Abstract
Background and Aims
About 6 % of an estimated total of 240 000 species of angiosperms are dioecious. The main precursors of this sexual system are thought to be monoecy and gynodioecy. A previous angiosperm-wide study revealed that many dioecious species have evolved through the monoecy pathway; some case studies and a large body of theoretical research also provide evidence in support of the gynodioecy pathway. If plants have evolved through the gynodioecy pathway, gynodioecious and dioecious species should co-occur in the same genera. However, to date, no large-scale analysis has been conducted to determine the prevalence of the gynodioecy pathway in angiosperms. In this study, this gap in knowledge was addressed by performing an angiosperm-wide survey in order to test for co-occurrence as evidence of the gynodioecy pathway.
Methods
Data from different sources were compiled to obtain (to our knowledge) the largest dataset on gynodioecy available, with 275 genera that include at least one gynodioecious species. This dataset was combined with a dioecy dataset from the literature, and a study was made of how often dioecious and gynodioecious species could be found in the same genera using a contingency table framework.
Key Results
It was found that, overall, angiosperm genera with both gynodioecious and dioecious species occur more frequently than expected, in agreement with the gynodioecy pathway. Importantly, this trend holds when studying different classes separately (or sub-classes, orders and families), suggesting that the gynodioecy pathway is not restricted to a few taxa but may instead be widespread in angiosperms.
Conclusions
This work complements that previously carried out on the monoecy pathway and suggests that gynodioecy is also a common pathway in angiosperms. The results also identify angiosperm families where some (or all) dioecious species may have evolved from gynodioecious precursors. These families could be the targets of future small-scale studies on transitions to dioecy taking phylogeny explicitly into account.
Keywords: Dioecy, gynodioecy, angiosperms, reproductive systems
INTRODUCTION
The evolution from hermaphroditism to dioecy (separate sexes) is considered one of the most important evolutionary transitions in the reproductive history of angiosperms (Barrett, 2010). Although relatively rare (approx. 6 % of species; Renner and Ricklefs, 1995), dioecy is a widespread reproductive system in angiosperms (reported in approx. 38 % of families; Renner and Ricklefs, 1995). For this reason, dioecy is thought to have evolved independently many times. These transitions may have been favoured by the advantage of avoiding self-pollination and the associated inbreeding depression, by the advantage of sexual specialization or by a combination of both (Lloyd, 1975; Charnov et al., 1976; Charlesworth and Charlesworth, 1978, 1981; Barrett, 2002). The evolutionary route used by species to transition from hermaphroditism to dioecy is the subject of ongoing debate. Although a direct transition has sometimes been suggested, the transition probably involved intermediate steps in most cases, as it seems unlikely that male and female sterility mutations arise simultaneously in hermaphroditic species (Charlesworth and Charlesworth, 1978).
Several potential pathways to dioecy have been suggested. In a first group of pathways, dioecy arises from a population of cosexual individuals that are predisposed to the evolution of separate sexes. In the monoecy pathway, all individuals carry a mixture of unisexual male and female flowers; the evolution of dioecy thus involves only a change in the flower type ratio within individuals. This change is driven by disruptive selection, but no alteration in floral morphology occurs (Lloyd, 1972). Other pathways involve the pre-existence of two categories of individuals within the ancestral cosexual population, which ultimately become specialized in one of the two sexual functions: thrum vs. pin morphs in distylous species, or protandrous vs. protogynous morphs in heterodichogamous species (Pannell and Verdù, 2006, and references therein). A second group of pathways involves, as a first step, the establishment of one category of unisexual individuals within a hermaphroditic population, either females (gynodioecy pathway) or males (androdioecy pathway). Because they co-occur with females (or males, respectively), hermaphrodites should be selected to increase maleness (or femaleness, respectively), which may ultimately lead to dioecy (Charlesworth and Charlesworth, 1978).
These pathways have received unequal attention from evolutionary biologists. To date, no transition from androdioecy to dioecy has been documented. Although gender specialization and/or dissortative mating have been reported in a few heterodichogamous species (Pendleton et al., 2000; Gleiser et al. 2008), no direct evidence has been found for the heterodichogamy pathway. The evolution to dioecy from distyly has been inferred in some lineages (Pailler et al., 1998, and references therein; Rosas and Dominguez, 2008). However, the rareness of these reproductive systems (distyly, heterodichogamy and androdioecy have been documented in 28, 11 and 12 families, respectively; Renner 2001; Barrett, 2002; Pannell, 2002) suggests that other pathways to dioecy may be more common. For this reason, the monoecy and gynodioecy pathways have received much more attention.
The gynodioecy pathway has been thoroughly investigated theoretically, probably because gynodioecy is one of the few reproductive systems for which the genetic determination is well known. Nuclear mutations and cytoplasmic male sterility (CMS) mutations are known to cause male sterility; both can be counteracted by nuclear genes that restore male function (Saumitou-Laprade et al., 1994; Chase, 2007; Spigler et al., 2011). Nuclear or nuclear–cytoplasmic gynodioecy can be stably maintained as theoretical models suggest (Lewis, 1941; Charlesworth and Charlesworth, 1978; Gouyon et al., 1991; Dufay et al., 2007). However, gynodioecy can evolve to dioecy. Although the conditions for that transition are different between nuclear and nuclear–cytoplasmic gynodioecy, the general principle is the same: once male sterility mutations have been established in a hermaphroditic population, partial or total female sterility mutations increasing male fitness can invade the population (Charlesworth and Charlesworth, 1978; Maurice et al., 1994; Schultz, 1994).
The gynodioecy pathway is supported by some data. First, the expected reallocation from female to male functions in hermaphrodites co-occurring with females has been documented in several sub-dioecious (anatomically cosexual but functionally male or female) and gynodioecious species (reviewed in Spigler and Ashman, 2012). Secondly, the existence of functionally male morphological hermaphrodites in many gynodioecious species or populations suggests an ongoing transition to dioecy (e.g. Delph and Carroll, 2001; Miller and Venable, 2002). Finally, some studies focusing on a particular group, such as the Silene genus, have documented an evolution of dioecy through gynodioecy using a phylogenetic approach (Desfeux et al., 1996; Rautenberg et al., 2010; Marais et al., 2011). All this suggests that the gynodioecy has been a route to dioecy, but we currently miss a broad view on how often it has, and in which taxonomic groups.
