Summary
Chorionic villus sampling (CVS) or amniocentesis for fetal sex determination is generally the first step in the prenatal diagnosis of X-linked genetic disorders such as Duchenne muscular dystrophy (DMD). However, non-invasive prenatal diagnostic (NIPD) techniques such as measurement of cell-free fetal DNA (cffDNA) in maternal plasma are preferable given the procedure-related miscarriage rate of CVS. We determined fetal sex during the first trimester using a quantitative real-time polymerase chain reaction (PCR) assay of cffDNA in pregnant carriers of DMD. The fetal sex was confirmed by amniocentesis karyotype analysis and multiplex ligation-dependent probe amplification (MLPA) at 16 weeks. This procedure may avoid unnecessary CVS or amniocentesis of female fetuses.
Keywords: Cell-free fetal DNA (cffDNA), non-invasive prenatal diagnostic (NIPD), Duchenne muscular dystrophy (DMD), fetal sex determination
1. Introduction
The first step in the prenatal diagnosis of X-linked genetic disorders like Duchenne muscular dystrophy (DMD) or hemophilia is the determination of fetal sex. Chorionic villus sampling (CVS) and amniocentesis have long been used to determine sex. A female fetus may have a wild-type genotype or be a carrier of DMD, but further genetic analysis is crucial for a male fetus because a male fetus has a 50% change of having DMD. Pregnant carriers risk miscarriage when undergoing an invasive prenatal diagnosis (IPD). Non-invasive prenatal diagnosis (NIPD) is preferable for fetal sex determination during the first trimester since it avoids the unnecessary risks of IPD in pregnant female DMD carriers.
Cell-free fetal DNA (cffDNA) was found in maternal plasma in 1997 (1) and its measurement represents a potential form of NIPD. The cffDNA in maternal plasma can be detected as early as 7 weeks. cffDNA comprises about 3–6% of the total cell-free DNA in maternal plasma (2). Detecting the sex-determining region on the Y chromosome (SRY) or other Y chromosome-specific sequences based on cffDNA from maternal plasma is one technique for non-invasive fetal sex determination during the early trimester of pregnancy (3–5). Fetal sex can be diagnosed before CVS can be performed. In the current study, cffDNA was measured for fetal sex determination during prenatal diagnosis of DMD. The fetal sex was determined at about 9 weeks of gestation by means of quantitative real-time polymerase chain reaction (PCR) with a taqman probe to detect SRY with cffDNA in maternal plasma. If the fetus was male, CVS was performed at 12 weeks followed by DMD and multiplex ligation-dependent probe amplification (MLPA) analysis. If the fetus was female, CVS was avoided. Fetal sex was later confirmed by ultrasound at 16 weeks. Whether a female fetus is a carrier or not can be determined after delivery.
2. Materials and Methods
2.1. Materials
All study protocols were approved by the Ethics Committee of He'nan Provincial People's Hospital. All of the pregnant women and their partners gave written informed consent and received genetic counseling. MLPA analysis was performed on the fetuses of 15 pregnant women who were DMD carriers.
2.2. Sampling and extraction of cffDNA in maternal plasma
EDTA blood samples (8 mL) were taken at about 9 weeks of gestation. The blood samples were centrifuged twice at 3,000 g and then at 12,000 g to obtain cell-free plasma. Cell-free DNA was extracted from 2 mL of maternal plasma using the QIAamp blood mini kit (Qiagen, Hilden, Germany). DNA was eluted into 40 µL of solution buffer.
2.3. Sex determination using quantitative real-time PCR
Quantitative real-time PCR analysis was performed using an Applied Biosystems 7500 Fast Real-Time PCR System. TaqMan amplification reactions were set up in a reaction volume of 20 µL by use of components (except TaqMan probes and amplification primers) supplied in the TaqMan® Fast Universal PCR Master Mix (2×) (Applied biosystems). TaqMan probes and PCR primers were synthesized by Sangon Biotech (Shanghai, China) Co., Ltd. (Primer1: 5′-TGGCGATT AAGTCAAATTCGC-3′; Primer2: 5′-CCCCCTAGTA CCCTGACAATGTATT-3′; Probe: 5′-(FAM)AGCAGT AGAGCAGTCAGGGAGGCAGA(TAMRA)-3′). Each reaction included 12.5 µL of TaqMan® Fast Universal PCR Master Mix (2×), 300 nM of each amplification primer, and 200 nM of the TaqMan probe. Eight µL of the extracted plasma DNA was used for amplification. Each sample was analyzed twice. The 500 genome equivalent (GE, 6.6 pg of DNA per genome equivalent), 100 GE, and 10 GE were used as positive controls to confirm the sensitivity of the PCR assay. Thermal cycling was initiated with a first denaturation step of 20 s at 95°C and then 40 cycles of 95°C for 3 s and 60°C for 30 s.
