Abstract
Background
The p73 protein, a paralogue of the p53 tumor suppressor, is essential for normal development and survival of neurons. TP73 is therefore of interest as a candidate gene for Alzheimer's disease (AD) susceptibility. TP73 mRNA is transcribed from three promoters, termed P1 – P3, and there is evidence for an additional complexity in its regulation, namely, a variable allelic expression bias in some human tissues.
Methods
We utilized RT-PCR/RFLP and direct cDNA sequencing to measure allele-specific expression of TP73 mRNA, SNP genotyping to assess genetic associations with AD, and promoter-reporter assays to assess allele-specific TP73 promoter activity.
Results
Using a coding-neutral BanI polymorphism in TP73 exon 5 as an allelic marker, we found a pronounced allelic expression bias in one adult brain hippocampus, while 3 other brains (two adult; one fetal) showed approximately equal expression from both alleles. In a tri-ethnic elderly population of African-Americans, Caribbean Hispanics and Caucasians, a G/A single nucleotide polymorphism (SNP) at -386 in the TP73 P3 promoter was weakly but significantly associated with AD (crude O.R. for AD given any -386G allele 1.7; C.I. 1.2–2.5; after adjusting for age and education O.R. 1.5; C.I. 1.1–2.3, N= 1191). The frequency of the -386G allele varied by ethnicity and was highest in African-Americans and lowest in Caucasians. No significant differences in basal P3 promoter activity were detected comparing -386G vs. -386A promoter-luciferase constructs in human SK-NSH-N neuroblastoma cells.
Conclusions
There is a reproducible allelic expression bias in mRNA expression from the TP73 gene in some, though not all, adult human brains, and inter-individual variation in regulatory sequences of the TP73 locus may affect susceptibility to AD. However, additional studies will be necessary to exclude genetic admixture as an alternative explanation for the observed associations.
Background
Genes functioning in neurons could modulate AD susceptibility either by acting during development to determine the number of cortical neurons at birth, thereby determining "cognitive reserve", or by acting during adult life to determine the cell death threshold of neurons when faced by external insults, such as amyloid-beta peptide. In principle, the protein encoded by TP73 could play a role in both processes. This protein is related to p53, which triggers apoptotic cell death as an effector mechanism in tumor surveillance. p73 knockout (KO) mice are born with disorganized hippocampal cyto-architecture, and they show degeneration of sympathetic neurons [1,2]. Moreover, these mice, which survive to adulthood, showed progressive cerebral atrophy [3].
Gene regulation at the TP73 locus is complex. The mRNA transcripts are initiated from two alternative promoters, leading to the production of two different protein isoforms. "Full-length" p73 mRNA, initiating from the P1 promoter, encodes a protein with an N-terminal transcriptional activation domain, while the shorter "delta-N" (ΔNp73) isoform, initiating from the major downstream P3 promoter, lacks this domain. The full-length p73 protein is pro-apoptotic, while the ΔNp73 acts as a natural dominant-negative, and protects cells from apoptosis [4]. The fact that the ΔNp73 isoform is preferentially expressed in neurons explains the net anti-apoptotic function of p73 in the peripheral nervous system, and probably in cortical neurons of the brain.
Some evidence suggests that there may be a second kind of complexity at the p73 locus – allelic expression bias. Monoallelic expression of TP73 was reported in neuroblastoma cells in the first publication describing this gene [5]. Monoallelic gene expression affects loci that are subject to parental imprinting, and it was initially suggested that TP73 might be imprinted. The significance of these observations were called into question by the finding of lack of imprinting of this gene in studies of the p73 KO mice [1], and the finding of biallelic expression of TP73 in some normal human tissues. But biased allelic expression can have other explanations in addition to imprinting, and a number of subsequent studies have continued to report monoallelic or biased allelic representation of TP73 mRNA, albeit in a tissue-specific pattern, and with inter-individual variability [5-10]. Such variation suggests that there may be functional genetic polymorphisms in regulatory sequence (promoters/enhancers) of this gene. Here we assess the allele frequencies of two TP73 SNPs in a case-control analysis of AD in a tri-ethnic elderly population sample, and we test for an allelic expression bias at the TP73 locus in human brains.