In contrast to gynodioecy, the underlying genetics of the monoecy pathway are only starting to be understood (Boualem et al., 2008; Martin et al., 2009). However, phenotypic models have shown that unisexual individuals can invade a monoecious population if the investment in one sexual function, particularly the male function, provides increasing fitness returns (e.g. Charnov, 1982). Moreover, some empirical evidence supporting the monoecy pathway is currently available. Several studies using a phylogenetic approach have shown evidence for a transition from monoecy to dioecy in some groups (Acer genus, Sapindaceae: Renner et al., 2007; Asteraceae: Torices et al., 2011) or for an association between monoecy and dioecy without inferring the direction of the transition (e.g. Bryonia genus, Cucurbitaceae: Volz and Renner, 2008). At a larger scale, Renner and Ricklefs (1995) analysed the statistical association between monoecy and dioecy at the family level and concluded that ‘the single most important predictor of a group's tendency to acquire dioecy is the presence of monoecy in the group’. This study is widely considered good evidence that the monoecy pathway is common. A similar study on the gynodioecy pathway is currently missing.
In this study, we investigated whether there is an association between dioecy and gynodioecy in angiosperms as predicted by the gynodioecy–dioecy pathway, following the work done on the monoecy–dioecy pathway. As dioecy is rare in angiosperms, and also because the aim of this study was to obtain a very general picture of the gynodioecy pathway, we do not focus on one family or taxonomic group but conduct our analysis angiosperm wide. As a result, we could not use methods for studying character evolution within a phylogeny (see the Materials and Methods); instead, we had to use a more classical statistical approach in which we nevertheless tried to take phylogeny into account. Importantly, we have revised the list of gynodioecious species found in Delannay (1978), making it – to our knowledge – the largest dataset on gynodioecy to date. Our study provides new evidence for the gynodioecy pathway, suggesting that both the gynodioecy and the monoecy pathways are common in angiosperms. These results are discussed in light of theoretical predictions about the evolution of reproductive systems in angiosperms.
MATERIALS AND METHODS
Datasets
Our data on reproductive systems come from two sources: the list of genera including dioecious species from S. S. Renner's group (www.umsl.edu/~renners) and the list of gynodioecious species from Delannay (1978). The data on gynodioecy were updated and augmented by a literature review encompassing a series of books from Darwin (1877) to Harder and Barrett (2006), and including Knuth (1906) as the main source, all publications we could find from the 1970s and 1980s, and more recent articles referenced in PubMed and ISI Web of Science. We supplemented these sources by looking at floras and books on plant taxonomy from the 18th century to the present in order to identify and confirm species for which females have been observed. This increased the number of genera containing gynodioecious species from about 125 (in Delannay, 1978) to 275.
Based on these two lists of genera containing dioecious and gynodioecious species, we built a dataset containing all angiosperm genera. The number of genera in each family was extracted from the Angiosperm Phylogeny Website (www.mobot.org/MOBOT/research/APweb). As cosexuality is the most common reproductive system in angiosperms (hermaphroditism, approx. 90 %; monoecy, approx. 5 %), we considered a genus without any documented dioecious and/or gynodioecious species to be cosexual (i.e. with hermaphroditic and/or monoecious species only). In doing so, we neglected the unidentified dioecious and/or gynodioecious species possibly present in that genus, making our approach conservative. Each genus was then assigned to one of the following categories: (1) cosexual species only, ‘Co’; (2) dioecious species only, ‘D’; (3) gynodioecious species only, ‘G’; and (4) both dioecious and gynodioecious species, ‘GD’. Note that all four categories can include cosexual species.
Because species/genera for which no information was available were assumed to be cosexual, we obviously underestimate the frequency of dioecy and gynodioecy. This bias is probably much stronger for gynodioecious species. This is because in many gynodioecious species, female plants are rare and/or absent from some populations or regions, and sexual polymorphism can be overlooked (e.g. Asikainen and Mutikainen, 2003; Nilsson and Agren, 2006; Alonso et al., 2007; Caruso and Case, 2007; Dufay et al., 2009) whereas dioecious species are relatively easy to identify. This frequency of unidentified gynodioecious species is probably higher in non-European flora as they have been searched less intensely for gynodioecious species than the European flora. The main available sources about gynodioecious species (including Delannay's work) focus indeed on the European flora. Dioecy is more frequent in the tropics (Renner and Ricklefs, 1995; Vamosi et al., 2003). The possible higher frequency of unidentified gyndioecious species in the tropics data may artificially weaken a statistical association between dioecy and gynodioecy. To avoid this and any other biases arising from gynodioecious species being less well studied outside Europe, we built an additional dataset (‘Europe’) that includes only genera found in Europe (we selected a sub-set of genera based on the information about geographic distribution in S. S. Renner's dataset, and on the Flora Europea). All the analyses described below were conducted on both datasets.
Statistical analyses
The contingency table framework
To test for an association between dioecy and gynodioecy in angiosperms, we prepared 2 × 2 contingency tables (D, not D, vs. G, not G) and performed a Fisher's exact test on the observed vs. expected numbers of genera in the four categories listed above (Co, ‘not G, not D’; D, ‘not G, D’; G, ‘not D, G’; and GD, ‘G and D’). The null hypothesis is that both reproductive systems are independent and that the observed numbers of cells in the 2 × 2 contingency table should match the expected numbers computed by summing the cells of the contingency tables. The null hypothesis is rejected when the observed and expected numbers differ using a Fisher's exact test, which means that the reproductive systems are either found in the same genera or tend to occur in different genera. Looking at the ratio of observed vs. expected GD genera (hereafter called GDo/e) allows one to distinguish between these two explanations. The expected number of GD genera, assuming independence between dioecy and gynodioecy, is obtained from the contingency tables and is equal to the total number of gynodioecious genera multiplied by the total number of dioecious genera divided by the total number of genera. For instance, among the 112 genera in the Apiales order, no genus has dioecious species (D), six genera are ‘G’, two are ‘GD’ and 104 have neither dioecious nor gynoecious species, leading to an expected number of ‘GD’ genera of 8 × 2/112 = 0·143 and a GDo/e ratio of 2/0·143 = 14. The Fisher's exact test on contingency tables can be performed only on taxa (families and higher taxonomic levels) including both dioecy and gynodioecy, and was performed using R (R Development Core Team, 2010).
The problems of using phylogenetic methods for studying character evolution
The approach described above is known to be sensitive to phylogenetic inertia (Weiblen et al., 2000), which has stimulated the development of methods such as Bayestraits, BiSSE and MuSSE for studying character evolution on a phylogeny (Pagel et al., 2004; Pagel and Meade, 2006; Maddison et al., 2007; FitzJohn et al., 2009). Bayestraits, however, assumes equal diversification rates in a phylogenetic tree, an assumption that is very often not fulfilled (Magallon and Sanderson, 2010). This may be problematic when studying the evolution of dioecy, since dioecious and non-dioecious lineages exhibit different rates of diversification (Heilbuth, 2000; Heilbuth et al., 2001; Vamosi and Otto, 2002; Vamosi et al., 2003; Käfer and Mousset, 2014; Käfer et al., 2014). This suggests that Bayestraits is not suitable for studying the evolution of dioecy. Methods including rates of speciation and extinction in their models, such as BiSSE and MuSSE, have been recently developed (Maddison et al., 2007; FitzJohn et al., 2009).
