMed] [Google Scholar]
- 20. Zhang HL, Yang LF, Zhu Y, Yao XD, Zhang SL, Dai B, Zhu YP, Shen YJ, Shi GH, Ye DW. Serum miRNA-21: Elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011; 71:326-331 [DOI] [PubMed] [Google Scholar]
- 21. Wang ZX, Bian HB, Wang JR, Cheng ZX, Wang KM, De W. Prognostic significance of serum miRNA-21 expression in human non-small cell lung cancer. J Surg Oncol. 2011; 104:847-851 [DOI] [PubMed] [Google Scholar]
- 22. Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006; 580:4214-4217 [DOI] [PubMed] [Google Scholar]
- 23. Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: Regulation during differentiation and by canonical Wnt signaling. J Cell Biochem. 2009; 108:216-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J Biol Chem. 2009; 284:15676-15684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mizuno Y, Tokuzawa Y, Ninomiya Y, Yagi K, Yatsuka-Kanesaki Y, Suda T, Fukuda T, Katagiri T, Kondoh Y, Amemiya T, Tashiro H, Okazaki Y. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009; 583:2263-2268 [DOI] [PubMed] [Google Scholar]
- 26. Arai A, Mizoguchi T, Harada S, Kobayashi Y, Nakamichi Y, Yasuda H, Penninger JM, Yamada K, Udagawa N, Takahashi N. c-Fos plays an essential role in the up-regulation of RANK expression in osteoclast precursors within the bone microenvironment. J Cell Sci. 2012. 10.1242/jcs.099986 [DOI] [PubMed] [Google Scholar]
- 27. Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011; 117:3648-3657 [DOI] [PMC free article] [PubMed] [Google Scholar]


