Abstract
The electrophoretic mobility shift assay (EMSA) can be used to study proteins that bind to DNA structures created by DNA-damaging agents. UV-damaged DNA-binding protein (UV-DDB), which is involved in nucleotide excision repair, binds to DNA damaged by ultraviolet radiation or the anticancer drug cisplatin. Ku, XRCC4/Ligase IV, and DNA–PKcs, which are involved in the repair of DNA double-strand breaks by nonhomologous end joining, assemble in complexes at DNA ends. This chapter will describe several EMSA protocols for detecting different DNA repair protein–DNA complexes. To obtain additional information, one can apply variations of the EMSA, which include the reverse EMSA to detect binding of 35S-labeled protein to damaged DNA, and the antibody supershift assay to detect the presence of a specific protein in the protein–DNA complex.
Keywords: UV-DDB, Global genomic repair, Nucleotide excision repair, Ku, DNA-PKcs, XRCC4/Ligase IV, DNA-PK, Nonhomologous end joining, Double-strand break repair, V(D)J recombination
1. Introduction
DNA repair pathways must include proteins that recognize and bind to DNA lesions. The search for such proteins has been facilitated by the use of electrophoretic mobility shift assays (EMSAs), which were first used to detect transcription factors that bind to specific DNA sequences (1, 2). To study DNA repair, EMSAs have been adapted to detect proteins that bind to specific DNA lesions or structural features.
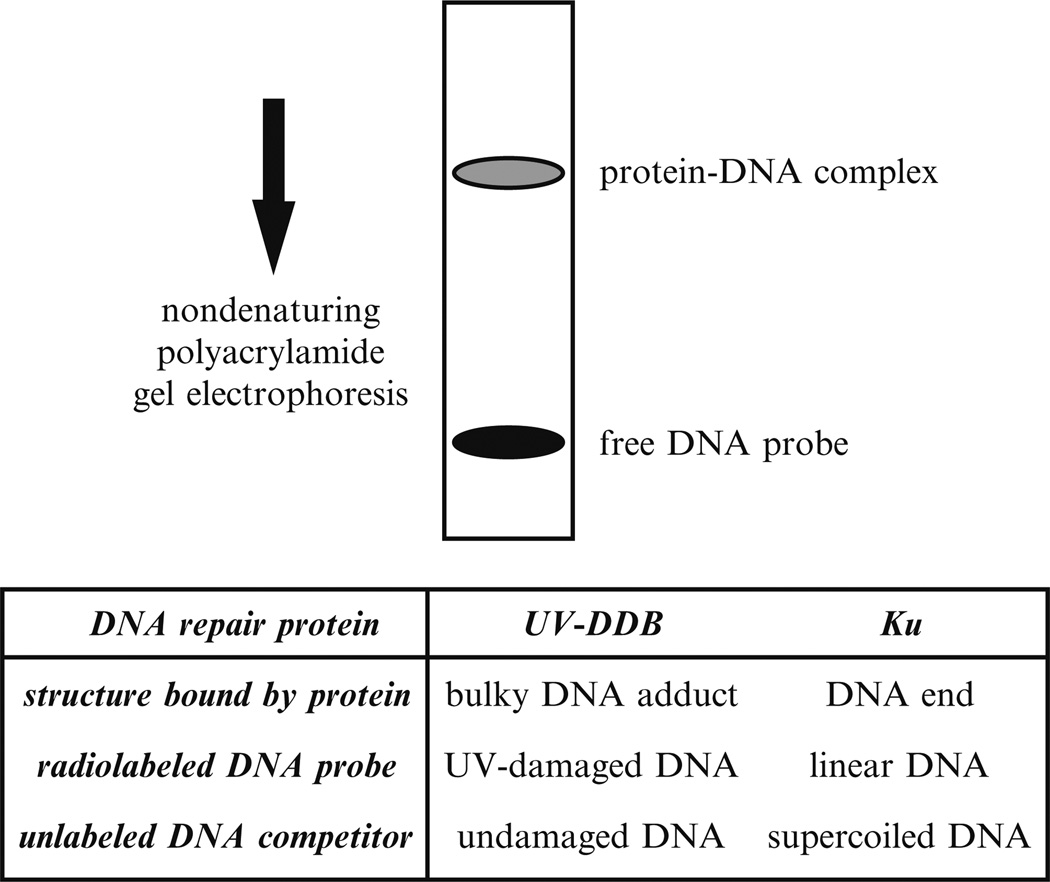
In an EMSA, non-denaturing polyacrylamide gel electrophoresis resolves protein–DNA complexes. A radiolabeled DNA probe contains the DNA lesion or DNA structure of interest. The protein–DNA complexes assemble by incubation of the DNA probe with either crude protein extract or purified proteins (Fig. 1).
Fig. 1.
EMSA for studying UV-DDB and Ku. UV-damaged DNA probe for UV-DDB or linear DNA probe for Ku may be incubated with cell extracts in the presence of the competitor DNA to block nonspecific binding. After incubation, the protein–DNA complex is resolved from the free DNA probe by non-denaturing gel electrophoresis.
EMSA experiments must address the specificity of protein binding. With crude extracts, extraneous proteins may bind to the DNA probe, rather than to the DNA lesion. With purified proteins, some DNA repair proteins may have a weaker but detectable affinity for undamaged regions of the DNA. To eliminate nonspecific binding events, the DNA probe can be mixed with a molar excess of undamaged competitor DNA.
For any EMSA, several general principles ensure proper detection of the protein–DNA complex. The procedure for extracting proteins from the cell nuclei must be optimized for the target protein. A cocktail of protease inhibitors must be present, particularly for high molecular weight proteins that may be vulnerable to degradation. The salt concentration of the extraction buffer must be high enough to dissociate the target protein from genomic DNA, but not so high that interfering components are also extracted. The electrophoresis conditions must be optimized to obtain a well-defined mobility shift: in particular, the salt concentration of the electrophoresis buffer should permit stable formation of the specific protein–DNA complex while minimizing the formation of nonspecific complexes. Use of a mini-gel apparatus permits resolution of the desired complexes in about 30 min; end labeling of the probe DNA with 32P or 33P permits detection of the complexes in a few hours. In our laboratory, gels are poured in lots of 13 and stored for up to 3 weeks at 4 °C so that experiments can be done with minimal setup time.
Commercially prepared gels have provided acceptable results for many experiments. However, in some cases involving the detection of large protein–DNA complexes, we have been unable to replicate earlier results even though the commercially prepared gels were ostensibly manufactured with the same reagents in the same concentrations. In such cases, we were able to replicate our results only after preparing the gels in our own laboratory.
Once a complex is detected, its specificity for the DNA probe should be verified. If the mobility shift of the DNA probe is due to formation of a protein–DNA complex rather than binding by a nonprotein molecule, complex formation should be sensitive to protease digestion. If the protein binds specifically to damaged DNA, a complex of the same mobility should not form with undamaged probe DNA. Alternatively, unlabeled competitor DNA with and without damage can be compared for its ability to compete away the labeled complex. The spectrum of DNA structures recognized by the protein can be determined by testing different competitor DNAs.
Of course, the discovery of a protein that binds to damaged DNA does not prove that it is involved in DNA repair. To establish biological significance, we have found it fruitful to screen mutant cell lines for abnormalities in binding activity. Successful screening depends on procedures described below for rapidly making cell extracts from small numbers of cells.
EMSAs have been used to study a number of DNA repair proteins. In this chapter, we describe our protocols for studying protein complexes involved in nucleotide excision repair and nonhomologous end joining. For nucleotide excision repair, UV-DDB recognizes UV-induced damage in both transcribed and nontranscribed DNA, thus facilitating global genomic repair (3). UV-DDB consists of two subunits, DDB1 and DDB2. For nonhomologous end joining, Ku binds DNA ends and subsequently recruits other components of the nonhomologous end joining repair complex, including DNA–PKcs, XRCC4/Ligase IV, and XLF (Cernunnos) (4).