2.4. MLPA analysis of male fetuses
For male fetuses, DNA was isolated from amniotic fluid cells using the TIANamp Genomic DNA kit (TIANGEN, Beijing, China), and the quality and quantity of DNA was checked using a Nano-Drop 2000 Spectrophotometer. Salsa MLPA kits (P034A2 and P035A2, MRC Holland, Amsterdam, Netherlands) were used to determine whether the fetus had DMD or not. In accordance with the manufacturer's instructions, amplification products were electrophoresed on an ABI 3130 Genetic Analyzer (6). Products were then analyzed using Coffalyser v9.2 software.
3. Results and Discussion
3.1. Fetal sex determination using cffDNA in maternal plasma
The 15 fetuses studied included 6 males and 9 females, and sex was later confirmed by CVS or ultrasound. Fetal sex was determined by means of quantitative real-time PCR after 9 weeks (Figure 1).
Figure 1.
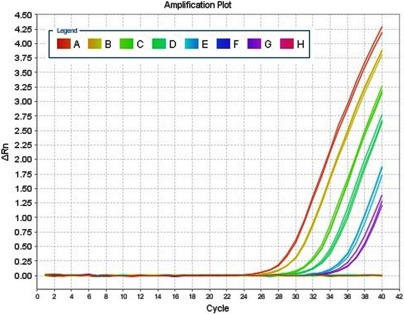
Quantitative real-time PCR. Amplification plots obtained using quantitative real-time PCR for the SRY gene. A: 500GE; B: 100GE; C, D, and E: positive samples; F: negative controls; G: 10GE; H: negative samples. The X-axis denotes the cycle number of a quantitative PCR reaction. The Y-axis denotes ΔRn, which is the fluorescence intensity over the background.
3.2. MLPA analysis of male fetuses
As shown in Figure 2, MLPA revealed deletion of exons 3–20 (A, B) and duplication of exons 13–43 (C, D) in male fetuses. Details on the mutations identified are shown in the figure.
Figure 2.
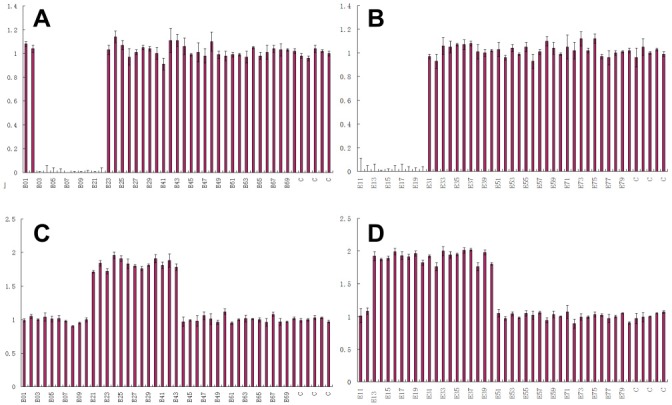
MLPA analysis of DMD. (A), The result for 034 indicates deletion of exons 3–10, 21, and 22; (B), The result for 035 indicates deletion of exons 11–20; (C), The result for 034 indicates duplication of exons 21–30 and 41–43; (D), The result for 035 indicates duplication of exons 13–20 and 31–40.
The main advantage of measuring cffDNA as part of prenatal diagnostic is that sampling can avoid unnecessary risks associated with conventional techniques of prenatal diagnosis (including CVS and amniocentesis). The mean concentration of cffDNA in maternal plasma was more than 20 times higher than that in the cellular fraction of maternal blood at the same gestational stage. The cellular fraction may be present in maternal plasma for years. cffDNA cannot be detected by enzymolysis a few hours after delivery (1), so false-positive results from women who had previously carried a fetus with DMD can be avoided.
cffDNA can be detected in the first trimester of pregnancy. Its measurement is necessary for fetal sex determination since a positive result can allow pregnant women to avoid suffering by terminating a pregnancy early. Non-invasive fetal sex determination using cffDNA can avoid unnecessary CVS and amniocentesis of female fetuses for the prenatal diagnosis of X-linked genetic disorders (4,7–9). DMD is one of the most common genetic muscular dystrophies. The onset of symptoms in affected individuals is generally before the age of 5 and most die in the course of the second or third decade of life due to respiratory or heart failure (10). Thus, prenatal diagnosis is crucial for families of carriers.
Fetal DNA analysis using maternal plasma would be most useful in the detection of paternally-inherited fetal mutations or autosomal recessive genetic disorders where the father and mother carry different mutations (11–14). Increased amounts of fetal DNA may also be found in instances of conditions associated with placental damage, such as pre-eclampsia (15,16). Recent studies showed that cffDNA could be used in prenatal screening for fetal chromosomal disorders (trisomy 18, trisomy 13, and Down syndrome) (17,18).