Methods
Participants and diagnosis
Selection of participants and diagnosis was previously described [11], and key aspects are repeated here. Subjects were individuals over the age of 65 residing in the Washington Heights-Inwood neighborhood of Manhattan. For those who agreed to participate, an in-person interview and a standardized assessment including a medical history, physical and neurological examination and neuropsychological battery [12] was completed. Individuals who qualified for initial inclusion in the community study (n = 1401) all had at least one subsequent follow-up evaluation. 282 (20.1%) were non-Hispanic Whites, 462 (33%) were African-American, 646 (46.1%) were Caribbean Hispanic and 11 (0.8%) were from other ethnic groups. We excluded individuals with other forms of dementia or Parkinson's disease. DNA for genotyping was available from most of these individuals. For AD cases, the diagnosis was established at a consensus conference of physicians and neuropsychologists and required evidence of cognitive deficit on the neuropsychological battery as well as evidence of impairment in social or occupational function. A Clinical Dementia Rating (CDR) score of 1.0 was the cut-off for the diagnosis of AD. These studies were approved by the Columbia University Institutional Review Board.
DNA and RNA isolation
Peripheral blood was obtained with informed consent. DNA for genotyping was isolated from blood using the FlexiGene™ kit (Qiagen). To prepare DNA and RNA from frozen brain the tissue was first pulverized under liquid nitrogen. DNA was then isolated by standard SDS/proteinase-K lysis and phenol/chloroform extraction; RNA was isolated using Trizol™ reagent (Sigma) according to the protocol provided by the manufacturer.
TP73 SNP genotyping and mutational screening
SNP locations, PCR primers and conditions, and diagnostic restriction enzymes for genotyping of TP73 polymorphisms are in Table 1. Mutational analysis of the P3 promoter was done from 12 individuals using the same 748 bp PCR product that was used for genotyping. For mutation analysis of the DNA-binding domain of p73, genomic PCR products were amplified corresponding to exons 4 through 8 (primer sequences available on request) and subjected to direct DNA sequencing.
Table 1.
PCR conditions and primers for analyzing TP73 SNPs
| Analysis | SNP Location | PCR Primers (Forward; Reverse) | Anneal. Temp. | Size (bp) | Restriction Enzyme |
| Promoter construct | P3 promoter | (F) GCACCACTGCAGAGCCCCTCCC (R) AACAACAAAACCCGCGGCCCACC | 60°C | 675 | N/A |
| Genotyping | P3 promoter | (F) AGAGCCCCTCCCTGGCTGTGCC (R) CGAGGATGCTGGGGAACATGC | 65°C | 748 | SpeI |
| Genotyping | Exon 5 | (F) ACAGGGGTGCAGTTGGGAC (R) CTGGGAAAATGGGGGCTGGTG | 60°C | 403 | BanI |
| Allelic Expression | Exon 4–6 RT-PCR | (F) CTTTCCAGCAGTCCAGCACGG (R) CGTGGTTGGGGCAGCGTTTCA | 60°C | 203 | BanI |
| Control for Allelic Expression | Exon 5 Genomic PCR | (F) TTGGGACCACTGGTCTCA (R) GTGGTTGGGGCAGCGTTTCA | 60°C | 235 | BanI |
Analysis of TP73 allele-specific mRNA expression
Allele-specific expression of TP73 mRNA in human brains was analyzed by reverse transcription-PCR (RT-PCR) using primers located in exons 4 and 6; while genomic PCR with primers flanking exon 5 served as a control (Table 1). First strand synthesis was with SuperScript (Invitrogen), using oligo dT primer. Typically, 2 μg of total RNA was used in a 20 ul reaction. After digestion with RNaseH, 1 ul of the RT product was used for PCR. Non-radioactive PCR products (40 cycles) were directly sequenced, and independent radiolabeling PCR reactions, done for a total of 35 cycles in the presence of 32PdCTP, were digested with BanI and the fragments resolved on 8% non-denaturing polyacrylamide gels.