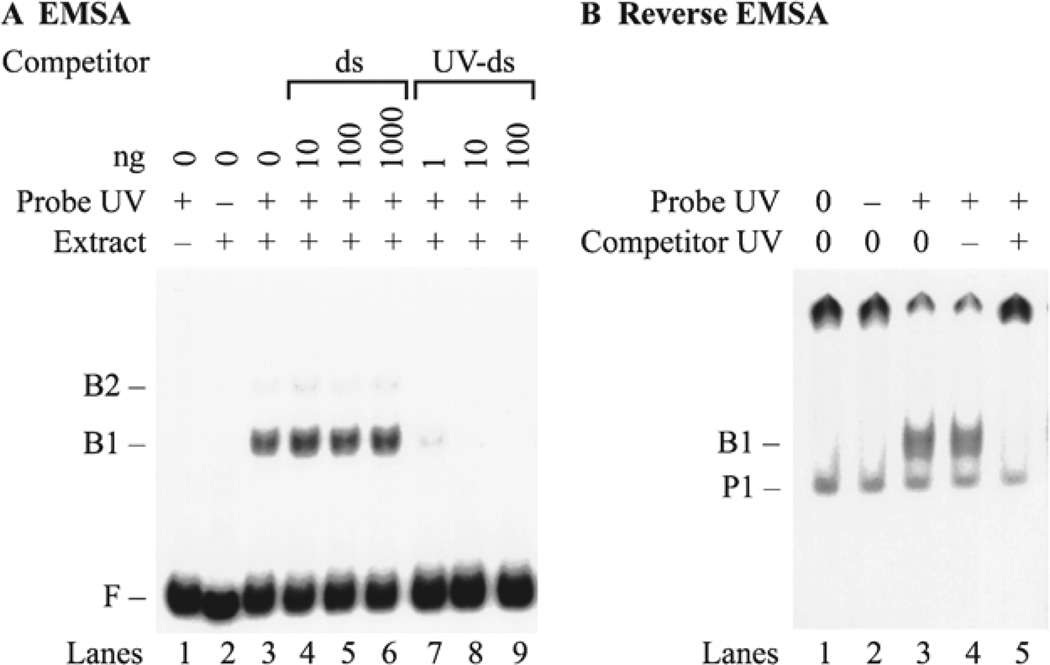
In the case of UV-DDB, the probe DNA was a linear DNA fragment damaged by exposure to UV-radiation, and the competitor DNA consisted of unlabeled linear DNA that was left undamaged. A complex with a reasonably well-defined mobility was detected even though UV-radiation introduced DNA lesions throughout the length of the DNA probe (5). This phenomenon was previously reported for the binding of a sequence-specific DNA-binding protein to a series of DNA probes containing the binding sequence at different sites. In that case, the mobility of the protein–DNA complex was relatively insensitive to the position of the binding sequence (6). The specificity of UV-DDB was established by showing that the binding activity was competed away by the addition of unlabeled UV-damaged competitor DNA to the binding reaction (Fig. 2a). UV-DDB was then purified (7) and partially sequenced in order to isolate a cDNA encoding a polypeptide of the expected molecular weight. To demonstrate that this polypeptide bound specifically to UV-damaged DNA, the cDNA was transcribed and translated in vitro so that the protein product was labeled with 35S-labeled methionine (8). A reverse EMSA was then used to show that the mobility of the labeled protein was specifically shifted by the addition of damaged DNA, but not be undamaged DNA (Fig. 2b).
Fig. 2.
EMSA and reverse EMSA for UV-DDB. (a) EMSA. Crude protein extracts from HeLa cells were incubated with f148 probe DNA in the presence of different amounts of unlabeled competitor DNA. The probe DNA was UV irradiated (lanes 1 and 3–9) or left intact (lane 2). Protein extract was omitted (lane 1) or added (lanes 2–9). Control binding reactions omitted competitor DNA (lanes 1–3). Competition was carried out with intact double-stranded DNA (ds) (lanes 4–6), UV-irradiated double-stranded DNA (UV-ds) (lanes 7–9). (b) Reverse EMSA. The fraction of 35S-labeled DDB1 protein that bound to a UV-DNA cellulose column was assayed in a binding reaction with unlabeled f148 DNA. Probe DNA was omitted (lane 1), added as undamaged DNA (lane 2), or added as UV-damaged DNA (lanes 3 – 5). Unlabeled plasmid competitor DNA was either omitted (lanes 1–3), added as undamaged DNA (lane 4), or added as UV-damaged DNA (lane 5). Some of the 35S-labeled DDB1 protein is retained in the well of the gel (P2) because it was purified from a UV-DNA cellulose column, so that some high molecular weight UV-damaged DNA co-eluted as a complex with 35S-labeled DDB1 protein.
To determine the biological relevance of UV-DDB, we screened mutant cell lines hypersensitive to UV radiation. A role in nucleotide excision repair was supported by the discovery that UV-DDB was absent in a subset of xeroderma pigmentosum group E cells, which are defective in this repair pathway (5). Furthermore, UV-DDB also recognized competitor DNA carrying intrastrand DNA crosslinks induced by the anticancer drug cisplatin, and levels of UV-DDB were increased in extracts from cells selected for resistance to cisplatin (9). Biological relevance was established by showing that microinjection of XPE cells with purified UV-DDB restored DNA repair to the cells (10). More specifically, the ectopic expression of UV-DDB in hamster cells enhanced the global genomic repair of cyclobutane dimers (11). Global genomic repair refers to the removal of DNA lesions from nontranscribed regions of the genome and from the nontranscribed strand in transcribed regions of the genome.
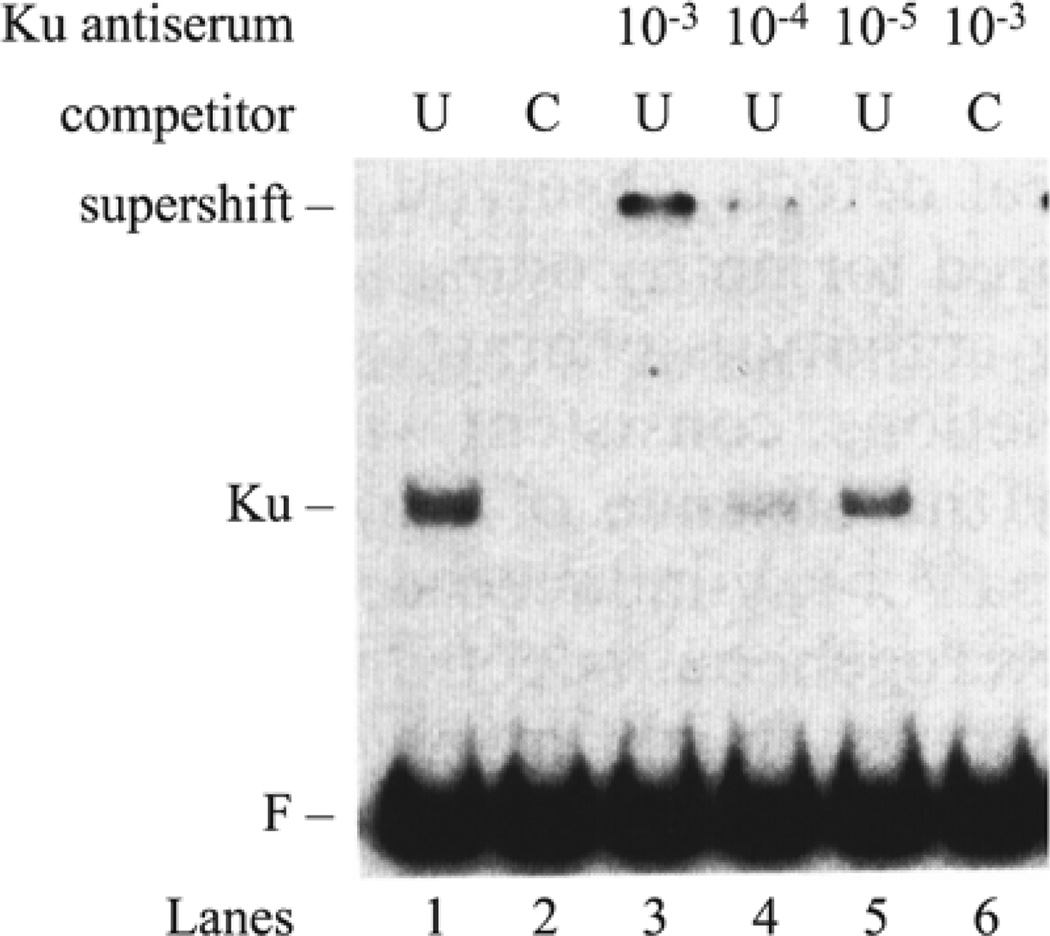
In the case of Ku, the probe was an undamaged linear DNA fragment. Supercoiled plasmid DNA served as competitor for nonspecific DNA-binding proteins, so that binding to the linear DNA probe was specific for the DNA ends (Fig. 3). This assay system detected a protein–DNA complex, which we originally denoted as DNA end-binding (DEB) factor (12). The specificity for DNA ends was established by showing competition for the protein–DNA complex by unlabeled competitor plasmid DNA cleaved with any one of several different restriction enzymes, producing DNA ends with 5′ or 3′ overhanging, or blunt, ends. The identification of DEB factor as Ku protein was first established by a useful adjunct to the EMSA, in which incubation of the binding reactions with anti-Ku antibodies produced a supershift of the original protein–DNA complex (13).