The main difficulty of using cffDNA in NIPD lies in the low concentration of cffDNA and presence of a larger quantity of background maternal DNA in plasma, so sensitive and specific techniques like the use of a real-time TaqMan system are needed. In conclusion, cffDNA could be used for fetal sex determination in the first trimester of pregnancy to screen for gender-specific inherited disorders while avoiding an unnecessary CVS.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 81170581).
References
- 1. Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997; 350:485-487 [DOI] [PubMed] [Google Scholar]
- 2. Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: Implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998; 62:768-775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lewis C, Hill M, Skirton H, Chitty LS. Fetal sex determination using cell-free fetal DNA: Service users' experiences of and preferences for service delivery. Prenat Diagn. 2012; 32:735-741 [DOI] [PubMed] [Google Scholar]
- 4. Miura K, Higashijima A, Shimada T, Miura S, Yamasaki K, Abe S, Jo O, Kinoshita A, Yoshida A, Yoshimura S, Niikawa N, Yoshiura K, Masuzaki H. Clinical application of fetal sex determination using cell-free fetal DNA in pregnant carriers of X-linked genetic disorders. J Hum Genet. 2011; 56:296-299 [DOI] [PubMed] [Google Scholar]
- 5. Kim SY, Lim JH, Park SY, Kim MY, Choi JS, Ryu HM. Non-invasive prenatal determination of fetal gender using QF-PCR analysis of cell-free fetal DNA in maternal plasma. Clin Chim Acta. 2012; 413:600-604 [DOI] [PubMed] [Google Scholar]
- 6. Verma PK, Dalal A, Mittal B, Phadke SR. Utility of MLPA in mutation analysis and carrier detection for Duchenne muscular dystrophy. Indian J Hum Genet. 2012; 18:91-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolialexi A, Tounta G, Apostolou P, Vrettou C, Papantoniou N, Kanavakis E, Antsaklis A, Mavrou A. Early non-invasive detection of fetal Y chromosome sequences in maternal plasma using multiplex PCR. Eur J Obstet Gynecol Reprod Biol. 2012; 161:34-37 [DOI] [PubMed] [Google Scholar]
- 8. Jin S, Lin XM, Law H, Kwek KY, Yeo GS, Ding C. Further improvement in quantifying male fetal DNA in maternal plasma. Clin Chem. 2012; 58:465-468 [DOI] [PubMed] [Google Scholar]
- 9. Hill M, Compton C, Lewis C, Skirton H, Chitty LS. Determination of foetal sex in pregnancies at risk of haemophilia: A qualitative study exploring the clinical practices and attitudes of health professionals in the United Kingdom. Haemophilia. 2012; 18:575-583 [DOI] [PubMed] [Google Scholar]
- 10. Emery AE. The muscular dystrophies. Lancet. 2002; 359:687-695 [DOI] [PubMed] [Google Scholar]
- 11. Long XJ, Long GF, Lin WX. Noninvasive prenatal diagnosis of Hb Bart's hydrops fetus using cell-free fetal DNA in maternal plasma. Zhonghua Xue Ye Xue Za Zhi. 2009; 30:175-178 [PubMed] [Google Scholar]
- 12. Tounta G, Vrettou C, Kolialexi A, Papantoniou N, Destouni A, Tsangaris GT, Antsaklis A, Kanavakis E, Mavrou A. A multiplex PCR for non-invasive fetal RHD genotyping using cell-free fetal DNA. In Vivo. 2011; 25:411-417 [PubMed] [Google Scholar]
- 13. Paterlini Bréchot P, Mouawia H, Saker A. Non-invasive prenatal diagnosis of cystic fibrosis. Arch Pediatr. 2011; 18:111-118 [DOI] [PubMed] [Google Scholar]
- 14. Yan TZ, Mo QH, Cai R, Chen X, Zhang CM, Liu YH, Chen YJ, Zhou WJ, Xiong F, Xu XM. Reliable detection of paternal SNPs within deletion breakpoints for non-invasive prenatal exclusion of homozygous α-thalassemia in maternal plasma. PLoS One. 2011; 6:e24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jakobsen TR, Clausen FB, Rode L, Dziegiel MH, Tabor A. High levels of fetal DNA are associated with increased risk of spontaneous preterm delivery. Prenat Diagn. 2012; 32:840-845 [DOI] [PubMed] [Google Scholar]
- 16. Hahn S, Rusterholz C, Hösli I, Lapaire O. Cell-free nucleic acids as potential markers for preeclampsia. Placenta. 2011; 32 (Suppl):S17-20 [DOI] [PubMed] [Google Scholar]
- 17. Palomaki GE, Deciu C, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, van den Boom D, Bombard AT, Grody WW, Nelson SF, Canick JA. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: An international collaborative study. Genet Med. 2012; 14:296-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ashoor G, Syngelaki A, Wagner M, Birdir C, Nicolaides KH. Chromosome-selective sequencing of maternal plasma cell-free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012; 206:322.e1-5 [DOI] [PubMed] [Google Scholar]