TP73 P3 promoter-reporter plasmids
The P3 promoter from -675 to +1 relative to the transcriptional start of exon 3bis (see Fig. 1) was amplified by genomic PCR from a -386G/A heterozygote using high-fidelity Taq polymerase (Roche) and the primers in Table 1 (promoter construct), cloned in the TA vector (Invitrogen), excised with KpnI and BglII restriction enzymes, and the fragment ligated into the pGL3 vector (Promega) upstream of the luciferase reporter gene. SK-NSH-N neuroblastoma cells, maintained in DMEM with 10% fetal calf serum, were transfected in triplicate with 0.5 micrograms of the promoter-LUC plasmids, plus 0.5 micrograms of the Renilla luciferase control plasmid. Luciferase assays were done 30 hours post-transfection (Dual Luciferase Assay System, Promega).
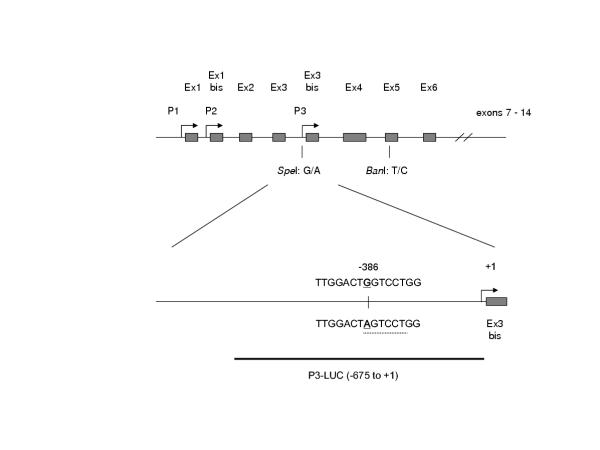
Figure 1.
Map of the 5' TP73 exons showing the SNPs utilized in this study. The arrows represent promoter sequences and the rectangles exons. The region of DNA containing the -386G/A SNP is magnified in the lower panel. The SNP is underlined and the dotted line indicates the variant progesterone receptor binding site (AGTCCT, consensus sequence A/TGTYCT) that overlaps the SNP and becomes non-consensus in the G-allele.
APOE genotyping
Subjects were genotyped for the APOE coding polymorphisms, using genomic PCR followed by CfoI digestion of the PCR products and acrylamide gel electrophoresis, as previously described [13,14].
Statistical analysis
Allele frequencies were determined by counting each allele and by calculating sample proportions. For comparison of cases and controls within and across ethnic groups, allele frequencies were calculated for all subjects and compared by the chi square analysis. Logistic regression was used to compute the odds ratio (O.R.) for the association between AD and the TP73 polymorphisms. Data were stratified by the presence or absence of an APOE ε4 allele and by adjusting for differences in age and education. Logistic regression analyses were conducted separately for each ethnic group. We tested for Hardy-Weinberg equilibrium using a chi-square analysis. Multivariate logistic regression was used to compute the O.R. for the association between AD and TP73 promoter polymorphisms, adjusting for age, sex, and education.
Results
The locations of the TP73 SNPs analyzed in this study are diagrammed in Figure 1. We first genotyped all AD cases and controls for the known BanI polymorphism in exon 5 of TP73. We observed ethnic differences in allele frequencies between Caucasians, Caribbean Hispanics and African-Americans in our population sample, with the highest frequency of the BanI+ allele in African-Americans, but using stringent diagnostic criteria we did not find a consistent association with AD (Table 2). After segregating the subjects by APOE genotypes, we observed a weak but significant association of the BanI +/- genotype with AD (reference genotype BanI -/-), but no allele dosage effect was observed, i.e. the BanI+/+ genotype was not positively correlated with AD (Table 3).
Table 2.