Such methods have been successfully applied to questions such as the transition from self-incompatibility to self-compatibility in Solanaceae (Goldberg et al., 2010). However, dioecy is rare in angiosperms, and simulations have revealed that these methods perform poorly when a derived character state is rare (Davis et al., 2013). Differences in speciation/extinction rates among plant families may make parameter estimation difficult, as these methods will assume one rate of speciation and one rate of extinction per character state for all angiosperms (Magallon and Sanderson, 2010). Working on a very large phylogenetic tree, such as that of the angiosperm genera (13 000 leaves), also raises practical methodological problems, not to mention the problem of obtaining such a phylogenetic tree. Using the BiSSE and MuSSE methods for addressing the question of the transition to dioecy in angiosperms does not seem feasible at the moment, justifying the use of other methods.
Strategies to account for phylogenetic biases
Phylogeny could bias our analysis in two ways. First, very strong associations between dioecy and gynodioecy in some parts of the angiosperm phylogeny (i.e. some taxa with very high GDo/e) may drive the angiosperm global GDo/e above 1 and wrongly give the impression that the association between dioecy and gynodioecy is widespread in angiosperms. To control for this effect, we first performed Fisher's exact tests at different taxonomic levels, according to the classification of the APG III (2009). At the level of all angiosperms, only one contingency table was built. When analyses were performed at lower levels (i.e.,‘classes’, ‘sub-classes’, orders and families), several contingency tables were built, one for each class, sub-class, and so on. Classes include dicots, monocots, magnoliids and early angiosperms (including Amborellales, Austrobaileyales, Nympheales, Ceratophyllales and Chloranthales). Sub-classes include the same, but dicots are split into basal eudicots (including Proteales, Buxales, Ranunculales, Sabiales and Trochodendrales), other core eudicots (including Gunnerales, Vitales, Saxifragales, Santalales, Caryophyllales, Dilleniales and Berberopsidales), fabids (formerly eurosids I), basal malvids (including Geraniales, Myrtales and Crossomatales), other malvids (including Malvales, Brassicales, Huertales, Sapindales and Picramniales), basal asterids (including Cornales and Ericales), lamiids (formerly euasterids I) and campanulids (formerly euasterids II), following the APG III nomenclature.
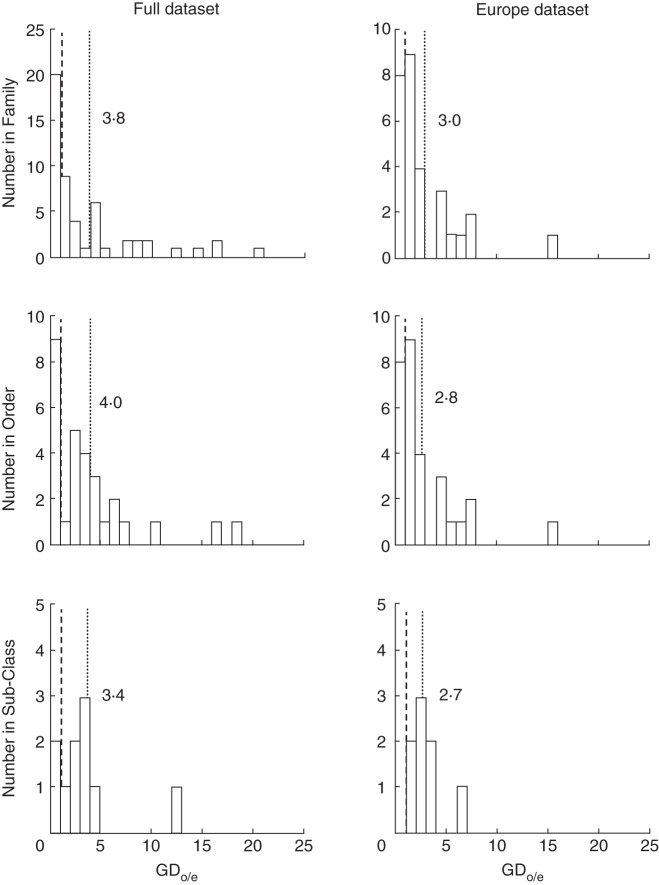
To control further for the effect of phylogeny on our results (i.e. the existence of very few taxa showing high GDo/e), we also performed a sign test on the number of taxa with a GDo/e >1. This was done for families, orders and sub-classes using R. This allowed us to test for a significant excess of genera with GDo/e >1 consistent with the gynodioecy–dioecy pathway in our dataset and at different taxonomic levels (the null hypothesis being that we have an equal number of GDo/e >1 and GDo/e <1). This test was not performed on classes or on all angiosperms due to small sample sizes. The mean GDo/e values were computed for different families, orders and sub-classes and are shown in Fig. 1.
Fig. 1.
Distribution of the ratio of observed vs. expected number of genera with both gynodioecious and dioecious species (GDo/e) at different taxonomic levels. Only taxa for which the GDo/e can be computed were included. The results are shown for ‘Full’ and ‘Europe’ datasets, with the mean GDo/e indicated by a dotted line. GDo/e = 1 is indicated by a dashed line.
The third correction for the phylogenetic bias mentioned above was a resampling approach (permutation test). The presence/absence of dioecious (D) and gynodioecious (G) species in a genus was coded as two characters with binary states (0/1). We generated 1000 new datasets of genera by shuffling independently (1) the presence/absence of dioecious species and (2) the presence/absence of gynodioecious species among the genera of a family. This was done for all families, and a Fisher's exact test was performed on the resulting contingency tables. We then compared the observed P-value from the Fisher's exact test on the original dataset with the distribution of P-values from the Fisher's exact tests on 1000 resampled datasets for each family. The new P-value is the proportion of resampled datasets with lower P-values than the observed P-values. This was also done for sub-classes, classes and at the angiosperm level.
The second possible problem with phylogeny is phylogenetic inertia. If gynodioecy and dioecy co-occurred by chance in an ancestral species, all the species that evolved from this ancestor may show the same association just because they ‘inherited’ it. A significant Fisher's exact test may result not because of a consistent association between dioecy and gynodioecy during evolution, but because of the phylogenetic inertia of a very few or maybe even a single past event. However, phylogenetic inertia should not affect our results too strongly because hermaphroditism is the ancestral state in most angiosperm orders; gynodioecy/dioecy are derived states. Although a detailed study dating the transitions to dioecy is currently lacking, it is commonly accepted that those transitions (and probably also the transitions to gynodioecy) are mostly recent (e.g. Charlesworth, 2002). To our knowledge, only one order (Apiales) and very few families include a large number of dioecious species, which may represent cases where the transition to dioecy is old. Those old transitions are probably extremely rare in angiosperms. Because the analysis here was done at the family level and at higher taxonomic levels, we expect little inertia in our dataset.