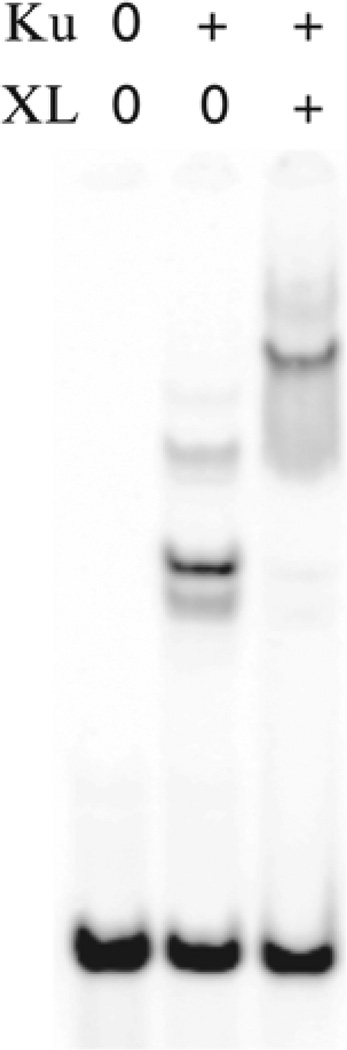
Fig. 3.
EMSA and antibody supershift for Ku. Nuclear extract from the hamster cell line AA8 was incubated in the presence of radiolabeled f148 probe and uncut plasmid (U) DNA competitor in lane 1, or cut plasmid (C) in lane 2. The Ku antiserum was included in lanes 3–5 in dilutions ranging from 10−3 to 10−5 in the presence of uncut plasmid (U). The supershifted complex is specific for DNA ends as shown in lane 6, where cut plasmid (C) is the competitor.
To determine the biological relevance of DEB factor, we screened a series of mutant cell lines hypersensitive to ionizing radiation, reasoning that a DEB protein might be involved in the repair of the double-strand breaks produced by ionizing radiation. This hypothesis was confirmed by the discovery that DEB factor was absent in three cell lines from X-ray complementation group 5, which is defective in double-strand break repair (13). Strikingly, these cells were also defective in V(D)J recombination, the pathway that generates immunological diversity by cleaving and rearranging the immunoglobulin genes in B cells and the T cell receptor genes in T cells. Biological relevance was established by showing that transfection of the mutant cells with a cDNA expression vector for the 86 kDa subunit of Ku restored both ionizing radiation resistance and V(D)J recombination (14, 15), establishing a role for Ku in nonhomologous end joining.
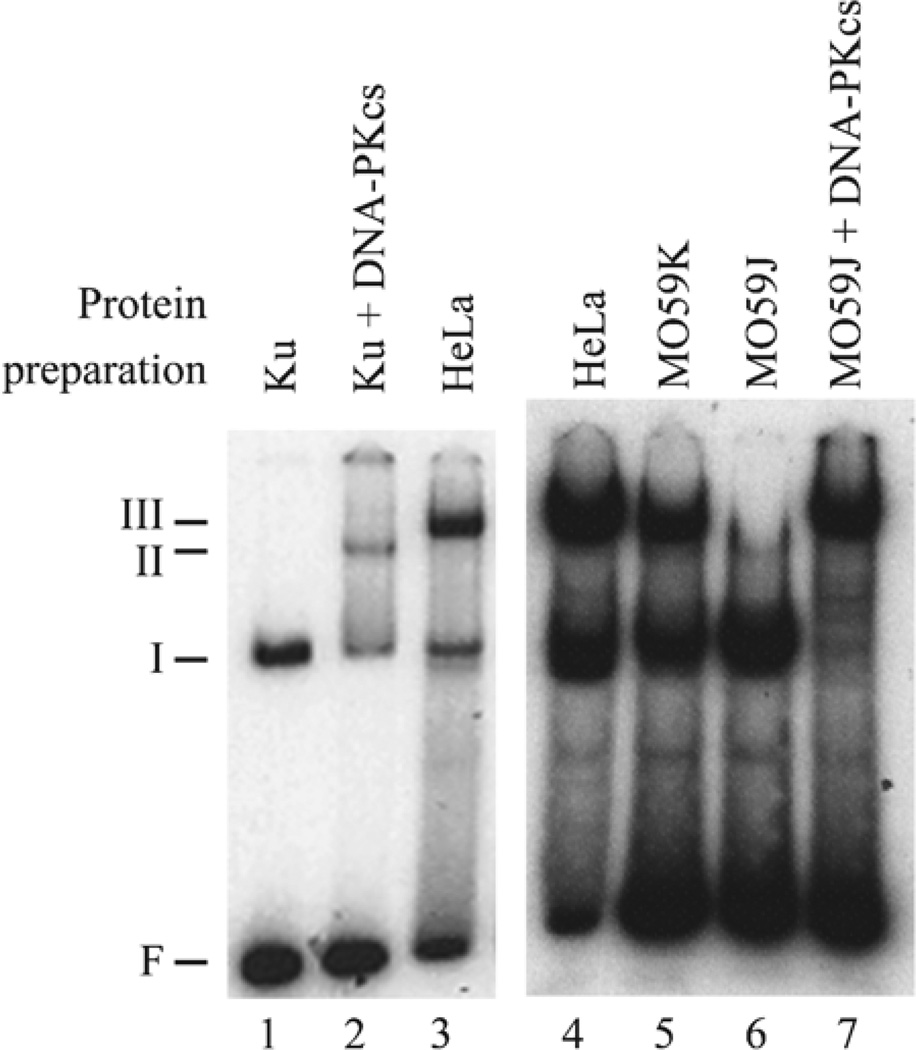
Upon binding to DNA ends, Ku is capable of translocating inward to allow binding of additional Ku molecules. Ku also recruits the DNA-dependent protein kinase catalytic subunit (DNA–PKcs) to the DNA ends, thus forming a higher-order complex with DNA (Fig. 4). (DNA-PK consists of its catalytic subunit DNA–PKcs and the heterodimeric Ku protein.) To detect binding by DNA–PKcs, we modified the EMSA by shortening the length of the radiolabeled DNA probe to 32 base pairs and eliminating competitor DNA. Indeed, higher-order complexes were detected with either purified proteins or cell extracts. Interestingly, cell extracts generate a higher-order complex with a slightly lower mobility than seen with purified proteins (compare lanes 2 and 3 in Fig. 4). Supershift assays with antibodies demonstrate that this complex contains both Ku and DNA–PKcs (data not shown). Extracts from M059J cells, which lack DNA–PKcs, do not support the formation of the higher-order complex, but supplementation of the mutant extracts with purified DNA–PKcs restores the higher-order complex (compare lanes 6 and 7 in Fig. 4). Finally, DNA–PKcs is capable of binding to DNA ends in the absence of Ku. Indeed, when the proteins are incubated with a short 32 bp oligonucleotide in the absence of competitor DNA, the EMSA is capable of detecting complexes containing DNA–PKcs. These complexes include DNA bound to DNA–PKcs, or Ku in a complex with DNA–PKcs (16).
Fig. 4.
EMSA for complex of DNA–PKcs and Ku bound to DNA. Radiolabeled f32 probe was incubated with purified protein consisting of 11 ng of Ku (lane 1), or 11 ng of Ku plus 35 ng of DNA–PKcs (lane 2). In addition, the probe was incubated with 500 ng of nuclear extract prepared from HeLa cells (lanes 3 and 4), M059K cells derived from a human glioma (lane 5), or M059J cells lacking DNA–PKcs derived from the same glioma (lanes 7 and 8). In lane 8, the M059J extract was supplemented with 50 ng of purified DNA–PKcs. The gel shows migration of the free probe (F) and protein–DNA complexes containing Ku (I), Ku and DNA–PKcs (II), and Ku, DNA–PKcs, and an unidentified factor (III).
In addition to recruiting DNA–PKcs, Ku also recruits XRCC4/Ligase IV to DNA ends (17). Ku and XRCC4/Ligase IV form a complex on DNA that migrates with a slower mobility than Ku alone (Fig. 5). Furthermore, antibodies against XRCC4 or Ku supershift the complex to an even higher position in the gel (17), indicating that Ku and XRCC4 are present in the complex. XRCC4 and Ligase IV form a stable dimeric complex under the conditions used for the binding reaction, suggesting that the protein–DNA complex contains all three proteins, Ku, XRCC4, and Ligase IV.
