Abstract
Background and Aims
The integrity and evolution of lichen symbioses depend on a fine-tuned combination of algal and fungal genotypes. Geographically widespread species complexes of lichenized fungi can occur in habitats with slightly varying ecological conditions, and it remains unclear how this variation correlates with symbiont selectivity patterns in lichens. In an attempt to address this question, >300 samples were taken of the globally distributed and ecologically variable lichen-forming species complex Tephromela atra, together with closely allied species, in order to study genetic diversity and the selectivity patterns of their photobionts.
Methods
Lichen thalli of T. atra and of closely related species T. grumosa, T. nashii and T. atrocaesia were collected from six continents, across 24 countries and 62 localities representing a wide range of habitats. Analyses of genetic diversity and phylogenetic relationships were carried out both for photobionts amplified directly from the lichen thalli and from those isolated in axenic cultures. Morphological and anatomical traits were studied with light and transmission electron microscopy in the isolated algal strains.
Key Results
Tephromela fungal species were found to associate with 12 lineages of Trebouxia. Five new clades demonstrate the still-unrecognized genetic diversity of lichen algae. Culturable, undescribed lineages were also characterized by phenotypic traits. Strong selectivity of the mycobionts for the photobionts was observed in six monophyletic Tephromela clades. Seven Trebouxia lineages were detected in the poorly resolved lineage T. atra sensu lato, where co-occurrence of multiple photobiont lineages in single thalli was repeatedly observed.
Conclusions
Low selectivity apparently allows widespread lichen-forming fungi to establish successful symbioses with locally adapted photobionts in a broader range of habitats. This flexibility might correlate with both lower phylogenetic resolution and evolutionary divergence in species complexes of crustose lichen-forming fungi.
Keywords: Adaptation, algal culture, crustose lichen, Lecanoromycetes, lichenized fungi, morphology, mycobiont, photobiont, phylogeny, Tephromela atra, Trebouxia
INTRODUCTION
The self-sustaining lichen symbiosis of fungi and algae is characterized by typical vegetative structures, the lichen thalli, which evolved >415 million years ago (Honegger et al., 2012). Since then major lineages of ascomycetes have diversified in the lichen symbiosis, with >18 000 currently recognized lichen species (Nash, 2008). In most lichen-forming fungi, characteristic vegetative thalli and fungal reproductive structures can develop only after suitable strains of fungal and algal species have met. While the relationships of the fungal partners, for which lichens are named, have been well studied since the early 1990s by molecular approaches, knowledge about the patterns of associations between mycobionts and photobionts is still lagging behind, particularly in crustose species.
Crustose lichens are numerically very diverse, yet their highly homoplastic morphological characters often make species difficult to classify. Many taxa have been gathered into species complexes where morphologically discrete groups can hardly be circumscribed. The ambiguities in identifying lichen species within species complexes still concerns not only morphological traits but also patterns of secondary chemistry and habitat specialization (Grube and Kroken, 2000). In the past years, many morphological species complexes have been disentangled by molecular phylogenetic analyses, and hitherto cryptic taxa have been characterized by a phylogenic species concept, e.g. in the large family Parmeliaceae (Crespo et al., 2010) and in the genus Rhizoplaca (Leavitt et al., 2013a). However, many lichen species complexes still remain unresolved, especially those with representatives occurring in a broader range of environmental conditions. In particular, it is still not well known how ecological specialization or local selectivity leads to speciation and diversification and what role the symbiotic partners could have in this process.
In the largest monophyletic group of lichen-forming fungi, the Lecanoromycetes, preferences for certain types of photobionts, either cyanobacteria or various lineages of green algae, have been recognized in the major lineages (Miadlikowska et al., 2006). Within this general framework, three categories can be distinguished according to the range of photobionts with which they are able to lichenize: photobiont specialists, intermediates and generalists (Yahr et al., 2006). Specialists accept single algal lineages, whereas generalists show broader preference for their algal partners and can associate with different strains according to ecological conditions. Examples of generalists have been detected in lichens with wide geographical range and ample ecological niches. These lichens may associate with locally adapted photobionts in climatically different regions (Blaha et al., 2006; Fernandez-Mendoza et al., 2011; Vargas Castillo and Beck, 2012).
Recently we studied the lichen-forming fungus Tephromela atra in a wide sense as a model for the evolution of cosmopolitan lichen species complexes (Muggia et al., 2014). The genus Tephromela is composed of crustose lichens growing on various substrates (siliceous and calcareous rocks, including concrete, as well as bark), at different elevations and under different ecological conditions. The type species, Tephromela atra (Huds.) Hafellner, has a rather broad ecological range and displays subtle morphological and chemical variations which have caused uncertainty about its taxonomic delimitation and local adaptation (Kalb and Hafellner, 1992; Grube et al., 2001; Nimis and Martellos, 2003). Alternatively, few well-resolved monophyletic clades within T. atra are also well supported by other characters including morphology, chemistry and/or ecology (Muggia et al., 2014). In the present work we analysed the photobionts of the same worldwide sampling used by Muggia et al. (2014) to investigate whether the selectivity of the mycobionts towards the photobionts is correlated with fungal genetic variation. We therefore studied the genetic diversity of the photobionts using internal transcribed spacer (ITS) and rbcL sequence data from small fragments of thalli and from isolated photobionts maintained in axenic cultures. Potential algal heterogeneity within entire thalli was additionally studied by single strand conformation polymorphism (SSCP) fingerprint analysis. Using a substantial sampling of Trebouxia, we recognized different patterns of photobiont selectivity in the T. atra complex and further discuss the delimitation of lichen-associated Trebouxia species.
MATERIALS AND METHODS
Sampling
Lichen thalli of Tephromela atra and of closely related species T. grumosa, T. nashii and T. atrocaesia were collected between 2009 and 2013 from six continents, across 24 countries and 62 localities, in Alpine, Arctic, Mediterranean, oceanic and continental regions; samples growing on different substrates were considered (Supplementary Data Table S1). We selected >300 samples for molecular analyses. One to three thallus samples from different localities and growth substrates were used for isolating the photobionts in culture.