RESULTS
An augmented dataset on gynodioecy
Using our revised dataset on gynodioecy, we found a low proportion of genera with gynodioecious species (275 genera, i.e. around 2 % of all 13 208 angiosperm genera accepted by P. Stevens, http://www.mobot.org/MOBOT/Research/APweb/welcome.html; see Supplementary Data Table S1), of which 59 also include dioecious species. Gynodioecy was found in 81 families, i.e. >18 % of all 449 angiosperm families (P. Stevens, http://www.mobot.org/MOBOT/Research/APweb/welcome.html) of which 52 families also include dioecious species. Families with gynodioecy were found in magnolids, monocots and all ‘sub-classes’ of dicots (Table 1). By comparing our data with the dataset published by Delannay (1978), we extended our knowledge of gynodioecy by identifying 150 new genera and 45 new families in which this reproductive system occurred. Using a small fraction of angiosperm species (the flora of France and Belgium), Delannay (1978) estimated that 7·5 % of species are gynodioecious. In contrast, we report a much lower proportion of genera with gynodioecious species at a larger geographic scale. However, focusing only on our European dataset, we also find an increased percentage of genera that include gynodioecious species (12 %). In contrast, the percentage of genera that include dioecious species was similar in the Full dataset (7 %) and the European dataset (8 %). Whether this trend is a by-product of unequal sampling efforts among geographical regions or a reflection of truly higher gynodioecy frequency in Europe remains an open question.
Table 1.
List of families including gynodioecious species
| Class | Sub-class | Order | Family | Dioecy in the family | Contains GD genera |
|---|---|---|---|---|---|
| Magnolids | Laurales | Lauraceae | * | * | |
| Piperales | Aristolochiaceae | * | * | ||
| Piperaceae | * | ||||
| Monocots | Alismatales | Alismataceae | * | ||
| Araceae | |||||
| Asparagales | Alliaceae | ||||
| Asparagaceae | * | * | |||
| Asphodelaceae | |||||
| Hyacinthaceae | |||||
| Iridaceae | |||||
| Liliales | Colchicaceae | * | * | ||
| Liliaceae | |||||
| Melanthiaceae | * | ||||
| Poales | Cyperaceae | * | * | ||
| Juncaceae | * | ||||
| Poaceae | * | * | |||
| Zingiberales | Zingiberaceae | ||||
| Dicots | Basal eudicots | Ranunculales | Papaveraceae | ||
| Ranunculaceae | * | * | |||
| Other core eudicots | Caryophyllales | Cactaceae | * | ||
| Caryophyllaceae | * | * | |||
| Didiereaceae | * | ||||
| Plumbaginaceae | |||||
| Polygonaceae | * | * | |||
| Portulaceae | |||||
| Rosales | Amaranthaceae | * | * | ||
| Santalales | Loranthaceae | * | * | ||
| Saxifragales | Grossulariaceae | * | * | ||
| Saxifragaceae | * | ||||
| Fabids | Celastrales | Celastraceae | * | ||
| Curcubitales | Corynocarpaceae | ||||
| Cucurbitaceae | * | * | |||
| Fabales | Fabaceae | * | |||
| Malpighiales | Clusiaceae | * | * | ||
| Erythroxylaceae | |||||
| Euphorbiaceae | * | * | |||
| Rhizophoraceae | * | ||||
| Violaceae | * | ||||
| Rosales | Moraceae | * | * | ||
| Rosaceae | * | * | |||
| Zygophyllales | Zygophyllaceae | * | |||
| Basal malvids | Crossomatales | Stachyuraceae | |||
| Geraniales | Geraniaceae | ||||
| Myrtales | Myrtaceae | * | |||
| Onagraceae | |||||
| Other malvids | Brassicales | Brassicaceae | * | ||
| Caricaceae | * | * | |||
| Limnanthaceae | |||||
| Resedaceae | |||||
| Malvales | Malvaceae | * | |||
| Thymelaeaceae | * | * | |||
| Sapindales | Anacardiaceae | * | * | ||
| Rutaceae | * | ||||
| Basal asteroids | Cornales | Hydrangaeaceae | * | ||
| Ericales | Ericaceae | * | * | ||
| Polemoniaceae | |||||
| Primulaceae | * | * | |||
| Sapotaceae | * | * | |||
| Styraceae | |||||
| Lamiids | Gentianales | Gentianaceae | |||
| Loganiaceae | |||||
| Rubiaceae | * | * | |||
| Lamiales | Gesneriaceae | ||||
| Lamiaceae | * | * | |||
| Oleaceae | * | * | |||
| Orobanchaceae | |||||
| Plantaginaceae | |||||
| Scrophulariaceae | |||||
| Verbenaceae | * | ||||
| Solanales | Convolvulaceae | ||||
| Solanaceae | * | * | |||
| Unplaced | Boraginaceae | * | |||
| Campanulids | Apiales | Apiaceae | * | * | |
| Pittosporaceae | * | * | |||
| Asterales | Asteraceae | * | * | ||
| Campanulaceae | * | * | |||
| Menyanthaceae | * | * | |||
| Rousseaceae | |||||
| Stylidiaceae | |||||
| Dipsaccales | Dipsaccaceae | ||||
| Valerianaceae | * | * |
Families that also include dioecious species and that include genera with both gynodioecious and dioecious species (GD) are labelled with an asterisk.
An association between gynodioecy and dioecy
Table 2a shows the results of our contingency analysis. We found a significant association (significant Fisher's exact test) between gynodioecy and dioecy in angiosperms as a whole. The overall ratio of the number of observed vs. expected genera with both gynodioecious and dioecious species (= GDo/e) was >1, in agreement with the gynodioecy–dioecy pathway. We tested for this association at different taxonomic levels and found taxa with significant Fisher's exact tests (all with a GDo/e >1) at family, order, sub-class and class levels (Table 2a). Both dicots and monocots showed a significant association between gynodioecy and dioecy, and within dicots most sub-classes also showed such an association. This suggests that our results are not biased by a few taxa with very high GDo/e ratios. This was confirmed by resampling (see the Materials and Methods), which gave very similar results (Table 2a).
Table 2.