Fig. 5.
EMSA for complex of Ku and XRCC4/Ligase IV bound to DNA. Probe DNA f298 labeled with 33P was incubated in the absence of protein (lane 1), together with recombinant Ku alone (lane 2), or together with both recombinant Ku and recombinant XRCC4/ Ligase IV (lane 3). The mobility of the protein–DNA complex was not due to DNA ligation, because the reaction conditions permitted only negligible ligation, as confirmed by gel electrophoresis (data not shown). The 33P labeling produced sharper bands, allowing for greater resolution compared to the 32P labeling (Figs. 2 – 4).
EMSAs have been used to identify other structure-specific proteins. The randomly damaged UV-irradiated DNA probe, which was used to detect UV-DDB in human extracts, failed to detect a corresponding factor in yeast extracts, but instead detected photolyase, which repairs UV-induced cyclobutane pyrimidine dimers by photoreactivation (18). Like UV-DDB, yeast photolyase recognized both UV-damaged and cisplatin crosslinked DNA. Although UV-DDB expression was associated with resistance to cisplatin, yeast photolyase conferred sensitivity to cisplatin (19). DNA probes with other structures have detected additional proteins. An oligonucleotide containing a single GT mismatch was used to identify the GT binding protein that has now been shown to be involved in mismatch repair (20). An oligonucleotide containing a single intrastrand cisplatin crosslink was used to search extracts for damage-specific DNA-binding proteins that proved to be the HMG-1 and HMG-2 proteins (21, 22). In this case, the proteins were not involved in DNA repair, but rather interfered with the nucleotide excision repair machinery and conferred sensitivity to cisplatin (23).
In summary, this chapter describes EMSAs for studying several protein–DNA complexes. Variations in probe length and labeling, extract preparation, protein and DNA concentrations, binding buffer, and gel electrophoresis conditions re fl ect the need to optimize conditions for each protein–DNA complex.
2. Materials
2.1. Probe DNA
2.1.1. Preparation of f148 and f32 Probe
f148: pRSVcat plasmid.
f32: oligonucleotides f32-1, 5′ -GTAGTTGTACCCGATGGT GGACGCGTGC-3′ and f32-2, 5′ -GGCCGCACGCGTCCA TCGGGTACAACTAC3′.
Hind III and Pvu II: 10 units/ µL (New England Biolabs, Beverly, MA).
Agarose.
1× TBE: 89 mM Tris–HCl, pH 8.0, 89 mM borate, and 2 mM EDTA.
Ethidium bromide solution: 2 µg/mL, dissolved in distilled water and filtered through 0.4 µm filter.
Long-wavelength UV light source (366 nm): Model UVL-56 (Ultra-violet products, Inc., San Gabriel, CA).
IBI-electroeluter: Model 46000 (IBI-Kodak, Rochester, NY).
Polyethylene tubing: I.D. 1.6 mm.
Salt solution: 10 µL of 0.5 % bromophenol blue and 1 mL of 10 M ammonium acetate.
18-gauge needle.
Razor blade.
Isobutanol.
Ethanol, 100 %.
Ethanol, 80 %.
TE: 10 mM Tris–HCl, pH 8.0, and 1 mM EDTA.
2.1.2. Preparation of f298 Probe
f298: any plasmid containing the Amp® gene for the PCR template; forward primer 5′ -CATACCAAACGACGAGCGTG AC-3′ and reverse primer 5′ -ATCCATAGTTGCCTGACT CCCC-3′.
Herculase II Fusion DNA Polymerase (Agilent, Santa Clara, CA).
5× Herculase II reaction buffer.
dNTPs.
Thermal cycler for PCR amplification.
QIAquick PCR purification Kit (Qiagen, Valencia, CA).
2.1.3. Labeling of Probe DNA (Klenow Method)
10× Klenow buffer: 100 mM Tris–HCl, pH 7.5, 50 mM MgCl2, and 75 mM dithiothreitol.
10 mM dATP, dGTP, and dTTP.
α-32P-dCTP: 3,000 Ci/mmol, 10 mCi/mL (GE Healthcare Life Sciences, Piscataway, NJ).
DNA polymerase I, Klenow fragment: 5 units/ µL (New England Biolabs, Beverly, MA).
2.1.4. Labeling of f148Probe (Exonuclease III/Klenow Method)
Items 1– 4 of Subheading 2.1.3.
Exonuclease III: 100 units/ µL, diluted in TE (New England Biolabs, Beverly, MA).
2.1.5. Labeling of f298 Probe (T4 PolynucleotideKinase Method)
10× T4 polynucleotide kinase reaction buffer: 700 mM Tris–HCl, pH 7.6, 100 mM MgCl2, 50 mM Dithiothreitol (DTT).
[γ-33P]ATP: 3,000 Ci/mmol, 10 mCi/mL (Perkin Elmer, Waltham, MA). See note 4 for comments on the use of 33P to label the DNA probe.
T4 polynucleotide kinase: 10 U/ µL (New England Biolabs, Beverly, MA).
Nucleotide-removal kit (Qiagen, Valencia, CA).
2.2. Cell Extracts
2.2.1. Whole Cell Extract
Phosphate-buffered saline: 1 g/L d-glucose, 36 mg/L sodiumpyruvate, 36 mg/L calcium phosphate, and 36 mg/L magnesiumphosphate.
Lysis buffer: 700 mM NaCl, 1 mM EGTA, 1 mM EDTA,10 mM β-glycerol-phosphate, 2 mM MgCl2, 10 mM KCl,1 mM sodium vanadate, 1 mM PMSF, 1 mM dithiothreitol,0.1 % NP-40, 10 µg/mL of each pepstatin, leupeptin, andaprotinin.
Bradford solution (BioRad, Hercules, CA).
2.2.2. Cytoplasmic and Nuclear Extract
Phosphate-buffered saline: 1 g/L d-glucose, 36 mg/L sodiumpyruvate, 36 mg/L calcium phosphate, and 36 mg/L magnesium phosphate.
Buffer A without NP-40: 10 mM Hepes-KOH, pH 7.4,1.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol, and 1 mM EDTA.
Buffer A with NP-40: 10 mM Hepes-KOH, pH 7.4, 1.5 mMMgCl2, 10 mM KCl, 1 mM dithiothreitol, 1 mM EDTA, and1.0 % (w/v) NP-40.
Buffer C: 20 mM Hepes-KOH, pH 7.4, 20 % glycerol (v/v),500 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, and 0.5 mM dithiothreitol.
100× Protease inhibitor cocktail: 100 mM PMSF, 10 mg/mL pepstatin A, 10 mg/mL leupeptin, 10 mg/mL aprotinin.
2.2.3. Non-denaturing Polyacrylamide Gel
Mini-gel plates: 0.75 mm thickness, 80 × 100 mm (Hoefer, Holliston, MA).
Gel caster: SE215 Mighty-Small multiple gel caster (Hoefer, Holliston, MA).
Acrylamide-bisacrylamide: 29:1, w/w (Molecular biology grade, Sigma-Aldrich, St. Louis, MO).
5× TGE buffer: 50 mM Tris–HCl, pH 8.5, 380 mM glycine, and 2 mM EDTA (see Note 1).
Ammonium persulfate: 10 % of fresh solution.
TEMED.
Gel combs: 10 or 15 lane combs with 0.75 mm thickness (Hoefer, Holliston, MA).
2.3. EMSA for Studying UV-DDB
2.3.1. UV-Damaged f148 DNA
Germicidal lamp: G15T8 (General Electric, Cleveland, OH).
Radiometer/photometer: Model IL1350 (International light Inc., Newburyport, MA).
2.3.2. Competitor DNA to Mask Nonspecific Binding
Poly(dI-dC) (GE Healthcare Life Sciences, Piscataway, NJ).
Salmon sperm DNA: Sheared by passing through 18-gauge needle with pressure.
Linear plasmid DNA: pRSVcat plasmid digested with Pvu II and extracted with phenol/chloroform, and precipitated in ethanol.
2.3.3. Competitor DNA to Test Binding Specificity
Supercoiled pRSVcat plasmid.
AP treatment buffer: 0.01 M sodium citrate, 0.1 M NaCl, pH 5.0.
Pt treatment buffer: 3 mM NaCl, 1 mM NaH2 PO4, pH 7.4.
Cis-diamminedichloroplatinum (cis-DDP, Sigma, St. Louis, MO): Stock solution was dissolved in Pt treatment buffer and stored at −20 °C in the dark.
Ethanol, 100 %.