Genotype frequencies of the TP73 exon 5 (BanI) polymorphism in AD cases and controls
| Genotype | AD Cases | Controls | Unadjusted Odds Ratio | Adjusted for age and education |
| BanI -/- | 200 (62.6%) | 380 (68.1%) | 1.0 | 1.0 |
| -/+ | 113 (33.8%) | 154 (27.6%) | 1.4; 0.9–1.8 | 1.4; 0.9–1.9 |
| +/+ | 12 (3.6%) | 24 (3.6%) | 0.9; 0.4–1.9 | 0.7 0.3–1.4 |
| N | 334 | 558 |
Table 3.
Odds ratios for AD as a function of the TP73 exon 5 (BanI) polymorphism stratified by the presence or absence of APOE ε4 alleles
| Genotype | APOE 4/4 | APOE -/4 | APOE -/- |
| BanI -/- | 1.0 | 1.0 | 1.0 |
| -/+ | 2.5; 0.2–29.2 | 0.9; 0.5–1.6 | 1.5; 1.1–2.2* |
| +/+ | - | 0.5; 0.2–1.8 | 1.4; 0.5–3.6 |
* p = 0.02
We therefore searched for additional sequence variants in TP73. As a screening panel, we chose genomic DNA from 12 people (24 chromosomes), including several individuals from each of the three ethnic groups. This analysis would not be expected to detect rare variants, but would detect common alleles. Sequencing of the exons encoding the DNA binding domain of p73 did not reveal any coding variants, although the BanI SNP, which is coding-neutral, was observed with a high frequency and served as a positive control. However, we did find a common SNP in the P3 promoter of TP73. This variant was a G→A transversion at position minus-386 relative to the transcriptional start site of the ΔN-p73 isoform. A transcription factor binding site database search [15]http://www.cbil.upenn.edu/tess/ showed that this SNP converted a variant consensus progesterone receptor binding site overlapping the -386A allele to a non-consensus sequence in the -386G allele (Fig. 1). This SNP also overlapped with a p53 consensus binding half-site, but no second nearby half-site was found, suggesting that p53 binding to the P3 promoter [16] is not likely to be affected by the -386G/A SNP.
To ask whether the -386G/A SNP might be associated with AD risk, we genotyped the series of cases and controls. These individuals were also genotyped for coding variants at the APOE locus, which are known modifiers of AD risk [17,18]. The results are shown in Table 4. The -386G/A allelic system was in Hardy-Weinberg equilibrium in the controls, and the -386G allele was significantly associated with AD in the group as a whole, both when the data were analyzed as unadjusted odds ratios, and after multivariate logistic regression analysis correcting for age and education.
Table 4.
Genotype frequencies of the -386G/A P3 promoter polymorphism in AD cases and controls
| Genotype | AD Cases | Controls | Unadjusted Odds Ratio | Adjusted for age and education |
| AA | 122 (30%) | 288 (37%) | 1.0 | 1.0 |
| AG | 196 (48%) | 380 (48%) | 1.2; 0.9–1.6 | 1.0; 0.7–1.3 |
| GG | 87 (22%) | 118 (15%) | 1.7; 1.2–2.5+ | 1.5; 1.1–2.3* |
| N | 405 | 786 |
+ p < 0.002 * p < 0.01
After stratifying by APOE genotype, the AD association of the -386G TP73 allele was seen in the groups with one or zero APOE ε4 alleles, but was not seen in the group of APOE ε4/ε4 homozygotes (Table 5). Since homozygosity for APOE ε4 in itself conveys a high risk of AD, this result is not surprising.
Table 5.
Odds ratios for AD as a function of the -386G/A P3 promoter polymorphism stratified by the presence or absence of APOE ε4 alleles
| -386G/A genotype | APOE 4/4 | APOE -/4 | APOE -/- |
| AA | 1.0 | 1.0 | 1.0 |
| AG | 0.2; 0.02–2.6 | 1.2; 0.6–1.9 | 1.2; 0.9–1.7 |
| GG | 0.8; 0.04–16.9 | 2.3; 1.2–4.8* | 1.6; 1.0–2.4* |
* p = 0.02 The APOE ε2 and ε3 alleles are designated as (-)
We next asked whether allele frequencies of the -386G/A SNP might differ by ethnicity. As shown in Table 6, there was indeed a difference among the three ethnic groups, with the highest frequency of the -386G allele in African-Americans and the lowest frequency in European Caucasians. Consistent with the known history of ethnic admixture in the Caribbean, Hispanics showed an intermediate allele frequency. When we reexamined the AD associations stratifying by ethnicity, we found a significant association of homozygosity for the -386G allele with AD in the Hispanics, and a similar direction of the effect, but failure to achieve significance, in the smaller number of African-Americans (Table 7). There were too few Caucasians with the -386G allele to allow conclusions.