Statistical association between dioecy and gynodioecy in angiosperms
| Phylogenetic level | Total no. of taxa | No. of taxa tested† | List of taxa with significant Fischer's exact test P-values | P-value | GDo/e |
|---|---|---|---|---|---|
| (a) Full dataset | |||||
| Family | 438 | 52 | Asteraceae | *** (***) | 20·8 |
| Ericaceae | *** (***) | 8 | |||
| Lamiaceae | * (*) | 5·4 | |||
| Loranthaceae | * (*) | 9·7 | |||
| Rosaceae | ** (**) | 5 | |||
| Rubiaceae | *** (***) | 9·4 | |||
| Order | 59 | 29 | Apiales | * (*) | 6 |
| Asterales | *** (***) | 18·2 | |||
| Brassicales | * (n.s.) | 6·5 | |||
| Ericales | ** (***) | 3·5 | |||
| Gentianales | *** (**) | 10·7 | |||
| Lamiales | ** (*) | 7·8 | |||
| Poales | n.s. (n.s.) | 3·3 | |||
| Rosales | * (**) | 2·5 | |||
| Santalales | * (*) | 4·9 | |||
| Sub-class | 11 | 10 | Basal asterids | * (***) | 3·3 |
| Campanulids | *** (***) | 12·2 | |||
| Fabids | * (**) | 2·5 | |||
| Lamiids | *** (***) | 4·5 | |||
| Monocots | * (*) | 3·2 | |||
| Other core eudicots | * (*) | 1·9 | |||
| Other malvids | * (n.s.) | 2·9 | |||
| Class | 4 | 3 | Dicots | *** (***) | 2·8 |
| Monocots | * (*) | 3·2 | |||
| Angiosperm | 1 | 1 | – | *** (***) | 2·9 |
| (b) ‘Europe’ dataset | |||||
| Family | 438 | 29 | Asteraceae | ** (**) | 6·1 |
| Ericaceae | n.s. (n.s.) | 2·8 | |||
| Lamiaceae | n.s. (n.s.) | 1·6 | |||
| Loranthaceae | NA | NA | |||
| Rosaceae | ** (**) | 4·7 | |||
| Rubiaceae | n.s. (n.s.) | 2·7 | |||
| Order | 59 | 21 | Apiales | ** (*) | 14 |
| Asterales | *** (***) | 6·3 | |||
| Brassicales | NA | NA | |||
| Ericales | n.s. (n.s.) | 1·6 | |||
| Gentianales | n.s. (n.s) | 3·2 | |||
| Lamiales | n.s. (n.s.) | 1·3 | |||
| Poales | * (n.s.) | 5·5 | |||
| Rosales | * (*) | 2·7 | |||
| Santalales | NA | NA | |||
| Sub-class | 11 | 8 | Basal asteroids | n.s. (n.s.) | 2·2 |
| Campanulids | *** (***) | 6·2 | |||
| Fabids | * (*) | 3 | |||
| Lamiids | n.s. (*) | 1·8 | |||
| Monocots | n.s. (n.s.) | 2·2 | |||
| Other core eudicots | n.s. (n.s.) | 1·6 | |||
| Other malvids | n.s. (n.s.) | 2·2 | |||
| Class | 4 | 2 | Dicots | *** (***) | 2·4 |
| Monocots | n.s. (n.s.) | 2·2 | |||
| Angiosperm | 1 | 1 | - | *** (***) | 2·4 |
Taxonomic groups with significant Fisher's test P-values are listed. The ratio of observed vs. expected genera with both dioecious and gynodioecious species (= GDo/e) is shown, as are P-values for Fisher's tests and P-values after resampling (in parentheses).
Results are shown at various taxonomic levels and for the full dataset (a) and the European genera only (b). To make comparison easier, all significant taxa in the analysis of the full dataset are also shown in the analysis of the European dataset, even when they are not significant.
*P < 0·05, **P < 0·005, ***P < 0·0005, n.s., non-significant. Symbols in parenthese are for P-values after resampling (see the Material and Methods). The underlined P-values are those that are still significant after Bonferroni correction for multiple tests.
†Number of groups for which the Fisher's test was possible (including both gynodioecous and dioecious genera).
The percentage of taxa with significant Fisher's exact tests decreased at order and family levels compared with class and sub-class levels, probably because the sample size (number of genera per taxa) decreased, limiting our statistical power. This was confirmed by our analysis of all taxa with GDo/e >1 (both with and without significant Fisher's exact tests) shown in Table 3. Indeed, we observed that almost all studied taxa, depending on the taxonomic level, have a GDo/e consistent with the gynodioecy–dioecy pathway (sign tests were found to be significant or marginally significant, i.e. P-value close to 0·05, see Table 3). Overall, the mean GDo/e is >1 at different taxonomic levels (see Fig. 1). These lines of evidence further suggest that the gynodioecy pathway is widespread in angiosperms and our results are not biased by a few taxa with strong gynodioecy–dioecy associations.
Table 3.
Number of groups with an excess of genera with both dioecious and gynodioecious species (GDo/e >1) at various taxonomic levels
| Taxonomic level | No. of groups with GDo/e >1/total no. groups |
|
|---|---|---|
| Full dataset | Europe | |
| Family | 32/52 (0·06) | 21/29 (0·01) |
| Order | 20/29 (0·03) | 15/21 (0·04) |
| Sub-class | 8/10 (0·05) | 8/8 (0·004) |
Statistical significance was assessed with a sign test. Only groups with both dioecious and gynodioecious genera are included.
P-values are shown in parentheses.
It is important to note that our gynodioecy dataset is enriched in European species. To control for this, we performed the same analysis on a dataset containing only European species, allowing for direct comparison between dioecy and gynodioecy data (see the Materials and Methods). As shown in Table 2b, the results of this analysis are quantitatively equivalent to those of the full dataset. Many of the taxa with significant Fisher's tests are common to both analyses; non-significant Fisher tests are probably the result of decreased sample size and loss of statistical power. Figure 1 shows that the mean GDo/e ratios are clearly >1 at different taxonomic levels, suggesting that the gynodioecy pathway is also widespread in the Europe dataset.
DISCUSSION
The frequency of gynodioecious species in angiosperms
Considerable debate exists with regard to the frequency of gynodioecious species in angiosperms. Some papers refer to gynodioecy as the second most frequent reproductive system in angiosperms after hermaphroditism (e.g. Collin et al. 2002; Bailey et al. 2003; Caruso et al. 2003; Chang, 2006), while others consider gynodioecy an extremely rare system (e.g. Charlesworth, 2002). Our new dataset on gynodioecy suggests that this reproductive system is rare in angiosperms, occurring in only 2 % of genera (vs. 7 % in genera with dioecious species; Renner and Ricklefs, 1995). In the original list of Delannay (1978), which focused on European species, gynodioecy occurred in >7 % of species (with no estimation at the genus level). It is difficult to ascertain whether these different estimates result from the well-known difficulty of identifying gynodioecious species/populations in nature (especially when the frequency of females is low), or from strong geographical biases that are known to exist at least for dioecy (dioecy is more frequent in the tropics; Renner and Ricklefs, 1995; Vamosi et al., 2003). For this reason, generalizing results from the European flora to angiosperms as a whole may be risky. On the other hand, the fraction of overlooked gynodioecious species/populations may be reduced for European species, which have been thoroughly investigated. Consequently, we suggest that our estimate is probably a lower bound for the frequency of genera with gynodioecious species in angiosperms.