Molecular analyses
The lichen material was examined under a dissecting microscope: one areole and one apothecium were taken from the saxicolous thalli; corticolous thalli were scraped off from a single area of approx. 0·5 cm2, avoiding the bark underneath. The dry material was transferred into a reaction tube, frozen, pulverized using a TissueLyserII (Retsch) and the DNA was extracted according to Cubero et al. (1999).
The identity and the phylogenetic relationships of the photobionts associated with Tephromela fungi were studied using sequences of the nuclear ribosomal internal transcribed spacer (ITS1, 5·8 rDNA, ITS2) and the large subunit of the protein-coding genes ribulose biphosphate carboxylase (rbcL). The ITS region was amplified using Trebouxia-specific primers ITS1T and ITS4T (Kroken and Taylor, 2000), and the rbcL locus was amplified with the algal-specific primers rbcL320 and rbcL803 (Nozaki et al., 1995). Polymerase chain reaction (PCR) conditions were as in previous studies (Muggia et al., 2008, 2010). The PCR products were visualized on a 1 % agarose gel stained with GelRed™ (Biotium, VWR) and subsequently purified using the QIAquick PCR purification kit (Qiagen, Vienna). Complementary strands were sequenced by Macrogen, Inc. (Amsterdam), assembled, and edited in BioEdit (Hall, 1999).
Culture isolation of photobiont
Sixty-five thalli of Tephromela spp. were selected for isolation of Trebouxia photobionts. A selected portion of the lichen thallus lacking fungal contamination and superficial algae was taken and washed three times by pipetting with a sterile 1 % dilution of Tween-80. Alternatively, the upper cortex of the lichen was removed with a sterile razor blade and tiny clumps of the algal layer were dissected and directly inoculated on Bold basal medium (BBM; Bold, 1949; Bishoff and Bold, 1963). Ampicillin was added to the medium to reduce bacterial growth. The Petri plates were incubated at 20 °C, with a light–dark regime (14:10 h), 60 % humidity and light intensity of 60–100 μmol photons m–2 s–1 for about 3–4 weeks. Established algal colonies were sub-cultivated on Trebouxia medium (TM; Ahmadjian, 1967); one part of the colony was prepared for DNA extraction. DNA was extracted according to Cubero et al. (1999), and ITS and rbcL loci were analysed as described above.
Light microscopy
Algal colonies were observed by light microscopy (Zeiss Axioscope) for morphological and anatomical structures using × 400 magnification. Colonies and algal cells were photographed with an AxioCam MRc5 (Zeiss) digital camera connected to the microscope, and digital images were documented with the program AxioVision (Axio VS40, Zeiss).
Transmission electron microscopy (TEM)
Cultured photobiont cells were fixed for 5 min in a 1 % aqueous solution of KMnO4 at room temperature, washed with distilled water and fixed again in a 1 % aqueous solution of KMnO4 for 25 min. Fixed cells were washed three times in distilled water and incubated in 0·5 % aqueous uranyl acetate overnight at 4 °C. Samples were then dehydrated for 20 min, in a graded series of 50, 70, 90 and 100 % ethanol each. Pure ethanol was then changed for propylene oxide, and cells were gradually infiltrated with increasing concentrations (30, 50, 70 and 100 %) of Agar 100 epoxy resin (Agar Scientific®) mixed with propylene oxide for a minimum of 3 h per step. Samples were embedded in pure, fresh Agar 100 epoxy resin and polymerized at 60 °C for 48 h. Ultrathin sections of 80 nm were stained for 3 min with lead citrate and viewed with a Philips CM 10 transmission electron microscope.
Alignment and phylogenetic analyses
The identity of the sequences was checked in GenBank by blast similarity search (Altschul et al., 1990). We decided to perform separate phylogenetic analyses of the ITS and rbcL loci because the lack of rbcL data in GenBank for the majority of the Trebouxia species would have introduced a considerable amount of missing data in the phylogenetic analysis.
We included in the phylogenetic analyses all our new sequences obtained from the original lichen thalli and from the culture isolates (Supplementary Data Table S1); 178 ITS and 53 rbcL sequences of Trebouxia spp. were additionally retrieved from GenBank (Supplementary Data Table S2, Fig. S1). In selecting the available sequences from GenBank we took care to include all the known Trebouxia species as well as all new recently discovered Trebouxia lineages (Guzow-Krzemińska, 2006; Muggia et al., 2010; Ruprecht et al., 2012; Pérez-Ortega et al., 2012; del Campo et al., 2013; Leavitt et al., 2013b). Nucleotide alignments were produced automatically with ClustalW as implemented in BioEdit 5.0.6 (Hall, 1999; http://jwbrown.mbio.ncsu.edu/BioEdit/bioedit.html) and manually adjusted. Ambiguous regions and short insertions (up to 10 bp) were excluded from the ITS alignment; the rbcL dataset did not contain ambiguous regions. The program jModelTest (Posada, 2008) was used to determine the nucleotide substitution models for the ITS locus and for the three codon positions of the rbcL locus using the Akaike Information Criterion and the maximum likelihood ratio test (Posada and Crandall, 1998).