Table 6.
Genotype frequencies of the -386G/A SNP in three ethnic groups
| -386G/A genotype | Caucasian | African-American | Hispanic |
| AA | 103(51%) | 96 (26%) | 206 (34%) |
| AG | 88(43%) | 179 (49%) | 301(50%) |
| GG | 13 (6%) | 92 (25%) | 99(16%) |
| N* | 204 | 367 | 606 |
The difference across ethnic groups is significant X2 = 52.4, p < 0.0001 * 14 subjects are excluded because of ethnicity reported as "other".
Table 7.
Odds ratios for AD as a function of the -386G/A P3 promoter polymorphism stratified by ethnicity
| -386G/A genotype | African-American | Hispanic | White |
| AA | 1.0 | 1.0 | 1.0 |
| AG | 1.1; 0.6–1.9 | 1.1; 0.8–3.1 | 1.6; 0.7–3.5 |
| GG | 1.6; 0.9–3.1 | 1.7; 1.1–2.9* | 0.6; 0.1–4.8 |
*p < 0.05
Although multiple interpretations of these results are possible, one hypothesis is that the -386G/A SNP, or closely linked polymorphisms, might influence AD risk via a functional effect on ΔNp73 expression in the brain. Small variations in the net expression of the ΔNp73 mRNA isoform in human brains would be hard to quantify, but examining allele-specific expression, in which one allele serves as an internal control for the other, is expected to be more sensitive. We therefore screened a series of 34 brains for heterozygosity at the TP73 exon 5 BanI polymorphism, which would allow mRNA from the two alleles to be distinguished by RFLP and direct sequencing. Since the collection of autopsy brains available to us was primarily, though not exclusively, from Caucasians, the allele frequency at this marker was low. Nonetheless, we found 4 brains, 1 fetal mid-gestation and 3 adult, which were heterozygous. Two of the adult brains were affected by AD and one was normal.
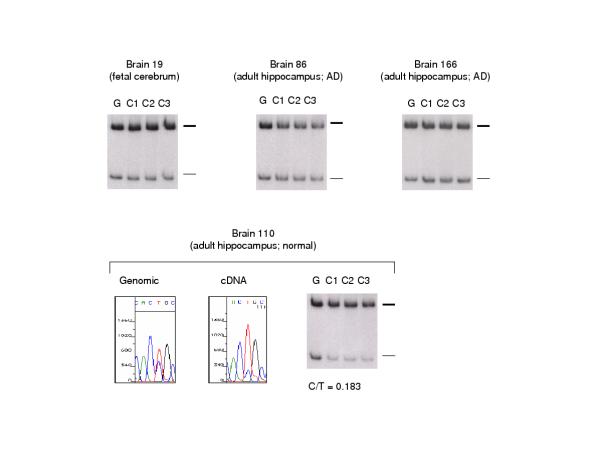
Among these 4 brains, 3 manifested approximately equal biallelic mRNA expression of TP73 mRNA, as indicated by comparison with genomic PCR controls, while one adult brain (the normal brain) showed a strong allelic expression bias (Figure 2). This bias was reproducible in multiple independent RT-PCR reactions (Fig. 2), and was also reproduced starting from two separate RNA preparations from this same brain. We utilized RFLP analysis since this method gives a simple read-out of the two alleles. Heteroduplex formation can cause artifacts in this method when the cDNA is over-amplified, so we kept the PCR cycle number to a minimum (35 cycles). Genomic controls were run in parallel using the same PCR conditions, and these gave highly reproducible RFLP results, which were taken as the standard for biallelic representation. Lastly, the results of direct sequencing, which is not sensitive to heteroduplex formation, were consistent with the RFLP results (Fig. 2). From this analysis, we conclude that some human brains manifest an allelic bias for TP73 mRNA expression. Lastly, we established the genotype at the -386G/A P3 SNP in the 4 brains (Table 8). The adult brain with strongly biased expression was indeed heterozygous for this marker, but heterozygosity was also found in the fetal brain, which showed approximately equal biallelic mRNA expression.