It is interesting to note that, although quite rare at the genus level, gynodioecy is found at a higher frequency at the family level (>18 % of angiosperm families). The same pattern has been reported for dioecy (found in 6 % of species, 7 % of genera and in >30 % of families). This suggests that dioecy has evolved many times independently (Charlesworth, 2002) but may, once evolved, lead to higher rates of extinction (Heilbuth, 2000; Heilbuth et al., 2001; Vamosi and Otto, 2002; Vamosi et al., 2003; Vamosi and Vamosi, 2004). Other explanations are, however, possible. In particular, recent studies suggest that a low transition rate and/or frequent reversions may better explain the rareness of dioecy in angiosperms than a high extinction rate (Käfer and Mousset, 2014; Käfer et al., 2014). A similar dynamic could affect gynodioecious species and explain our results. Alternatively, a high rate of transition from gynodioecy to dioecy could also explain this pattern.
The routes to dioecy
In angiosperms as a whole, we found that gynodioecy and dioecy co-occurred within the same genera significantly more often than expected if the evolution of gynodioecy and dioecy had always followed independent routes. This is, to our knowledge, the first study that reports an association between these reproductive systems at such a broad scale; this association is consistent with the gynodioecy pathway (but does not rule out the monoecy pathway; see below). Because we could not use phylogenetic methods to study character evolution, our results may also be explained by the existence of ‘hotspots’ of unrelated reproductive system transitions (Weiblen et al., 2000). Indeed, factors that favour dioecy, i.e. advantage of sexual specialization and of inbreeding avoidance, are similar to those that favour gynodioecy (Lloyd, 1975; Charlesworth and Charlesworth, 1978). Closely related species sharing life history traits or ecological constraints may thus independently evolve dioecy or gynodioecy as a result of the same selective pressures. Only a phylogenetic approach could help discriminate between these two hypotheses; our study provides a list of families that would be interesting to use in such an approach.
Importantly, the fact that gynodioecy and dioecy are associated does not rule out the monoecy–dioecy pathway. As shown in Table 2, our analyses were performed only on taxa having genera with dioecious and/or gynodioecious species (for which the GDo/e can be computed). At the family level, this represents only 52 out of the 438 angiosperm families, while 100 families contained dioecious but not gynodioecious species. This suggests that dioecy in these groups may have taken other pathways. It would be interesting to compare our results with those of Renner and Ricklefs (1995), who reported an association between monoecy and dioecy at the scale of all angiosperms. However, because their analysis was performed on families (not on genera), it is difficult to compare the two studies directly. Nevertheless, many families/orders/superorders have both dioecious and monoecious species (see table 13 in Renner and Ricklefs, 1995), which suggests that the monoecy–dioecy pathway applies in these cases. Moreover, some taxa have monoecy, dioecy and gynodioecy, which suggests that both pathways could occur in the same taxon. For instance, a statistical association between gynodioecy and dioecy was found in Asteraceae, but this family also includes many monoecious species. In their phylogenetic study of transitions in reproductive systems in Asteraceae, Torices et al. (2011) found that the number of transitions to dioecy was lower through the gynodioecy pathway as compared with the monoecy pathway. However, the rate of transition was higher in the former than in the latter (i.e. once evolved, gynodioecious species in this particular family quickly switch to dioecy). This suggests that both pathways to dioecy may occur in the same clade, possibly with different dynamics.
Our study provides a list of 52 families of potential interest for further investigations of the gynodioecy pathway. These genera include both gynodioecious and dioecious species (Table 1). When our statistical analysis was performed at the family level, a majority of these families showed no significant association (Table 2). This could suggest that gynodioecy and dioecy do not co-occur in the same genera and have independent evolutionary histories in these families. However, the analysis of the GDo/e (Table 3; Fig. 1) clearly suggests that a lack of statistical power is the explanation in most cases. This may be the case for the Caryophyllaceae family, where no significant association was found. Previous work strongly suggests that indeed some dioecious Caryophyllaceae species have evolved through the gynodioecy pathway (Desfeux et al., 1996; Rautenberg et al., 2010; Marais et al., 2011). Finally, future work should further explore the distribution and dynamics of reproductive systems in the six families listed in Table 2, as they are interesting candidates for the study of the gynodioecy pathway. Some have already been investigated: Asteraceae (Torices et al., 2011) and the Fragaria genus within the Rosaceae (Spigler et al., 2008, 2011). However, to our knowledge, the evolution of dioecy is poorly understood in the majority of genera and families highlighted here.
Does gynodioecy always lead to dioecy?
In 29 families, we found gynodioecy but not dioecy. A large number of theoretical studies have investigated the conditions required for the stable maintenance of gynodioecy (Charlesworth and Charlesworth, 1978; Gouyon et al., 1991; Bailey et al., 2003; Dufay et al., 2007) and have shown that, once established in a population, the co-occurrence of females and hermaphrodites does not necessarily lead to the invasion of female sterility mutations (and, thus, to dioecy). This should be particularly true in gynodioecious species that exhibit a low female frequency or variable frequency over time, in which case selection for maleness will be weaker. According to theory, the female advantage (increase in female fitness compared with hermaphrodites) should have a strong, positive impact on the female ratio in gynodioecious populations (e.g. Dufay et al., 2007). Thus, if in some groups this female advantage is small, females might not be frequent enough for dioecy to evolve. In species in which hermaphrodites do not self-pollinate, only resource reallocation can provide females with an advantage. This contrasts with species in which hermaphrodites self-pollinate and only females are obligate outcrossers. In this case, females will benefit from both resource reallocation and inbreeding depression avoidance, which will increase their fitness advantage over hermaphrodites (reviewed in Dufay and Billard, 2012). Accordingly, Karron et al. (2012) noted that gynodioecious species in which inbreeding avoidance plays a role in female advantage seem more likely to have dioecious relatives, compared with species in which that advantage stems only from resource reallocation. This stresses the need for large datasets that include various aspects of reproductive systems (i.e. gynodioecy/dioecy, selfing rates and/or self-incompatibility/self-compatibility) because these aspects very probably interact with each other. Our updated dataset on gynodioecy should help in preparing such datasets.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Pierre-Henri Gouyon for putting J.P.H. and the other authors in contact, and Susanne Renner for making her dataset on dioecy publicly available. We are also grateful to Susanne Renner in her role as Editor and to three anonymous referees for their comments, which were very helpful in improving the manuscript. This work was supported by the Agence Nationale de la Recherche (ANR-11-BSV7-013-03).