The phylogenetic hypotheses of both ITS and rbcL were estimated individually using Bayesian and maximum likelihood (ML) approaches. We applied a codon partition approach in analysing the rbcL marker in the Bayesian analysis, treating each codon position as separate and applying the corresponding selected model to each. Two sequences of Trebouxia galapagensis and T. higginsiae were selected as outgroups in the ITS analysis; six sequences of Asterochloris erici, A. excentrica, A. glomerata, A. italiana, A. magna and A. pyriformis represented the outgroups of the rbcL analysis. The outgroups were selected among species closely related to the Trebouxia ingroups as known from previous phylogenetic studies (Blaha et al., 2006; Muggia et al., 2010; Del Campo et al., 2013) and the availability of the selected molecular markers in the public database GenBank. The Bayesian Markov Chain Monte Carlo (B/MCMC) algorithm was run as implemented in MrBayes 3.1.2 (Huelsenbeck and Ronquist, 2003; Ronquist et al., 2005) with six chains simultaneously, each initiated with a random tree, for 10 million generations for the ITS analysis; trees were sampled every 100th generation for a total sample of 100 000 trees. The temperature was set at 0·05 and the nswap parameter at 2 for obtaining the best mixing between cold and heated chains. Burn-in was set at 5 million generations (the first 50 000 sampled trees) and a majority rule consensus tree was calculated from the posterior sample of 50 001 trees. The B/MCMC analysis of the rbcL locus comprised 2 million generations, trees were sampled every 100th generation for a total sample of 20 000 trees, the burn-in was set at 1 million generations (the first 10 000 sampled trees) and a majority rule consensus tree was calculated from the posterior sample of 10 001 trees. For both analyses, the log-likelihood scores were plotted against generation time using Tracer 1·4 (Rambaut and Drummond, 2007) to determine when the stationarity of likelihood values had been reached as a guide for where to set the burn-in stage (Ronquist et al., 2005). The convergence of the chains was confirmed by the convergent diagnostic of the Potential Scale Reduction Factor (PSRF), which approached 1 (Ronquist et al., 2005).
The program RAxML 7.0.4 (Stamatakis et al., 2005) was used for the ML analyses and estimation of bootstrap support. As only a single model of molecular evolution can be used across the partitions, the ML analysis was performed both for the ITS and rbcL datasets using the GTRMIX model, and 1000 bootstrap replicates were run; codon partitions were applied in the analysis of the rbcL locus.
The phylogenetic trees were visualized in TreeView (Page, 1996).
In assigning a name to the recovered clades, we used (1) the original name that clades received when they were first published [e.g. Trebouxia ‘TR1’, Trebouxia ‘TR9’ (del Campo et al., 2010), Trebouxia ‘sp.1’ (Blaha et al., 2006), Trebouxia ‘URa1–URa3’ (Ruprecht et al., 2012), Trebouxia ‘muralis I’ (Guzow-Krzemińska, 2006) and ‘BMP1–BMP5’ (Leavitt et al., 2013b)]; (2) the name of known Trebouxia species (e.g. T. impressa) which has been recognized in previous phylogentic studies (e.g. Beck et al., 1998; Kroken and Taylor, 2000; Hauck et al., 2007); or (3) the nomenclature clade I –clade V to indicate those Trebouxia lineages identified as new. Culture isolates are reported in italics to differentiate them from the original thallus samples; this format is used throughout text, figures and tables. Furthermore, sequences obtained from the axenically cultured photobionts are labelled with their own DNA extraction numbers so that they can be differentiated from those obtained from the original thallus. This helps to avoid incongruences whenever differences in photobiont identity would be recovered between thallus samples and culture isolates. We visually compared the Trebouxia photobiont clades with the Tephromela spp. mycobiont clades identified in the recent phylogenetic analysis of Muggia et al. (2014).
SSCP analysis
In order to test the possibility that unsampled variation in photobiont ITS sequences existed in Trebouxia, we applied the fingerprint analysis of SSCPs (Orita et al., 1989). After preliminary phylogenetic results, we selected 27 samples for the SSCP experiment: 17 original specimens with eight geographic origins, two individual culture isolates (L1664 and L1384), and four specimens (L1101, L1259, L1263 and L1408) together with their corresponding culture isolates (L1379, L1653, L1654 and L1660). The DNA samples were amplified using algal-specific primers for the ITS1: ITS2N (Beck et al., 1998) and nucSSU-1780-5' (Piercey-Normore and DePriest, 2001). The short ITS1 fragment obtained (300 bp) allows optimal separation of sequence variants in the SSCP analyses. The phosphorylated primer nucSSU-1780-5' was used for the exonuclease digestion to produce single-stranded fragments. PCR amplifications were performed as in Muggia et al. (2013). Amplicons were cleaned using Qiaquick spin columns (Qiagen, Vienna). Samples were prepared for SSCP experiments according to Schwieger and Tebbe (1998). Fragments were separated using the temperature gradient gel electrophoresis TGGE Maxi System (Biometra, Vienna, Austria) without a temperature gradient. The bands were visualized by silver staining, and the gel was scanned for documentation. Selected bands were excised from the gel with a sterile razor blade; the DNA was then extracted from the bands, purified and used as the template for re-amplification and sequencing using the original PCR primers (Muggia et al., 2013). PCR products were sequenced with the primer nucSSU-1780-5' (Macrogen, Inc., Amsterdam, The Netherlands) and sequences were compared with those obtained for the phylogenetic analysis.
RESULTS
Phylogenetic analyses
A total of 143 samples of Trebouxia from the original lichen thalli and 29 culture isolates could be successfully sequenced for either one or both loci. We obtained 133 ITS and 131 rbcL new sequences (Supplementary Data Table S1). The optimal models of nucleotide substitution were as follows: TIM2 + I + G for the ITS dataset, and GTR + G, TVMef + G and GTR + I + G, respectively, for the first, the second and the third codon positions of the rbcL locus. Bayesian and ML phylogenetic hypotheses were topologically congruent for both loci; the Bayesian topologies are presented separately (ITS in Fig. 1, rbcL in Supplementary Data Fig. S1).