Figure 2.
Allele-specific TP73 mRNA expression in human brains. In the BanI RFLP autoradiograms the T-allele (uncut) is indicated by the thick marker line and the C-allele (cut) by the thin marker. Additional smaller fragments ran at a lower position and are not shown. The genomic PCR (G) is a control for equal biallelic representation. The RT-PCR in each case was repeated three times from independent aliquots of cDNA (C). Direct sequencing confirms the pronounced allelic expression bias (stronger upper band) in adult brain 110, with the genomic sequence chromatogram serving as a control for biallelic representation. Less pronounced but reproducible biases are seen among the other brains (weaker upper band in RT-PCR products from brain 86).
Table 8.
Genotypes of the four informative brains examined for TP73 allele-specific mRNA expression
| Brain ID | Region | Age | Status | P3 -386 (SpeI) | Exon5 (BanI) | p73 allelic expression |
| 19 | cerebrum | 18 wk | normal | AG | TC | Biallelic |
| 110 | hippocampus | 62 yrs | normal | AG | TC | Strong allelic bias |
| 86 | hippocampus | 83 yrs | AD | AA | TC | Biallelic |
| 166 | hippocampus | 89 yrs | AD | AA | TC | Biallelic |
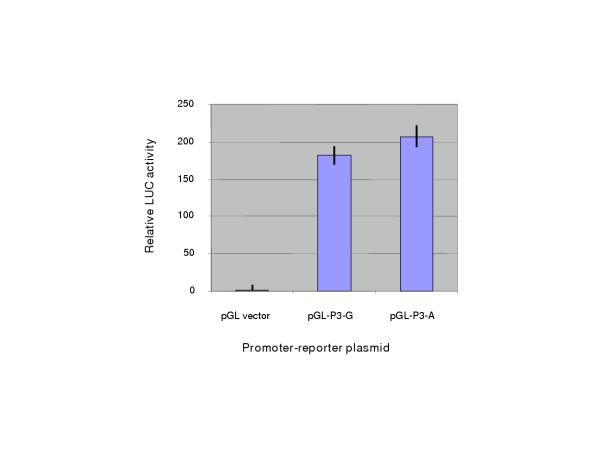
Lastly, we tested for major effects of the -386G/A SNP on P3 promoter activity by transfecting promoter-luciferase reporter plasmids into human SK-NSH-N neuroblastoma cells. No significant difference in basal promoter activity was seen in this assay (Fig. 3).
Figure 3.
Activity of allelic variants of the TP73 P3 promoter-luciferase constructs in SK-NSH-N neuroblastoma cells. Values are mean (bars) and standard deviation (vertical lines) of triplicate determinations of firefly luciferase activity, normalized to Renilla luciferase. The promoter confers a strong increase in luciferase expression compared to the vector control, but there is no significant difference between promoter activity of the -386G and -386A alleles in this assay.
Discussion
TP73 was originally identified in a common region of DNA deletion on chromosome 1p36 in neuroblastomas, suggesting that p73 could be a tumor suppressor. But further studies, notably a knockout mouse model, demonstrated that p73 is in fact required for the development, maintenance and survival of neurons. ΔNp73 is a dominant isoform in brain and it has anti-apoptotic function in neurons. This background motivated us to examine the possibility of an association between TP73 variants and the risk of AD. In the current study, a nucleotide polymorphism, -386G/A, in the P3 promoter of TP73 (the promoter that drives expression of the ΔNp73 isoform) showed a weak but statistically significant association with AD. Moreover, we found that 1 of 4 human brains manifested a strong allelic expression bias at the TP73 locus, consistent with the presence of functional SNPs in the regulatory sequences of this gene.