LITERATURE CITED
- Alonso C, Mutikainen P, Herrera CM. Ecological context of breeding system variation: sex, size and pollination in a (predominantly) gynodioecious shrub. Annals of Botany. 2007;100:1547–1556. doi: 10.1093/aob/mcm254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APG III. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121. [Google Scholar]
- Asikainen E, Mutikainen P. Female frequency and relative fitness of females and hermaphrodites in gynodioecious Geranium sylvaticum (Geraniaceae) American Journal of Botany. 2003;90:226–234. doi: 10.3732/ajb.90.2.226. [DOI] [PubMed] [Google Scholar]
- Bailey MF, Delph LF, Lively CM. Modeling gynodioecy: novel scenarios for maintaining polymorphism. American Naturalist. 2003;161:762–776. doi: 10.1086/374803. [DOI] [PubMed] [Google Scholar]
- Barrett SCH. The evolution of plant sexual diversity. Nature Reviews Genetics. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- Barrett SCH. Darwin's legacy: the forms, function and sexual diversity of flowers. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:351–368. doi: 10.1098/rstb.2009.0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem A, Fergany M, Fernandez R, et al. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science. 2008;321:836–838. doi: 10.1126/science.1159023. [DOI] [PubMed] [Google Scholar]
- Caruso CM, Case AL. Sex ratio variation in gynodioecious Lobelia siphilitica: effects of population size and geographic location. Journal of Evolutionary Biology. 2007;20:1396–1405. doi: 10.1111/j.1420-9101.2007.01361.x. [DOI] [PubMed] [Google Scholar]
- Caruso CM, Maherali H, Jackson RB. Gender-specific floral and physiological traits: implications for the maintenance of females in gynodioecious Lobelia siphilitica. Oecologia. 2003;135:524–531. doi: 10.1007/s00442-003-1199-2. [DOI] [PubMed] [Google Scholar]
- Chang SM. Female compensation through the quantity and quality of progeny in a gynodioecious plant, Geranium maculatum (Geraniaceae) American Journal of Botany. 2006;93:263–270. doi: 10.3732/ajb.93.2.263. [DOI] [PubMed] [Google Scholar]
- Charlesworth D. Plant sex determination and sex chromosomes. Heredity. 2002;88:94–101. doi: 10.1038/sj.hdy.6800016. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. A model for the evolution of dioecy and gynodioecy. American Naturalist. 1978;112:975–997. [Google Scholar]
- Charlesworth B, Charlesworth D. Allocation of resources to male and female functions in hermaphrodites. Biological Journal of the Linnean Society. 1981;15:57–74. [Google Scholar]
- Charnov ZL. The theory of sex allocation. Princeton, NJ: Princeton University Press; 1982. [Google Scholar]
- Charnov EL, Smith JM, Bull JJ. Why be an hermaphrodite? Nature. 1976;263:125–126. [Google Scholar]
- Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends in Genetics. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Collin CL, Pennings PS, Rueffler C, Widmer A, Shykoff JA. Natural enemies and sex: how seed predators and pathogens contribute to sex-differential reproductive success in a gynodioecious plant. Oecologia. 2002;131:94–102. doi: 10.1007/s00442-001-0854-8. [DOI] [PubMed] [Google Scholar]
- Darwin C. The different forms of flowers on plants of the same species. London: Murray; 1877. [Google Scholar]
- Davis MP, Midford PE, Maddison W. Exploring power and parameter estimation of the BISSE method for analyzing species diversification. BMC Evolutionary Biology. 2013;13:38. doi: 10.1186/1471-2148-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannay X. La gynodioecie chez les angiospermes. Naturalistes Belges. 1978;59:223–235. [Google Scholar]
- Delph LF, Carroll SB. Factors affecting the relative seed fitness and female frequency in a gynodioecious species, Silene acaulis. Evolutionary Ecology Research. 2001;3:487–505. [Google Scholar]
- Desfeux C, Maurice S, Henry JP, Lejeune B, Gouyon PH. Evolution of reproductive systems in the genus Silene. Proceedings of the Royal Society B: Biological Sciences. 1996;263:409–414. doi: 10.1098/rspb.1996.0062. [DOI] [PubMed] [Google Scholar]
- Dufay M, Billard E. How much better are females? The occurrence of female advantage, its proximal causes and its variation within and among gynodioecious species. Annals of Botany. 2012;109:505–519. doi: 10.1093/aob/mcr062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufay M, Touzet P, Maurice S, Cuguen J. Modelling the maintenance of male-fertile cytoplasm in a gynodioecious population. Heredity. 2007;99:349–356. doi: 10.1038/sj.hdy.6801009. [DOI] [PubMed] [Google Scholar]
- Dufay M, Cuguen J, Arnaud JF, Touzet P. Sex ratio variation among gynodioecious populations in sea beet: can it be explained by negative frequency-dependent selection. Evolution. 2009;63:1483–1497. doi: 10.1111/j.1558-5646.2009.00653.x. [DOI] [PubMed] [Google Scholar]
- FitzJohn RG, Maddison WP, Otto SP. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Systematic Biology. 2009;58:595–611. doi: 10.1093/sysbio/syp067. [DOI] [PubMed] [Google Scholar]
- Gleiser G, Verdu M, Segarra-Morages JG, Gonzalez-Martinez SC, Pannell JR. Dissortative mating, gender specialization, and the evolution of gender dimorphism in heterodichogamous Acer opalus. Evolution. 2008;62:1676–1688. doi: 10.1111/j.1558-5646.2008.00394.x. [DOI] [PubMed] [Google Scholar]
- Goldberg EE, Kohn JR, Lande R, Robertson KA, Smith SA, Igic B. Species selection maintains self-incompatibility. Science. 2010;330:493–495. doi: 10.1126/science.1194513. [DOI] [PubMed] [Google Scholar]
- Gouyon PH, Vichot F, Vandamme JMM. Nuclear–cytoplasmic male-sterility – single-point equilibria versus limit-cycles. American Naturalist. 1991;137:498–514. [Google Scholar]
- Harder LD, Barrett SCH. Ecology and evolution of flowers. Oxford: Oxford University Press; 2006. [Google Scholar]
- Heilbuth JC. Lower species richness in dioecious clades. American Naturalist. 2000;156:221–241. doi: 10.1086/303389. [DOI] [PubMed] [Google Scholar]
- Heilbuth JC, Ilves KL, Otto SP. The consequences of dioecy for seed dispersal: modeling the seed-shadow handicap. Evolution. 2001;55:880–888. doi: 10.1554/0014-3820(2001)055[0880:tcodfs]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Käfer J, Mousset S. Standard sister clade comparison fails when testing derived character states. Systematic Biology. 2014;63:601–609. doi: 10.1093/sysbio/syu024. [DOI] [PubMed] [Google Scholar]
- Käfer J, de Boer H, Mousset S, Kool A, Dufay M, Marais GAB. Dioecy is associated with higher diversification rates in flowering plants. 2014. Journal of Evolutionary Biology (in press) [DOI] [PubMed]