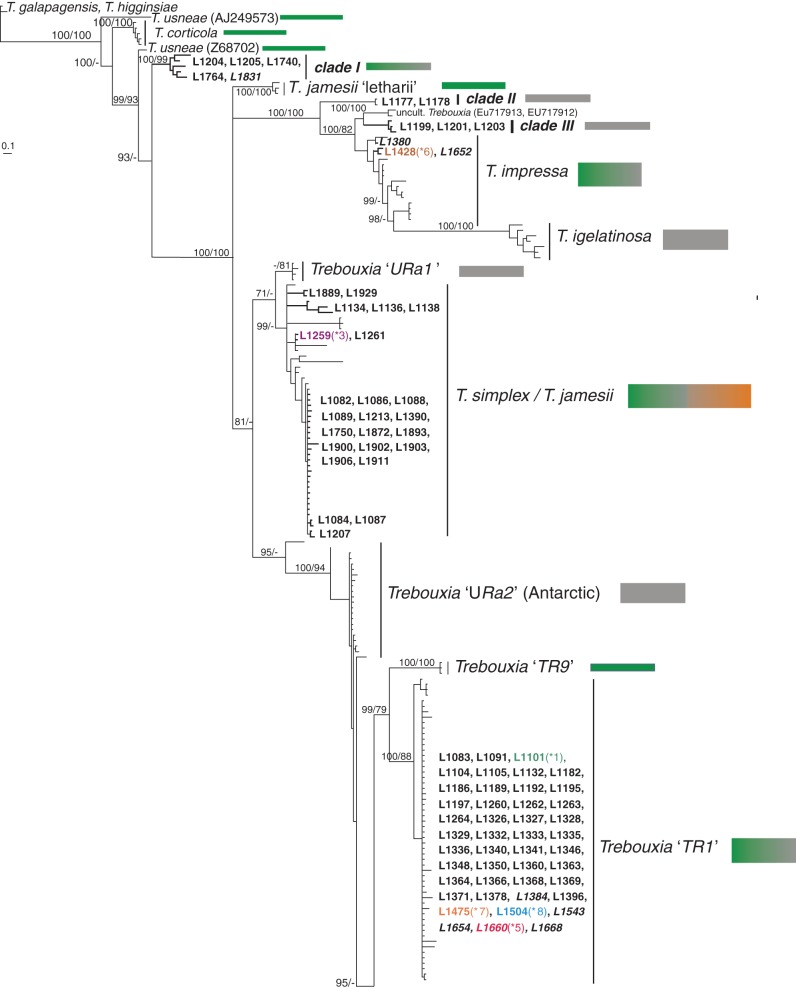
Fig. 1.
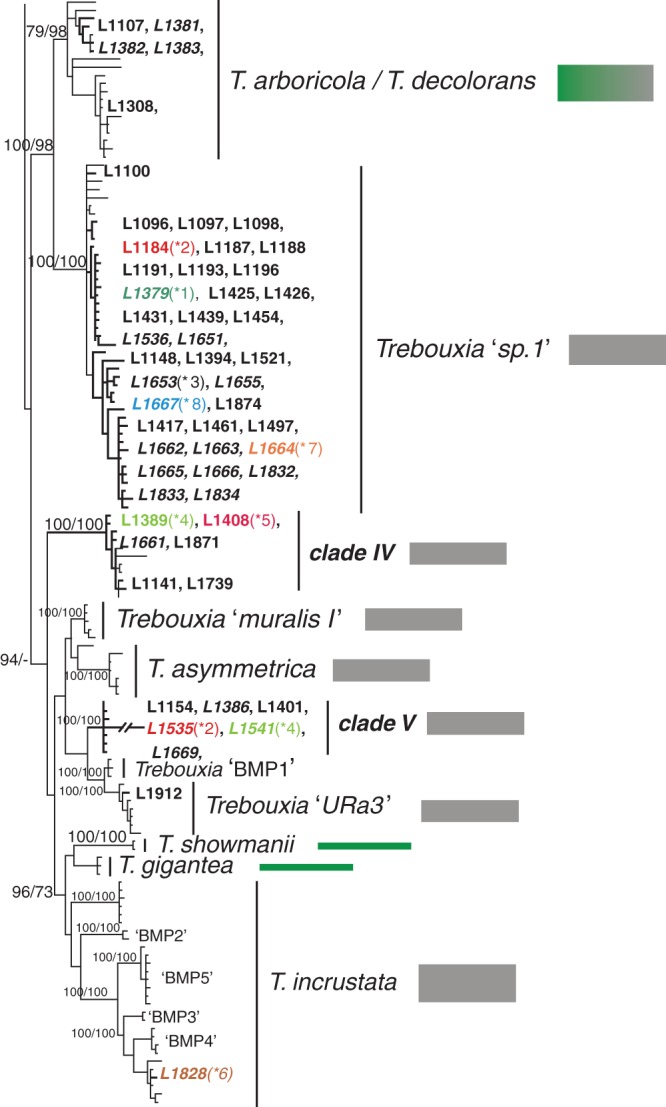
Phylogenetic hypothesis of the ITS locus of Trebouxia species: the 50 % majority rule consensus tree of the Bayesian analysis is presented; branch support (Bayesian PP >95 %/ML bootstrap values >70 %) is reported above or beside branches; branches and DNA extraction numbers of our new Trebouxia sequences (Supplementary Data, Table S1) are highlighted in bold. Incongruences (*1–8) of photobiont identities recovered between culture isolates and their original lichen thalli are colour-coded. DNA extraction numbers from cultures are in italics. Sequences retrieved from GenBank are reported in Supplementary Data Table S2. The growth substrate of lichens from which Trebouxia sequences were obtained is colour-coded beside the clades: green, bark, grey, rock (calcareous and siliceous), brown, soil.
The ITS phylogenetic hypothesis (Fig. 1) is highly supported and highly resolved. The majority of the known Trebouxia species were monophyletic. Trebouxia ‘URa2’, found in Antarctic epilithic lichens (Ruprecht et al., 2012), is the only previously recognized group which remains completely unresolved at the base of lineages Trebouxia ‘TR1’, Trebouxia ‘TR9’ and the closely related T. arboricola/T. decolorans and Trebouxia ‘sp.1’. The phylogenetic relationships are fully congruent with previous studies (Muggia et al., 2010; Ruprecht et al., 2012; Pérez-Ortega et al., 2012; del Campo et al., 2013; Leavitt et al., 2013b). Clades ‘BMP2–BMP5’ are nested within T. incrustata.