Our brain expression data add to the findings of one prior study, in which 3 of 4 informative fetal brains showed virtually monoallelic expression of TP73 mRNA, with one fetal brain showing equal biallelic expression [9]. While our current results are in a broad sense consistent with those findings, our experimental approach was different in two respects, which make the two sets of data complementary. First, we examined adult brains; and second, we utilized the exon 5 BanI RFLP as a marker, while the previous study utilized a different polymorphism (StyI RFLP) in exon 2, upstream of the P3 promoter. This second difference means that we have examined TP73 mRNA derived predominantly from the P3 promoter (the major isoform in neural tissues), while the prior study examined mRNA derived exclusively from the upstream P1/P2 promoters.
Explanations for an allelic expression bias include genomic imprinting, which seems unlikely at the TP73 locus based on the negative data from mice but has not been strictly excluded in humans, or the presence of functional sequence polymorphisms in the promoters/enhancers of this gene. The -386G/A SNP itself is one candidate. The adult hippocampus that showed an allelic expression bias in our study was indeed heterozygous for this SNP, but in the fetal cerebral cortex that we examined, heterozygosity for this SNP evidently did not result in biased expression. So either the -386G/A SNP is not the critical sequence variant, or alternatively the effect of this variant is manifested in the adult hippocampus but not in whole fetal cerebral cortex. Our promoter-reporter assays in SK-NSH-N neuroblastoma cells did not indicate a strong difference in basal P3 promoter activity in comparing the -386G and -386A alleles. However, this polymorphism affects a predicted progesterone receptor binding site and progesterone-dependent transcriptional activity remains to be tested.
The interpretation of the statistical association of the -386G allele with AD risk is amenable to several interpretations. Since the -386G allele is significantly more frequent in African-Americans and Caribbean Hispanics compared to Caucasians, and since we have observed an increased rate of AD in the former two ethnic groups in our population sample [19], the association of the -386G allele with AD might reflect genetic admixture. That is, this allele may associate with AD simply because it is more frequent in the two ethnic groups that, at least in the population that we have studied, have a higher frequency of AD. That the association of the -386G allele with AD remained significant when we confined our analysis only to the Hispanics, together with the fact that the direction of the observed effect was the same when the analysis was confined to African-Americans, argues against this explanation. However, these observations do not completely exclude ethnic admixture as an explanation for our findings. More interesting, of course, is the possibility that sequence variation in or near the TP73 P3 promoter may have a direct effect on ΔNp73 expression in the brain. Haplotype analysis is in progress to further investigate this question.
Authors' Contributions
Authors QL, ESA, EY, MW and SLR carried out the genotyping, promoter-reporter assays and allelic expression analysis. Author JPV organized and supervised the brain dissections and tissue banking and made the neuropathological diagnoses. Author BT conceived the study of TP73 polymorphisms and brain expression and supervised the laboratory studies, and author RPM conceived the community-based study of AD, recruited and diagnosed the subjects, carried out the statistical analyses and obtained grant funding for this work.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgments
The authors thank Katerina Mancevska for expert assistance with brain dissections and tissue banking and Antonio Iavarone for helpful advice and neuroblastoma cells. This work was supported by grant P01 AG07232 from the N.I.H. to R.P.M. and a grant from the N.I.H. supporting the Columbia University Alzheimer's Disease Research Center.
Contributor Information
Quanyi Li, Email: ql2106@columbia.edu.
Eleni S Athan, Email: esa2@columbia.edu.
Michelle Wei, Email: mw380@columbia.edu.
Eric Yuan, Email: yy2114@columbia.edu.
Samuel L Rice, Email: Samuel.L.Rice.Jr@Dartmouth.edu.
Jean-Paul Vonsattel, Email: jgv2001@columbia.edu.
Richard P Mayeux, Email: rpm2@columbia.edu.