N-Methyl- N′ -nitro- N-nitrosoguanidine (MNNG, Sigma) was dissolved to 15 mM in ethanol and stored at −20 °C.
MNNG treatment buffer: 10 mM Tris–HCl, pH 7.5 and 50 mM NaCl.
2.3.4. UV-DDB Binding Reaction
5× binding buffer: 12 mM Hepes-KOH, pH 7.9, 60 mM KCl, 5 mM MgCl2, 4 mM Tris, 0.6 mM EDTA, 1 mM dithiothreitol, and 12 % glycerol (v/v).
BSA: 10 mg/mL.
Plasmid DNA: pRSVcat, 1 mg/mL.
Poly(dI-dC)/salmon sperm DNA: 1/0.5 mg/mL.
Whole cell or nuclear extract: 0.1–0.5 mg/mL.
Loading dye: 0.025 % bromophenol blue in 5× binding buffer.
Whatman 3MM paper (GE Healthcare Life Sciences, iscataway, NJ).
X-ray film: Kodak XAR-5 (Sigma-Aldrich, St. Louis, MO).
2.4. Variation on EMSA: Reverse Electrophoretic Mobility Shift Assay
2.4.1. In Vitro Synthesis of 35S-Labeled DDB1
DDB1 plasmid: cloned into pCDNA3 vector (Invitrogen, Carlsbad, CA).
Rabbit reticulocyte lysate (Promega, Madison, WI).
35S-labeled methionine: 1,000 Ci/mmol, 50 mCi/mL (GE Healthcare Life Sciences, Piscataway, NJ).
T7 RNA polymerase buffer: 40 mM Tris–HCl, pH 7.9, 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, and 0.5 mM UTP.
T7 RNA polymerase: 10 units/ µL (Promega, Madison, WI).
Amino acid mixture minus methionine: 1 mM (Promega, Madison, WI).
RNasin ribonuclease inhibitor: 40 units/ µL (Promega, Madison, WI).
RNase-free water: treated with 0.1 % DEPC at 37 °C overnight and autoclaved at 120 °C for 20 min.
2.4.2. Purification of Active 35S-Labeled DDB1
UV-DNA cellulose resin: prepared as described previously (7).
200 µL pipetman tip.
Buffer D: 10 mM Hepes-KOH, pH 7.9, 2 mM EDTA, 2 mM dithiothreitol, 0.01 % NP-40 (v/v).
2.4.3. Reverse Electrophoretic Mobility Shift Assay
5× binding buffer: 12 mM Hepes-KOH, pH 7.9, 60 mM KCl, 5 mM MgCl2, 4 mM Tris, 0.6 mM EDTA, 1 mM dithiothreitol, and 12 % glycerol (v/v).
BSA: 10 mg/mL.
Plasmid DNA: pRSVcat, 1 mg/mL.
Poly(dI-dC)/salmon sperm DNA: 1/0.5 mg/mL.
35S-labeled DDB1.
Loading dye: 0.025 % bromophenol blue in 5× binding buffer.
50 % methanol and 10 % acetic acid.
Amersham Amplify™ Fluorographic Reagent (GE Healthcare Life Sciences, Piscataway, NJ).
Whatman 3MM paper (GE Healthcare Life Sciences, Piscataway, NJ).
Baby powder.
Whatman 3MM paper (GE Healthcare Life Sciences, Piscataway, NJ).
X-ray film: Kodak XAR-5 (Sigma-Aldrich, St. Louis, MO).
2.5. EMSA for Studying Nonhomologous End Joining Proteins
2.5.1. Probe DNA
Materials for purifying and radiolabeling the f148 and f32 probe are described in Subheading 2.1.
2.5.2. Competitor DNA to Mask Nonspecific Binding
Supercoiled pRSVcat plasmid: purified by CsCl centrifugation (24).
2.5.3. Competitor DNA to Test Binding Specificity
Poly(dA) or poly(dT) (GE Healthcare Life Sciences, Piscataway, NJ).
Double-stranded linear DNA: unlabeled f148 DNA.
Single-stranded circular DNA: phage M13 DNA (New England BioLabs, Beverly, MA).
Supercoiled pRSVcat plasmid: purified by CsCl centrifugation (24).
2.5.4. Ku Binding Reaction
5× binding buffer: 12 mM Hepes-KOH, pH 7.9, 60 mM KCl, 5 mM MgCl2, 4 mM Tris, 0.6 mM EDTA, 1 mM dithiothreitol, and 12 % glycerol (v/v).
Supercoiled plasmid DNA: pRSVcat, 1 mg/mL.
Protein extract: 0.1–0.5 mg/mL.
Loading dye: 0.025 % bromophenol blue in 5× binding buffer.
Polyacrylamide gel: 4 %.
Whatman 3 MM paper (GE Healthcare Life Sciences, Piscataway, NJ).
X-ray film: Kodak XAR-5 (Sigma-Aldrich, St. Louis, MO).
2.5.5. Ku Plus DNA–PKcs Binding Reaction
5× binding buffer: 50 mM Tris–HCl, pH 7.8, 5 mM EDTA, 500 mM NaCl, 25 % glycerol, 1 mM dithiothreitol.
Protein extract: 0.1–5.0 mg/mL.
Loading dye: 0.025 % bromophenol blue in 1× binding buffer.
Polyacrylamide gel: 4 %.
Whatman 3 MM paper (GE Healthcare Life Sciences, Piscataway, NJ).
X-ray film: Kodak XAR-5 (Sigma-Aldrich, St. Louis, MO).
2.5.6. Ku Plus XRCC4/Ligase IV Binding Reaction
2× binding buffer: 50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 145 mM KCl, 10 mM Mg2Cl, 4 mM DTT, 100 µg/mL BSA, 0.05 % Triton X-100, 10 % (v/v) Glycerol, 0.2 mM EDTA and 10 % (w/v) polyethylene glycol (MW > 8,000 Da).
Recombinant Ku and XRCC4/Ligase IV. For methods to express and purify the recombinant proteins, see ref. 4.
5 % Polyacrylamide TBE gel (Bio-Rad, Hercules, CA).
Whatman 3MM paper (GE Healthcare Life Sciences, Piscataway, NJ).
Storage phosphor screen (GE Healthcare Life Sciences, Piscataway, NJ).
Typhoon 9400 imager (GE Healthcare Life Sciences, Piscataway, NJ).
2.5.7. Variation on EMSA: Antibody Supershift
3. Methods
3.1. Probe DNA
3.1.1. Preparation of f148 Probe
The 148 bp DNA fragment (f148) was isolated from bacterial chloramphenicol acetyltransferase gene.
Digest pRSVcat plasmid with Hind III and Pvu II restriction enzymes.
Separate the digested DNA fragments on a 1.5 % agarose gel.
Stain the gel with ethidium bromide solution and visualize the 148 bp DNA fragment (f148) with long-wavelength UV light. Long-wavelength UV was used instead of short-wavelength UV to prevent damage to the DNA.
Excise the band containing the f148 fragment with a razor blade.
Elute the f148 DNA fragment from the gel slice by using an IBI-electroeluter, as described below.
Soak the gel slice containing f148 in 0.2× TBE buffer for 5–10 min.
Preclear the electroeluter chamber by electrophoresis in 0.2× TBE for 20 min at 150 V. Carefully remove air bubbles in the chamber.
Place the gel slice in a circular receptacle. Surround the gel slice with 0.2× TBE buffer but do not cover the gel slice. Keep valve up.
Flush V-channel with 0.2× TBE buffer by using 18-gauge needle on 1 mL syringe with polyethylene tubing to protect electroeluter device.
Underlay 125 µL of salt solution into the V-channel.
Close the cover to move the valve to the intermediate position.
Run the electroeluter at 150 V; watch for the migration of the DNA band out of the gel slice with the handheld long-wavelength UV light.
Stop the electroelution when all of the DNA fragments have left the gel slice (approximately 10–20 min).
Push the valve to the lowest position.
Withdraw the contents in the V-channel and rinse the channel with 100 µL of the salt solution.
Extract the eluate twice with 400 µL of isobutanol, which will remove the contaminated dyes and reduce the volume.
Measure the final volume of the eluate and add two volumes of 100 % ethanol and precipitate at −20 °C for 2 h.
Spin the ethanol precipitate at 13,000 × g for 20 min at 4 °C.
Wash the pellet with cold 80 % ethanol solution.
Suspend the pellet in TE solution and measure the concentration.
3.1.2. Preparation of f32 Probe
Mix 300 µL of 5 ng/ µL of f32-1 with 300 µL 5 ng/ µL f32-2 in TE buffer (materials are described in Subheading 2.1.1).