We recovered new Trebouxia sequences in 12 clades, five of which, clades I–V, are new, highly resolved lineages in Trebouxia. The lineages T. corticola, T. usneae, T. jamesii ‘letharii’, Trebouxia ‘TR9’, T. showmanii and T. gigantea include photobionts exclusively of epiphytic lichens (Fig. 1; Supplementary Data Table S2). In contrast T. simplex/T. jamesii includes photobionts from epiphytic, terricolous and epilithic lichens, and T. impressa, Trebouxia ‘TR1’ and T. arboricola/T. decolorans include photobionts from epiphytic and epilithic thalli. The remaining lineages include photobionts only from epilithic lichens. The majority of the Tephromela photobionts analysed here come from saxicolous thalli; photobionts of epiphytic specimens are found only in clade I (L1764 from Africa), in T. simplex/T. jamesii (L1134, L1136 and L1138 from Pacific North West North America; L1889 and L1929 from New Zealand) and in T. arboricola/T. decolorans (L1107 from Spain).
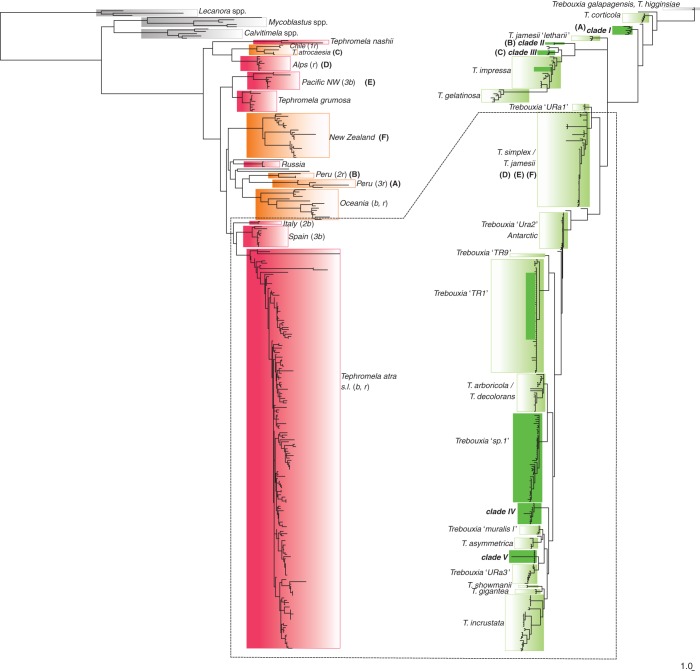
We observed six cases (A–F, Fig. 2) where Tephromela mycobiont lineages studied by Muggia et al. (2014) associate with certain Trebouxia lineages: (A) Trebouxia clade I associates with Tephromela thalli originating from Africa and Peru [fungal clade Peru (3r)]; (B, C) Trebouxia clade II and clade III are found only in Tephromela thalli from Peru, associating, respectively, with fungal lineages Peru (2r) and Tephromela atrocaesia; (D–F) Tephromela mycobionts from the Alps [fungal lineage Alps (r)], New Zealand (fungal lineage New Zealand) and the Pacific North West North America [fungal lineage Pacific NW (3b)] associate with representatives of the Trebouxia simplex complex. Trebouxia simplex also associates with mycobionts from northern European countries belonging to the fungal lineage T. atra sensu lato (s.l.; b, r) (Fig. 2; Supplementary Data Table S1). Trebouxia from Alaskan and Australian thalli represent separate lineages, respectively, named here clade IV and clade V. These Trebouxia lineages do not associate, however, with mycobionts from the lineages New Zealand and Oceania (Fig. 2) but with mycobionts found in lineage T. atra s.l. (b, r). Mycobionts of this latter lineage (Fig. 2) – the majority coming from temperate, boreal and Mediterranean regions – form symbioses mainly with photobionts belonging to Trebouxia ‘TR1’, T. arboricola/T. decolorans and Trebouxia ‘sp.1’. Three sequences from thalli and culture isolates (L1428, L1380 and L1652) are found within T. impressa and one culture isolate (L1828) within T. incrustata clades.
Fig. 2.
Comparison of Tephromela mycobiont (left) and Trebouxia photobiont (right) phylogenies. A high specificity of mycobiont–photobiont association is indicated by letters (A–F); low specificity between mycobionts and photobionts is highlighted by a dashed line. Mycobiont phylogeny is retrieved from Muggia et al. (2014).
Though differences in sequence datasets exist, the phylogenetic hypothesis based on the rbcL locus (Supplementary Data Fig. S1) recognizes the majority of the clades found in the ITS analysis. The backbone phylogeny is highly supported by Bayesian and ML values, but clades remain partly unresolved or paraphyletic (Supplementary Data Fig. S1).
Analyses of the cultured photobionts
Isolation of algal photobionts was successful for 30 samples from 11 geographic origins. According to sequence data, these isolates belong to eight Trebouxia lineages: T. impressa, Trebouxia ‘TR1’, T. arboricola/T. decolorans, Trebouxia ‘sp.1’, clade I, clade IV, clade V and T. incrustata (Fig. 1; Supplementary Data Table S3). The phylogenetic analysis (Fig. 1) revealed eight incongruences between the results obtained from the culture isolation and the direct sequencing approaches: (1) Trebouxia sequences from the lineage Trebouxia ‘TR1’ were amplified from the original lichen samples L1101 (Spain), L1475 (Italy) and L1504 (France) but Trebouxia ‘sp.1’ was isolated in culture (L1379, L1664 and L1667); (2) T. simplex/T. jamesii was amplified from L1259 (Finland) but the culture isolate L1653 represents Trebouxia ‘sp.1’; (3) Trebouxia sequences of clade V were amplified from samples L1389 and L1408 (Alaska) but corresponding culture isolates belong to clade VI (L1541) and to clade Trebouxia ‘TR1’ (L1660), respectively; (4) Trebouxia ‘sp.1’ was amplified from sample L1184 (Spain) but the culture isolate L1535 belongs to clade V; (5) T. impressa was amplified from sample L1428 but the culture isolate L1828 represents T. incrustata (Supplementary Data Table S3). The photobionts (culture isolates L1381, L1386, L1654, L1543, L1663 and L1655; Supplementary Data Table S3) isolated from six samples (L1107, L1154, L1263, L1369, L1461 and L1521) corresponded to those amplified directly from the original thalli. The comparison between culture and thallus sequences was not possible for 16 samples, due to unsuccessful amplification or sequencing either of the original thallus or of the culture isolate. Photobionts from Alpine, Chilean and Peruvian samples failed to grow in cultures even after repeated attempts.