Benjamin Tycko, Email: bt12@columbia.edu.
References
- Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- Pozniak CD, Barnabe-Heider F, Rymar VV, Lee AF, Sadikot AF, Miller FD. p73 is required for survival and maintenance of CNS neurons. J Neurosci. 2002;22:9800–9809. doi: 10.1523/JNEUROSCI.22-22-09800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll UM, Erster S, Zaika A. p53, p63 and p73--solos, alliances and feuds among family members. Biochim Biophys Acta. 2001;1552:47–59. doi: 10.1016/S0304-419X(01)00036-1. [DOI] [PubMed] [Google Scholar]
- Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/S0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- Mai M, Qian C, Yokomizo A, Tindall DJ, Bostwick D, Polychronakos C, Smith DI, Liu W. Loss of imprinting and allele switching of p73 in renal cell carcinoma. Oncogene. 1998;17:1739–1741. doi: 10.1038/sj.onc.1202099. [DOI] [PubMed] [Google Scholar]
- Kang MJ, Park BJ, Byun DS, Park JI, Kim HJ, Park JH, Chi SG. Loss of imprinting and elevated expression of wild-type p73 in human gastric adenocarcinoma. Clin Cancer Res. 2000;6:1767–1771. [PubMed] [Google Scholar]
- Nomoto S, Haruki N, Kondo M, Konishi H, Takahashi T. Search for mutations and examination of allelic expression imbalance of the p73 gene at 1p36.33 in human lung cancers. Cancer Res. 1998;58:1380–1383. [PubMed] [Google Scholar]
- Hu JF, Ulaner GA, Oruganti H, Ivaturi RD, Balagura KA, Pham J, Vu TH, Hoffman AR. Allelic expression of the putative tumor suppressor gene p73 in human fetal tissues and tumor specimens. Biochim Biophys Acta. 2000;1491:49–56. doi: 10.1016/S0167-4781(00)00017-8. [DOI] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Athan ES, Lee JH, Arriaga A, Mayeux RP, Tycko B. Polymorphisms in the Promoter of the Human APP Gene: Functional Evaluation and Allele Frequencies in Alzheimer Disease. Arch Neurol. 2002;59:1793–1799. doi: 10.1001/archneur.59.11.1793. [DOI] [PubMed] [Google Scholar]
- Stern Y, Andrews H, Pittman J, Sano M, Tatemichi T, Lantigua R, Mayeux R. Diagnosis of dementia in a heterogeneous population. Development of a neuropsychological paradigm-based diagnosis of dementia and quantified correction for the effects of education. Arch Neurol. 1992;49:453–460. doi: 10.1001/archneur.1992.00530290035009. [DOI] [PubMed] [Google Scholar]
- Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- Maestre G, Ottman R, Stern Y, Gurland B, Chun M, Tang MX, Shelanski M, Tycko B, Mayeux R. Apolipoprotein E and Alzheimer's disease: ethnic variation in genotypic risks. Ann Neurol. 1995;37:254–259. doi: 10.1002/ana.410370217. [DOI] [PubMed] [Google Scholar]
- Schug Jonathan, Overton G. Christian. TESS: Transcription Element Search Software on the WWW. Computational Biology and Informatics Laboratory School of Medicine University of Pennsylvania. 1997;Technical Report CBIL-TR-1997-1001-v0.0 [Google Scholar]
- Waltermann A, Kartasheva NN, Dobbelstein M. Differential regulation of p63 and p73 expression. Oncogene. 2003;22:5686–5693. doi: 10.1038/sj.onc.1206859. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeux R, Stern Y, Ottman R, Tatemichi TK, Tang MX, Maestre G, Ngai C, Tycko B, Ginsberg H. The apolipoprotein epsilon 4 allele in patients with Alzheimer's disease. Ann Neurol. 1993;34:752–754. doi: 10.1002/ana.410340527. [DOI] [PubMed] [Google Scholar]
- Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, Andrews H, Feng L, Tycko B, Mayeux R. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. Jama. 1998;279:751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]