- Karron JD, Ivey CT, Mitchell RJ, Whitehead MR, Peakall R. New perspectives on the evolution of plant mating systems. Annals of Botany. 2012;109:493–503. doi: 10.1093/aob/mcr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuth P. Handbook of flower pollination. Oxford: Clarendon Press; 1906. [Google Scholar]
- Lewis D. Male sterility in natural populations of hermaphrodite plants. New Phytologist. 1941;40:56–63. [Google Scholar]
- Lloyd DG. Breeding systems in Cotula L. (Compositae, Anthemideae) 1. Array of monoclinous and diclinous systems. New Phytologist. 1972;71:1181–1194. [Google Scholar]
- Lloyd DG. The maintenance of gynodioecy and androdioecy in angiosperms. Genetica. 1975;45:325–339. [Google Scholar]
- Maddison WP, Midford PE, Otto SP. Estimating a binary character's effect on speciation and extinction. Systematic Biology. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- Magallon S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2010;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- Marais GAB, Forrest A, Kamau E, Käfer J, Daubin V, Charlesworth D. Multiple nuclear gene phylogenetic analysis of the evolution of dioecy and sex chromosomes in the genus Silene. PLoS One. 2011;6:e21915. doi: 10.1371/journal.pone.0021915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Troadec C, Boualem A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- Maurice S, Belhassen E, Couvet D, Gouyon PH. Evolution of dioecy – can nuclear–cytoplasmic interactions select for maleness? Heredity. 1994;73:346–354. doi: 10.1038/hdy.1994.181. [DOI] [PubMed] [Google Scholar]
- Miller JS, Venable DL. The transition to gender dimorphism on an evolutionary background of self-incompatibilty: an example from Lycium (Solanaceae) American Journal of Botany. 2002;89:1907–1915. doi: 10.3732/ajb.89.12.1907. [DOI] [PubMed] [Google Scholar]
- Nilsson E, Agren J. Population size, female fecundity, and sex ratio variation in gynodioecious Plantago maritima. Journal of Evolutionary Biology. 2006;19:825–833. doi: 10.1111/j.1420-9101.2005.01045.x. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. American Naturalist. 2006;167:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Systematic Biology. 2004;53:673–84. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- Pailler T, Humeau L, Figier J, Thompson JD. Reproductive trait variation in the functionally dioecious and morphologically heterostylous island endemic Chassalia corallioides (Rubiaceae) Biological Journal of the Linnean Society. 1998;64:297–313. [Google Scholar]
- Pannell JR. The evolution and maintenance of androdioecy. Annual Review of Ecology and Systematics. 2002;33:397–425. [Google Scholar]
- Pannell JR, Verdu M. The evolution of gender specialization from dimorphic hermaphroditism: paths from heterodichogamy to gynodioecy and androdioecy. Evolution. 2006;60:660–673. doi: 10.1554/05-481.1. [DOI] [PubMed] [Google Scholar]
- Pendleton RL, Freeman DC, McArthur ED, Sanderson SC. Gender specialization in heterodichogamous Grayia brandegei (Chenopodiaceae): evidence for an alternative pathway to dioecy. American Journal of Botany. 2000;87:508–516. [PubMed] [Google Scholar]
- Rautenberg A, Hathaway L, Oxelman B, Prentice HC. Geographic and phylogenetic patterns in Silene section Melandrium (Caryophyllaceae) as inferred from chloroplast and nuclear DNA sequences. Molecular Phylogenetics and Evolution. 2010;57:978–991. doi: 10.1016/j.ympev.2010.08.003. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- Renner SS. How common is heterodichogamy? Trends in Ecology and Evolution. 2001;16:595–597. [Google Scholar]
- Renner SS, Ricklefs RE. Dioecy and its correlates in the flowering plants. American Journal of Botany. 1995;82:596–606. [Google Scholar]
- Renner SS, Beenken L, Grimm GW, Kocyan A, Ricklefs RE. The evolution of dioecy, heterodichogamy, and labile sex expression in Acer. Evolution. 2007;61:2701–2719. doi: 10.1111/j.1558-5646.2007.00221.x. [DOI] [PubMed] [Google Scholar]
- Rosas F, Dominguez CA. Male sterility, fitness gain curves and the evolution of gender specialization from distyly in Erythroxylum havanense. Journal of Evolutionary Biology. 2009;22:50–59. doi: 10.1111/j.1420-9101.2008.01618.x. [DOI] [PubMed] [Google Scholar]
- Saumitou-Laprade P, Cuguen J, Vernet P. Cytoplasmic male sterility in plants: molecular evidence and the nucleocytoplasmic conflict. Trends in Ecology and Evolution. 1994;9:431–435. doi: 10.1016/0169-5347(94)90126-0. [DOI] [PubMed] [Google Scholar]
- Schultz ST. Nucleo-cytoplasmic male sterility and alternative routes to dioecy. Evolution. 1994;48:1933–1945. doi: 10.1111/j.1558-5646.1994.tb02224.x. [DOI] [PubMed] [Google Scholar]
- Spigler RB, Ashman TL. Gynodioecy to dioecy: are we there yet? Annals of Botany. 2012;109:531–543. doi: 10.1093/aob/mcr170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spigler RB, Lewers KS, Main DS, Ashman TL. Genetic mapping of sex determination in a wild strawberry, Fragaria virginiana, reveals earliest form of sex chromosome. Heredity. 2008;101:507–517. doi: 10.1038/hdy.2008.100. [DOI] [PubMed] [Google Scholar]
- Spigler RB, Lewers KS, Ashman TL. Genetic architecture of sexual dimorphism in a subdioecious plant with a proto-sex chromosome. Evolution. 2011;65:1114–1126. doi: 10.1111/j.1558-5646.2010.01189.x. [DOI] [PubMed] [Google Scholar]
- Torices R, Mendez M, Gomez JM. Where do monomorphic sexual systems fit in the evolution of dioecy? Insights from the largest family of angiosperms. New Phytologist. 2011;190:234–248. doi: 10.1111/j.1469-8137.2010.03609.x. [DOI] [PubMed] [Google Scholar]
- Vamosi JC, Otto SP. When looks can kill: the evolution of sexually dimorphic floral display and the extinction of dioecious plants. Proceedings of the Royal Society B: Biological Sciences. 2002;269:1187–1194. doi: 10.1098/rspb.2002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamosi JC, Vamosi SM. The role of diversification in causing the correlates of dioecy. Evolution. 2004;58:723–731. doi: 10.1111/j.0014-3820.2004.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Vamosi JC, Otto SP, Barrett SCH. Phylogenetic analysis of the ecological correlates of dioecy in angiosperms. Journal of Evolutionary Biology. 2003;16:1006–1018. doi: 10.1046/j.1420-9101.2003.00559.x. [DOI] [PubMed] [Google Scholar]
- Volz SM, Renner SS. Hybridization, polyploidy and evolutionary transitions between monoecy and dioecy in Bryonia (Cucurbitaceae) American Journal of Botany. 2008;95:1297–1306. doi: 10.3732/ajb.0800187. [DOI] [PubMed] [Google Scholar]
- Weiblen GD, Oyama RK, Donoghue MJ. Phylogenetic analysis of dioecy in monocotyledons. American Naturalist. 2000;155:46–58. doi: 10.1086/303303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.