SSCP analysis
We excised and re-amplified 48 bands from the SSCP gel, and 23 of these were successfully sequenced (Fig. 3). Sequencing results were manually compared with the sequences of the whole ITS fragments. We succeed in sequencing up to three bands for one sample (e.g. L1259). The banding patterns between original samples and their corresponding algal isolates differ in intensity and presence of bands. There are cases in which bands are shared between samples (either original thalli or culture isolates) whose photobiont sequences were assigned to the same phylogenetic clade in the ITS analysis (Fig. 1): clade II (L1177 and L1178), Trebouxia ‘TR1’ (L1660, L1101, L1384, L1654, L1263, L1369, L1504 and L1182) and Trebouxia ‘sp.1’ (L1379, L1184, L1664 and L1461). Samples belonging to clade III (L1199 and L1203) have identical banding patterns (Fig. 3). Although single ITS fragments were expected from axenic culture isolates, only the isolate L1664 presents a single band, whereas L1653, L1660, L1379, L1384 and L1654 yielded multiple bands.
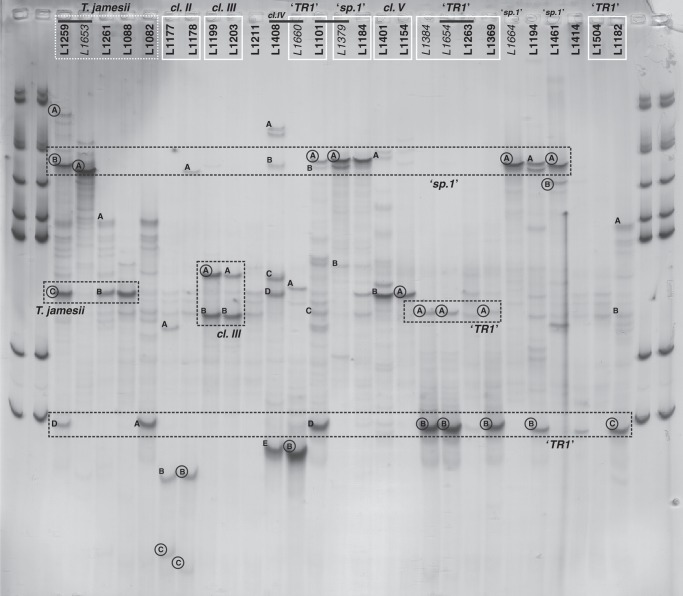
Fig. 3.
Single strand conformation polymorphism (SSCP) analysis of ITS1 fragments of Trebouxia photobionts. Selected samples of Tephromela spp. and cultured photobionts were analysed. Bands excised from the gel are marked by letters (A–D); successfully sequenced bands are additionally encircled. Black bars above the numbers join original lichen samples to their corresponding photobiont isolates. The affiliations of the photobiont samples – identified in the Trebouxia phylogeny of Fig. 1 – are reported above the sample numbers and grouped by white lines. The position of SSCP bands corresponding to Trebouxia ‘sp.1’ and Trebouxia ‘TR1’ sequences (as they are the most frequent bands) is shown by black dashed lines.
In 13 samples we recovered bands with ITS sequence identical to that obtained from either culture-based or direct amplification of corresponding thallus samples: (1) L1259C corresponds to T. simplex of thalli L1082, L1088 and L1261 (Fig. 1) – this band is also present in the SSCP samples L1261 and L1088 but was not sequenced from them; (2) L1653A corresponds to Trebouxia ‘sp.1’; (3) L1778B corresponds to the sequence of L1778 in clade II; (4) L1199A corresponds to the sequence of L1199 in clade III; (5) L1101A, L1379A, L1664A and L1461A correspond to the sequences L1379, L1664 and L1461 in Trebouxia ‘sp.1’; and (6) L1384B, L1654B, L1369B, L1194B and L1182C correspond to the sequences of L1384, L1654, L1369 and L1182 in Trebouxia ‘TR1’. The sample L1194 was not successfully sequenced for the phylogenetic analysis but the PCR amplification succeeded for the SSCP experiment. We cannot therefore compare sequences for this sample, but the sequence from the SSCP band L1194B is the same as L1384B, L1654B, L1369B and L1182C. In other cases, sequences recovered from bands differ to a variable degree from those obtained from culture-based or thallus samples, and bands at the same run length do not necessarily represent the same sequence. The sequence from L1660B differs by only 6 bp from that sequenced in the phylogeny corresponding to Trebouxia ‘TR1’. The sequence from L1154A neither matches the sequence in clade V from the original sample, nor corresponds to the sequenced L1259C at the same height in the SSCP gel (Fig. 3). The sequence L1259A is an uncultured Trebouxia and differs by seven nucleotides from L1653A corresponding to Trebouxia ‘sp.1’. Bands L1259B and L1653A differ in >15 nucleotides; bands L1384A and L1654A differ by 2 bp from L1384B and L1654B; and bands L1461A and L1461B differ by six nucleotides. Asterochloris sequences have been obtained from the two bands L1177C and L1178C (Fig. 3).