Heat the mixture of oligonucleotides to 100 °C for 2 min.
Anneal the oligonucleotides by turning off the heat source and allowing the mixture to cool to room temperature.
3.1.3. Preparation of f298 Probe
- Set up reaction mixture at room temperature as follows:
(a) 5× Herculase II reaction buffer 10 µL (b) dNTPs (25 mM each) 0.5 µL (c) Template DNA 10 ng (d) Forward primer (10 µM) 1.25 µL (e) Reverse primer (10 µM) 1.25 µL (f) Nuclease-free water to 49.5 µL Add 0.5 µL of Herculase II fusion DNA polymerase.
Amplify the DNA by PCR for 35 cycles, with annealing temperature 60 °C and extension time of 20 s.
To verify successful amplification of the probe, run 1 µL of the reaction on a 1 % agarose gel prior to purification.
Purify the PCR product using Qiaquick PCR purification kit (Qiagen, Valencia, CA)
3.1.4. Labeling of f148 or f32 Probe (Klenow Method) (See Note 2)
- Set up reaction mixture at room temperature as follows:
(a) 10× Klenow buffer 1 µL (b) f148 or f32 DNA (5 ng/µL) 4 µL (c) 10 mM dATP 0.5 µL (d) 10 mM dGTP 0.5 µL (e) 10 mM dTTP 0.5 µL (f) α-32P-dCTP (10 µCi/µL) 1 µL (g) Klenow (5 units/µL) 1 µL (h) Distilled water to 10 µL Incubate at room temperature for 20 min.
Inactivate the Klenow by incubating the reaction mixture at 65 °C for 10 min. This step may be omitted if step 4 is performed immediately.
Purify the labeled f148 with nucleotide-removal kit (Qiagen, Valencia, CA).
3.1.5. Labeling of f148 Probe (Exonuclease III/Klenow Method) (See Note 3)
- Set up exonuclease III digestion reaction as follows:
(a) 10× Klenow buffer 2 µL (b) f148 DNA fragment (5 ng/µL) 4 µL (c) Exonuclease III (0.2 units/µL, diluted in TE) 1 µL (d) Distilled water to 10 µL Incubate at room temperature for 10 min.
Inactivate the exonuclease III by incubating the reaction mixture for 10 min at 65 °C
- Add the following to the cooled reaction mixture at room temperature:
(a) 10 mM dATP 1 µL (b) 10 mM dGTP 1 µL (c) 10 mM dTTP 1 µL (d) α-32P-dCTP (10 µCi/µL) 1 µL (e) Klenow (5 units/µL) 1 µL (f) Distilled water to 20 µL Incubate at 37 °C for 30 min.
Inactivate the Klenow by incubating the reaction mixture at 65 °C for 10 min. This step may be omitted if step 7 is performed immediately.
Purify the labeled f148 with nucleotide-removal kit (Qiagen, Valencia, CA).
3.1.6. Labeling of f298 Probe (T4 Polynucleotide Kinase Method)(See Note 4)
- Set up reaction mixture at room temperature as follows:
(a) 10× kinase buffer 2 µL (b) f298 DNA 10 pmole (c) γ-33P-dATP (3,000 Ci/mmole, 10 mCi/mL) 20 pmole (6 µL) (d) Nuclease-free water to 19 µL Heat the mixture to 70 °C for 5 min and put on ice.
Add 1 µL of T4 polynucleotide kinase and incubate at 37 °C for 30 min.
Purify the labeled f298 DNA with nucleotide-removal kit (Qiagen, Valencia, CA).
3.2. Preparation of Cell Extracts
3.2.1. Whole Cell Extract
Harvest 2 × 106 cells from culture dishes in 1 mL of ice-cold phosphate-buffered saline.
Pellet the cells by centrifugation for 1 min at 13,000 × g.
Resuspend the pellet in 30 µL of the lysis buffer.
Incubate the lysates at 4 °C for 30 min with gentle shaking.
Centrifuge the lysates at 13,000 × g for 30 min at 4 °C.
Save the supernatant at −80 °C (see Note 5).
Measure the protein concentration by a modification of the Bradford method (25).
3.2.2. Cytoplasmic and Nuclear Extract
Harvest 2 × 106 adherent cells from culture dishes in 1 mL of ice-cold phosphate-buffered saline.
Wash the cells once with 500 µL of 1× phosphate-buffered saline.
Add 10 µL protease inhibitor cocktail to 1 mL buffer A without NP-40. Wash the cells once with 500 µL of Buffer A without NP-40.
Add 1 µL protease inhibitor cocktail to 100 µL buffer A with NP-40. Resuspend the pellets in 20 µL of Buffer A with NP-40.
Incubate the suspensions at 4 °C for 10 min with gentle shaking.
Spin down the lysates at 13,000 × g for 5 s at 4 °C.
Save the supernatant as cytoplasmic extract at −80 °C (see Note 5).
Wash the pellet in 300 µL buffer A without NP-40 by gentle pipetting. Microfuge at 13,000× g for 5 s.
Add 10 µL of protease inhibitor cocktail to 1 mL buffer C. Resuspend the pellet from step 8 in 75 µL of Buffer C (see Notes 6 and 7).
Incubate the suspensions at 4 °C for 20 min with gentle shaking.
Spin down the nuclear lysates at 13,000 × g for 15 min at 4 °C.
Save the supernatant as nuclear extract at −80 °C (see Note 5).
Measure the protein concentration by the method of Bradford (25)
3.3. Preparation of Non-denaturing Gel
Assemble 13 sets of 0.75 mm thick mini-gels in a gel caster.
- Prepare 4 % polyacrylamide gel solution as follows (see Note 8):
(a) Acrylamide-bisacrylamide (29:1) 14 mL (b) 5× TGE buffer 20 mL (c) Distilled water 65 mL (d) Ammonium persulfate (10 %) 1 mL (e) TEMED 120 µL Gently mix the gel solution and immediately pour to the top of the assembled gel caster with caution to avoid bubbles.
Tap the gel caster gently several times to remove the bubbles in the gel caster.
Insert the comb into the top of each gel.
Leave at room temperature for at least 4 h.
Disassemble the gel caster and wrap each gel with Saran Wrap and store at 4 °C until use. Be careful to remove residual polyacrylamide, which adheres to the outside of the individual gels, since this can interfere with efficient electrophoresis (see Note 9).
3.4. EMSA for Studying UV-DDB
3.4.1. UV Irradiation of the f148 Probe
The labeled f148 was damaged by UV radiation at a DNA concentration of 0.2 µg/mL with a germicidal lamp at a fl ux of 10.4 J/m2/s for total doses of 100–5,000 J/m2 (see Notes 10 and 11).
3.4.2. Competitor DNA Against Nonspecific Binding
Crude cellular extracts have nonspecific DNA-binding proteins including DEB proteins (see Note 12). Thus, excess unlabeled linear double-stranded DNA was included in the reaction mixture. The amount of the competitor DNA was determined empirically because different extracts require different amounts of competitor DNA. A mixture of poly(dI-dC) and salmon sperm DNA (2:1, w/w) was generally used.
3.4.3. Competitor DNA to Test Specificity of Binding
The binding specificity for UV-damaged DNA and single-stranded DNA was measured by competition assay. The single-stranded competitor DNA was prepared by heating linear double-stranded DNA at 100 °C for 5 min and then cooling rapidly in ice water. UV-damaged single-stranded DNA was prepared by exposing the single-stranded DNA to UV radiation.
The chemically damaged competitor DNA was prepared as described previously (26) (see Note 13). Apurinic DNA was prepared by incubating DNA (300 µM DNA phosphate) in AP treatment buffer at 70 °C for different times. Approximately one purine base is released every 4 min under these reaction conditions. Cisplatin damaged DNA was prepared by incubating supercoiled plasmid (300 µM DNA phosphate) with cis-DDP at 37 °C for 12–18 h in the dark. DNA was purified by ethanol precipitation. MNNG was incubated with DNA (300 µM DNA phosphate) for 24 h at 37 °C in MNNG treatment buffer. DNA was purified by ethanol precipitation.
3.4.4. Binding Reaction
- Assemble the reaction mixture as follows:
(a) 5× binding buffer 2 µL (b) UV-32P-f148 probe 0.2 or 0.02 ng (c) BSA (10 mg/mL) 0.3 µL (d) Plasmid DNA (1 mg/mL) 2 µL (e) Poly(dI-dC)/salmon sperm DNA (1.0/0.5 mg/mL) 0.5–2 µL (f) Distilled water to 8 µL (g) Whole cell or nuclear extract (0.1–0.5 mg/mL) 2 µL Add whole cell or nuclear extract last.