Morphological and anatomical analysis of cultured photobionts
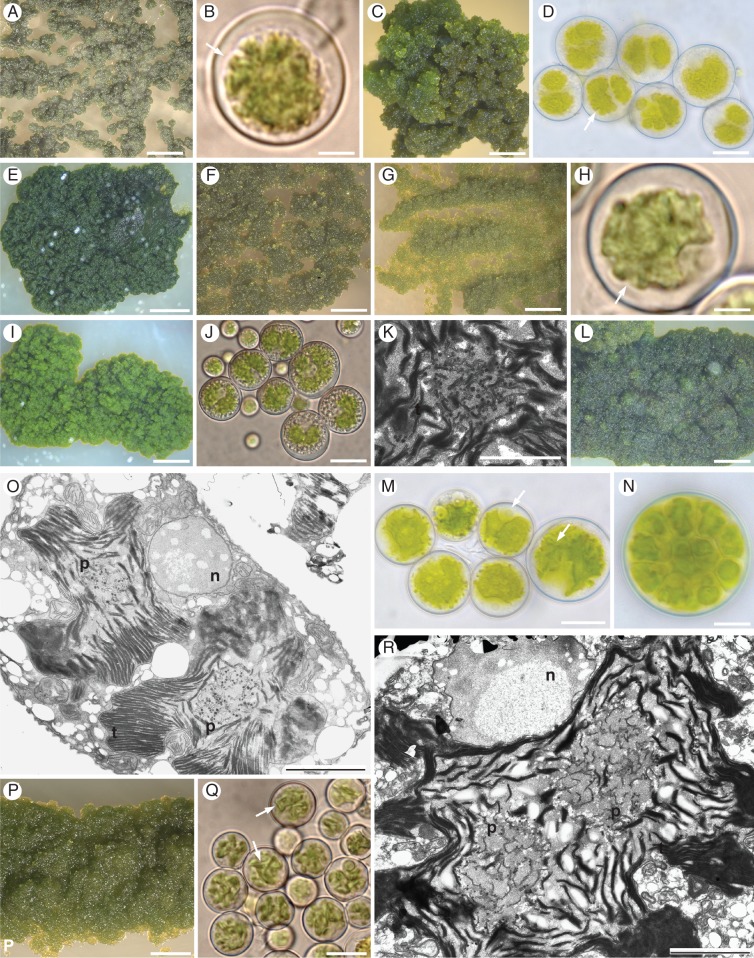
Cultured algal colonies differed in growth rate and type (Supplementary Data Table S3), and many required substantial time to develop. ‘Fast growing’ photobionts had well-developed colonies in <2 months, while ‘slow growing’ algae required twice as long to reach the same size. Some colonies had a brittle morphology and presented rough surfaces (Fig. 4A, C, F, G), whereas others were rather slimy to glossy (Fig. 4E, I, L). Variation in growth rate and type is not correlated with the phylogenetic assignments of the isolates: different morphologies are observed among isolates belonging to the same lineage but coming from different lichen thalli (Supplementary Data Table S3). Zoospores were never observed in photobionts; up to 32 daughter cells were counted in autospores. Chloroplasts with short, indistinct lobes, characteristics of T. impressa type sensu Friedl (1989), support the molecular identification of the strains as T. impressa (Fig. 4D). Chloroplast morphology studied in strains from the lineages Trebouxia ‘sp.1’ and Trebouxia ‘TR1’ – from seven different origins – showed high similarity to the T. arboricola type and T. decolorans type, characterized by numerous, splitting and finely divided lobes (Fig. 4M, Q). The chloroplast morphology of the strain of clade I (Fig. 4B) is highly similar to the T. impressa type; chloroplast of clade IV and clade V with wider lobes resembling the T. gigantea-type (Fig. 4H, J). The pyrenoid type was studied only for Trebouxia ‘sp.1’ and clade V, as these clades turned out to be the sole new clades in our primary phylogenetic analysis. The pyrenoid type of these strains strongly corresponds to the gigantea type sensu Friedl (1989) (Fig. 4K, O, R).
Fig. 4.
Morphology and anatomy of Trebouxia species isolated in axenic culture. The cultures are named according to the DNA extraction numbers (Supplementary Data Tables S1 and S3); the name of the corresponding Trebouxia lineage of Fig. 1 is reported in parentheses. (A, C, E–G, I, L, P) Algal colonies on TM medium. (B, D, H, J, M, N, Q) Algal cells observed by light microscopy; arrows indicate the lobes of the chloroplast (central green body) and the nucleus; (N) Autospore; 12 daughter cells are visible. (K, O, R) Transmission electron microscopy (TEM) microphotographs of algal cells: nucleus (n), pyrenoid (p) – in detail in (K) – and thylakoid (t); the presence of two pyrenoids in (O) and (R) is probably due to the first duplication that chloroplasts undergo before the first cell division in generating autospores. (A, B) L1831 (clade I); (C, D) L1380 (T. impressa); (E) L1382 (T. arboricola); (F) L1828 (T. incrustata); (G, H) L1661, (clade IV); (I–K, O) L1535 (clade V); (L–N, R) L1379 (Trebouxia ‘sp.1’), (P, Q) L1654 (Trebouxia ‘TR1’). Scale bars: P = 0·5 mm; C = 1 mm, A, F, G, L = 2 mm; E, I = 3 mm; D, J = 25 μm; M, N, Q = 10 μm; B, H = 4 μm; K, O, R = 2 μm.
DISCUSSION
Diversity of Trebouxia as symbiont of Tephromela
According to our present analysis, the photobionts in Tephromela belong to at least 12 lineages within Trebouxia. A total of 113 photobionts could clearly be assigned to known lineages, whereas five clades apparently represent newly recovered lineages in Trebouxia. New monophyletic lineages in lichenized Trebouxia species have also been found in the past few years by other authors (Casano et al., 2011; Leavitt et al., 2013b); additional diversity is expected in future studies, especially from poorly sampled regions. The newly recovered Trebouxia lineages in this study originate from geographic regions (e.g. Alaska, Russia and the Southern Hemisphere) which are in fact still poorly explored. Further analysis of coccoid green algal photobionts in these regions will make it possible to partition out the effects of geographical distribution vs. selectivity in the future.