Incubate the reaction mixture at room temperature for 30 min.
Add 2 µL of loading dye to the reaction mixture and gently mix together.
Resolve the protein–DNA complexes by non-denaturing gel electrophoresis at 10 V/cm at room temperature.
Dry the gel on Whatman 3 MM paper and expose to X-ray film at −80 °C (Fig. 2a).
3.5. Variation on EMSA: Reverse Electrophoretic Mobility Shift Assay
Specific binding of UV-DDB to UV-damaged DNA probe was also visualized by a reverse mobility shift assay with labeled UV-DDB and unlabeled UV-f148 probe (Fig. 2b).
3.5.1. In Vitro Synthesis of 35S-Labeled DDB1
UV-DDB consists of a 125 kDa subunit, DDB1, and a 48 kDa subunit, DDB2. Labeled DDB1 protein was synthesized by transcribing the DDB1 cDNA from a T7 promoter with T7 RNA polymerase and translating the mRNA in a rabbit reticulocyte lysate in the presence of (35S) methionine.
- Assemble the in vitro transcription/translation reaction mixture as follows:
(a) pCDNA3 (DDB1) plasmid (1 mg/mL) 1 µL (b) Rabbit reticulocyte lysate 25 µL (c) (35S) methionine 4 µL (d) T7 RNA polymerase buffer 2 µL (e) T7 RNA polymerase 1 µL (f) Amino acid mixture minus methionine 1 µL (g) RNasin ribonuclease inhibitor 1 µL (h) RNase-free water to 50 µL Incubate at 30 °C for 120 min.
Store the in vitro translation products at −80 °C.
3.5.2. Purification of Active 35S-Labeled DDB1 Protein
Not all of the DDB1 protein synthesized in vitro is active in binding UV-damaged DNA. Thus, the active fraction was purified through UV-DNA cellulose affinity chromatography.
Pack 10 µL of UV-DNA cellulose resin in a 200 µL Pipetman tip and equilibrate the column with Buffer D.
Load the in vitro translated 35S-labeled DDB1 proteins onto the column (50 µL of the reticulocyte lysate).
Wash the column extensively with 600 µL of Buffer D containing 200 mM NaCl.
Elute the bound 35S-labeled DDB1 protein with Buffer D containing 1 M NaCl.
Store the fractions at −80 °C.
3.5.3. Reverse Electrophoretic Mobility Shift Assay
- Assemble the reaction mixture as follows:
(a) 5× binding buffer 2 µL (b) UV-f148 probe 1 ng (c) BSA (10 mg/mL) 0.3 µL (d) Plasmid DNA (1 mg/mL) 2 µL (e) Poly (dI-dC)/salmon sperm DNA (1/0.5 mg/mL) 0.5 µL (f) Distilled water to 8 µL (g) 35S-labeled DDB1 2 µL Incubate the reaction at room temperature for 30 min.
Add 2 µL of loading dye to the reaction mixture and gently mix together.
Resolve the protein–DNA complex by non-denaturing gel electrophoresis at 10 V/cm at room temperature.
Fix the gel in 50 % methanol and 10 % acetic acid for 2 h.
Soak the gel in the Amersham Amplify™ Fluorographic Reagent for 30 min.
Dry the gel on Whatman 3 MM paper.
Treat the sticky surface of the dried gel with baby powder, which allows direct contact of the gel with the X-ray film.
Expose the gel to X-ray film at −80 °C.
3.6. EMSA for Studying Ku
3.6.1. Probe DNA
Prepare the f148 probe as described in Subheading 3.1.1 and radiolabel the probe as described in Subheading 3.1.4 (see Notes 14 and 15).
3.6.2. Competitor DNA Against Nonspecific Binding
Crude nuclear extracts have proteins that may bind nonspecifically to the labeled f148 probe without binding to the DNA ends. To minimize the effects of these proteins on the EMSA, supercoiled plasmid is included in the binding reaction. Because the super-coiled pRSVcat plasmid contains the f148 fragment but has no free ends, it is a good nonspecific competitor (see Note 16). In general, human cell extracts require 2 µg of nonspecific competitor in a 10 µL reaction. Rodent cell extracts, however, require only 50–100 ng of nonspecific competitor.
3.6.3. Competitor DNA to Test Binding to Different Types of DNA Ends
Competition assays are performed using single-stranded linear DNA (poly(dA) or poly(dT)), double-stranded linear DNA (unlabelled f148), single-stranded circular DNA (phage M13), or supercoiled double-stranded DNA (pRSVcat). These competitor DNAs are included in the binding reaction in concentrations ranging from 0.2 to 200 ng. Competitors containing free DNA ends (unlabelled f148) or DNA containing single-stranded to double-stranded DNA transitions (phage M13) compete for Ku binding activity.
3.6.4. Binding Reaction
- Prepare the reaction mixture as follows:
(a) 5× binding buffer 2 µL (b) f148 probe 0.2 ng (c) Supercoiled plasmid 2 µg (human extracts) or 50–100 ng (rodent extracts) (d) Distilled water to 8 µL (e) Nuclear extract 0.5 µg Incubate the reaction for 5 min at room temperature.
Add 2 µL of loading dye to the reaction and mix by gentle pipetting.
Resolve the protein–DNA complexes by non-denaturing gel electrophoresis at 10 V/cm at room temperature.
Dry the gel on 3 MM Whatman paper and expose to X-ray film at −80 °C.
3.6.5. Antibody Supershift
Anti-Ku antibodies may be added to the binding reaction to supershift the Ku/DNA complex. Serial dilutions of the antibodies are made in 1 % BSA and added to the reaction mix above, prior to adding the protein extract (Fig. 3).
3.7. EMSA for Studying Ku and DNA–PKcs Bound to DNA
3.7.1. Probe DNA
Prepare the f32 probe as described in Subheading 3.1.2 and radiolabel the probe as described in Subheading 3.1.4.
3.7.2. Competitor DNA
We have found that shortening the length of the DNA probe allows a complex containing DNA–PKcs to form on DNA ends in the absence of competitor DNA (16) (see Note 17). Additionally, certain competitor DNAs, including poly dA and supercoiled plasmid, generate a supershift of the DNA–PKcs-containing complex. This may be due to the ability of DNA–PKcs to synapse two DNA molecules (27).
3.7.3. Binding Reaction (See Notes 18 and 19)
- Prepare the reaction mixture as follows:
(a) 5× binding buffer 2 µL (b) f32 probe 0.2 ng (c) Distilled water to 8 µL (d) Nuclear extract (human) 0.5 µg (rodent) 2.0 µg (e) NaCl to a fi nal concentration of 200 mM (human extracts) 400 mM (rodent extracts) Incubate the reaction for 5 min at room temperature.
Add 2 µL of loading dye to the reaction and mix by gentle pipetting.
Resolve the protein–DNA complexes by non-denaturing gel electrophoresis at 10 V/cm at room temperature.
Dry the gel on 3 MM Whatman paper and expose to X-ray film at −80 °C.
3.7.4. Antibody Supershift
Anti-DNA–PKcs antibodies may be added to the binding reaction to supershift or disrupt the DNA–PKcs containing complexes. Serial dilutions of the antibodies are made in 1 % BSA and added to the reaction mix above, prior to adding the protein extract.
3.8. EMSA for Studying Ku and XRCC4/Ligase IV Bound to DNA
3.8.1. Probe DNA
Prepare the f298 probe as described in Subheading 3.1.3 and radiolabel the probe as described in Subheading 3.1.6.
3.8.2. Binding Reaction
- Prepare the reaction mixture as follows:
(a) 2× binding buffer 5 µL (b) 33P-labeled f298 probe 20 fmole (c) Recombinant Ku 10 fmole (d) Recombinant XRCC4/Ligase IV 40 fmole (e) Nuclease-free water to 10 µL Incubate the reaction at 37 °C for 30 min.
Pre-run the gel for 1.5 h in 0.5× TBE buffer at 60 V.
Load at least one well with 0.01 % bromophenol blue in 1× binding buffer to provide a tracking dye. No dyes are included in the binding reactions.
Load binding reactions and run the 5 % polyacrylamide gel in fresh and chilled 0.5× TBE buffer at 60 V for 3 h.
Remove the gel from the cassette and attach it to 3MM Whatman paper. Cover the gel with Saran wrap. To dry the gel, apply vacuum with heat for 30 min, and then turn off heat to let cool for 30 min.
Expose the gel to a storage phosphor screen. Image with Typhoon scanner.
Footnotes
The high-salt buffers TGE and TBE were superior to low-salt buffers in these EMSAs because the high-salt buffers reduced nonspecific binding more efficiently.
When labeling the DNA probe with Klenow fragment, the radiolabeled nucleotide used to fill in the 5′ overhang of the DNA should be complementary to an internal base of the 5′ overhang. Inefficient labeling may occur if the radiolabeled nucleotide is complementary to the last base of the 5′ overhang because of 3′ to 5′ exonuclease activity in the Klenow fragment.
The exonuclease III/Klenow method increases the specific activity of the probe (Subheading 3.1.5). This can be helpful in detecting UV-DDB in rodent cells, where levels are substantially lower than in human cells.
We have used both 32P and 33P to prepare radiolabeled DNA probes. Choice of 32P provides greater sensitivity, while choice of 33P provides greater resolution of the bands in the EMSA. For the analysis of very large protein–DNA complexes, which migrate only small distances during gel electrophoresis, we prefer to use 33P-labeled DNA probes.
Extracts may be stored at 4 °C for 1–3 days, but should be stored at −80 °C for longer periods of time. Extracts should be stored in aliquots to minimize repeated freeze–thaw steps. Thawing of the extracts should be done on ice to minimize protease activity, which may occur at higher temperatures.
The extraction procedure should be optimized by testing different salt concentrations in buffer C or changing the NP-40 concentration.
The presence of NP-40 in the extraction buffer (buffer C) can inhibit extraction of some proteins. Washing the nuclei following the treatment with buffer A containing NP-40 removes the NP-40 and overcomes this effect (Subheading 3.2.2, step 8). Additionally, larger proteins may be cleaved by proteases to a greater extent than smaller proteins, making it critical that protease inhibitors be present at all steps in the extraction procedure following the lysis in buffer A.
Lowering the acrylamide concentration and decreasing the length of the DNA probe can facilitate the detection of larger protein–DNA complexes by permitting the migration of these complexes into the gel. Examples include the complexes of Ku-DNA–PKcs (Fig. 4) and Ku-XRCC4/Ligase IV (Fig. 5) that assemble on DNA ends. In such cases, we have tested a range of acrylamide concentrations and acrylamide to bisacrylamide ratios in order to find conditions that best stabilize the protein–DNA complex, while permitting sufficient migration of the complex into the gel. Commercially prepared gels may vary from lot to lot. To ensure reproducibility, we recommend casting one’s own gels.
Conduction leaks in the gel can lead to aberrant migration of the free probe and protein–DNA complexes. Cleaning areas around the gel and gel box where salt buildup can occur ameliorates the problem.
In detecting UV-DDB, increasing the UV doses to the probe DNA will increase the number of lesions per DNA molecule. Higher-order complexes corresponding to multiple UV-DDB binding events can be visualized at these higher doses (7).
When cyclobutane pyrimidine dimers were removed from a UV-damaged DNA probe by treatment with purified photolyase, the binding was decreased compared to the untreated UV-damaged DNA probe. This suggests that UV-DDB recognizes at least some cyclobutane dimers (7).
Because these EMSAs rely on the structure-specific binding properties of the DNA repair proteins, it is important that the competitor DNA be free of contaminating structures that might compete for binding activity.
UV-DDB recognizes DNA damaged by several agents including UV irradiation, but direct binding was observed only to the UV-damaged double-stranded DNA probe. The binding of UV-DDB to apurinic sites, cisplatin adducts, nitrogen mustard adducts, and single-stranded DNA was detected by showing that these forms of damaged DNA could compete with UV-damaged DNA for binding to UV-DDB (7, 26). UV-DDB may have low affinities for these forms of DNA, so that the competition assay may be more effective than the direct binding assay in detecting binding activity.
Increasing the length of the radiolabeled DNA probe can be utilized to detect multiple Ku-binding events. This is possible because Ku is able to bind DNA and translocate along the molecule. Thus, longer DNA lengths permit more Ku molecules to bind and produce a “ladder” pattern in the EMSA (12).
Decreasing the length of the DNA probe can allow a decrease in the amount of competitor DNA used in the EMSA for Ku.
The competitor used in EMSA for Ku should be supercoiled DNA that is free of contaminating nicked or linear DNA. Ku binds strongly to free DNA ends and has been reported to bind nicks, so competitors with these structures will obscure Ku-binding activity.
There should not be competitor DNA in EMSAs to detect DNA–PKcs. Additionally, the probe length should be short. We have found 32 base pairs to be optimal in EMSAs to detect DNA–PKcs.
DNA–PKcs levels are lower in rodent extracts than human extracts, thus the concentration of extract used in an EMSA should be increased from 0.5 µg (human extract) to 2.0 µg (rodent extract).
Since DNA–PKcs EMSAs do not utilize competitor DNA, we have tested the effect of different salt concentrations on nonspecific probe binding and DNA–PKcs complex formation. DNA–PKcs complexes can be detected with NaCl concentrations as high as 500 mM, with higher salt decreasing nonspecific binding. For rodent extracts, which use more protein extracts, we recommend increasing the NaCl concentration to 400 mM.
References
- 1.Fried M, Crothers DM. Equilibrium and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garner MM, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to speci fi c DNA regions: application to the components of the E. coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3059. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang J, Chu G. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair. 2002;1:601–616. doi: 10.1016/s1568-7864(02)00052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci USA. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu G, Chang E. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science. 1988;242:564–567. doi: 10.1126/science.3175673. [DOI] [PubMed] [Google Scholar]
- 6.Singh H, et al. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 7.Hwang BJ, Chu G. Purification and characterization of a protein that binds to damaged DNA. Biochemistry. 1993;32:1657–1666. doi: 10.1021/bi00057a033. [DOI] [PubMed] [Google Scholar]
- 8.Hwang BJ, Liao J, Chu G. Isolation of a cDNA encoding a UV-damaged DNA binding factor defective in xeroderma pigmentosum group E cells. Mutat Res. 1996;362:105–117. doi: 10.1016/0921-8777(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 9.Chu G, Chang E. Cisplatin-resistant cells express increased levels of a factor that recognizes damaged DNA. Proc Natl Acad Sci USA. 1990;87:3324–3327. doi: 10.1073/pnas.87.9.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keeney S, et al. Correction of the DNA repair defect in xeroderma pigmentosum group E by injection of a DNA damage-binding protein. Proc Natl Acad Sci USA. 1994;91:4053–4056. doi: 10.1073/pnas.91.9.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang J, et al. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rathmell WK, Chu G. A DNA end-binding factor involved in double-strand break repair and V(D)J recombination. Mol Cell Biol. 1994;14:4741–4748. doi: 10.1128/mcb.14.7.4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rathmell WK, Chu G. Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc Natl Acad Sci USA. 1994;91:7623–7627. doi: 10.1073/pnas.91.16.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smider V, et al. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 15.Taccioli GE, et al. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 16.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nick McElhinny SA, et al. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson M, Chu G. Evidence that xeroderma pigmentosum cells from complementation group E are deficient in a homolog of yeast photolyase. Mol Cell Biol. 1989;9:5105–5112. doi: 10.1128/mcb.9.11.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox M, Feldman B, Chu G. A novel role for DNA photolyase: binding to drug-induced DNA damage is associated with enhanced cytotoxicity in yeast. Mol Cell Biol. 1994;14:8071–8077. doi: 10.1128/mcb.14.12.8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiricny J. Colon cancer and DNA repair: have mismatches met their match? Trends Genet. 1994;10:164–168. doi: 10.1016/0168-9525(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 21.Donahue BA, et al. Characterization of a DNA damage-recognition protein from mammalian cells that binds specifically to intrastrand d(GpG) and d(ApG) DNA adducts of the anticancer drug cisplatin. Biochemistry. 1990;29:5872–5880. doi: 10.1021/bi00476a032. [DOI] [PubMed] [Google Scholar]
- 22.Toney J, et al. Isolation of cDNAs encoding a human protein that binds selectively to DNA modified by the anticancer drug cisdiamminedichloroplatinum(II) Proc Natl Acad Sci USA. 1989;86:8328–8332. doi: 10.1073/pnas.86.21.8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown SJ, Kellet PJ, Lippard SJ. Ixr1, a yeast protein that binds to platinated DNA and confers sensitivity to cisplatin. Science. 1993;261:603–605. doi: 10.1126/science.8342024. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 26.Payne A, Chu G. Xeroderma pigmentosum group E binding factor recognizes a broad spectrum of DNA damage. Mutat Res. 1994;310:89–102. doi: 10.1016/0027-5107(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 27.DeFazio L, et al. Synapsis of DNA ends by the DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]