Ultrastructural traits of chloroplasts and pyrenoids from axenically cultured photobionts have been used to characterize Trebouxia species in the past (Friedl, 1989). However, attempts to use morpho-anatomical characters to relate the detected new lineages to described species revealed either homoplasy of such characters or their presence across closely related species (non-exclusivity). Until the significance of ultrastructural traits is better understood using molecular data, we avoid taxonomic implications and refrain from premature description of new species in Trebouxia. Clearly the species concepts in lichenized Trebouxia need to be revised as serious problems are evident in identifying and delimiting species within this genus of photobionts. Different informal naming schemes recently emerged for photobiont lineages and might result in inconsistencies of classification until any standardized approach is established. Recently, O'Brien (2013) compiled a large phylogenetic analysis of Trebouxia species and summarized association patterns between myco- and photobionts. He labelled five new Trebouxia clades (sp.1–sp.5), two of which, Trebouxia sp.1 and Trebouxia sp.2 (O'Brien, 2013), can be linked to the clades originally identified as Trebouxia ‘TR9’ (del Campo et al., 2010) and Trebouxia ‘TR1’ (del Campo et al., 2010), respectively. The other three clades O'Brien (2013) names (sp.3–sp.5) cannot be linked with any of our clades I–V. Furthermore, phylogenetic and morphological analyses previously suggested that Trebouxia ‘TR9’ (del Campo et al., 2010) corresponded to the lineage Trebouxia ‘sp.1’ (Blaha et al., 2006; Muggia et al., 2010). However, our present extended taxon sampling recognizes them as two distinct lineages. We further show that Trebouxia ‘TR1’ and Trebouxia‘sp.1’ are not restricted to the Mediterranean region. On the other hand, the clades named BMP2–BMP5 were originally suggested to be new Trebouxia clades (Leavitt et al., 2013b), but they are sub-clades of Trebouxia incrustata in our wider sampling. Irrespective of the status as separate species or intraspecific categories, we think that the resolution of the commonly used ITS marker is often insufficient to segregate ecologically adapted strains of Trebouxia. The taxonomic uncertainty in Trebouxia also has implications for ecological studies, as the definition of mycobiont–photobiont relationships (hence selectivity and specificity of the symbionts) rely on taxon concepts and may occur at low taxonomic levels (Pérez-Ortega et al., 2012; Werth, 2012).
Specificity and selectivity in cosmopolitan lichen symbioses
The worldwide sampling in the present work helps to distinguish between specificity (the range of possible photobionts) and selectivity (the selected photobionts from a pool of possible photobionts) and to assess if the T. atra species complex can be classified by the scheme of Yahr et al. (2004), which distinguished between partner generalists, specialists and intermediates. Our results indicated that lineages of Tephromela are variable regarding its symbiotic association patterns. Specific mycobiont–photobiont associations were observed in six geographically circumscribed and monophyletic Tephromela lineages, each with one of four Trebouxia lineages. The Tephromela species growing in the Alps, under oceanic conditions (such as the lineages Alps, Pacific NW and New Zealand) and at high elevations in Peru are highly specific towards their photobiont. They associate only with photobionts from the T. simplex (‘T. jamesii’) complex or with the newly recovered Trebouxia lineages clade I–III, respectively. Photobionts of the Trebouxia simplex complex were commonly assigned to T. jamesii until the type of the latter species was shown to be more closely related to T. arboricola by Beck (2002). Trebouxia simplex is commonly found in lichens from cool habitats (e.g. of arctic–alpine distribution patterns), which occur with acid substrate conditions (Blaha et al., 2006; Muggia et al., 2010; Domaschke et al., 2012). On the other hand, one worldwide distributed lineage, assigned to T. atra s.l. (Muggia et al., 2014), associates with a much wider spectrum of photobionts comprising up to seven different Trebouxia lineages, reflecting a low selectivity of the mycobionts. Interestingly mycobiont and photobiont lineages clearly demonstrate different levels of selectivity, and their associations correlate with both their geographic distribution and ecological conditions.
The high diversity of photobionts is recovered in the geographically widespread T. atra lineage, which thrives on a wide range of different substrates and ecological conditions, including sea shores and dry Mediterranean regions, but also in cold arctic habitats. This pattern agrees with previous studies of ecologically variable species of crust-forming lichens which associate with a broader range of algae (e.g. Lecanora muralis, Guzow-Krzemińska, 2006; Muggia et al., 2013). Blaha et al. (2006) discovered that Mediterranean and lowland samples of the widespread lichen species Lecanora rupicola associated with a range of photobionts, which differed from alpine habitats, where the regular association with Trebouxia simplex was found. Other lichens, usually not presenting the crustose growth type, may also be widely distributed, but when they are ecologically more restricted they often have narrower photobiont ranges, probably explained by habitat-dependent patterns. Cetraria aculeata, occurring on acid ground in arctic–alpine habitats, is a well-studied example for a narrow range of photobionts (Domaschke et al., 2012). Mycobionts in the temperate region are consistently associated with a specific photobiont lineage, yet a photobiont switch in the past apparently enabled C. aculeata to colonize temperate as well as polar habitats, as found by Fernandez-Mendoza et al. (2011). These authors suggest that rare photobiont switches expand the geographical and ecological range of lichen mycobionts by association with locally adapted photobionts in climatically different habitats. This can also be interpreted as an example of habitat-adapted symbiosis, a concept recently introduced to explain the ecologically correlated occurrence of different fungal endophytes in plants (Rodriguez et al., 2008). Similarly, patterns of local adaptation correlated with host associations are also widespread in insect–plant and coral–algae mutualistic symbioses (Fox and Morrow, 1981; Byler et al., 2013). Specialization to certain ecological settings mirrors the ability of a population or a community to respond to features of the surrounding environment, rather than the attribute of a species throughout its geographical range (Fox and Morrow, 1981). In coral–algae symbiosis, the association with a photobiont species may persist, but more advantageous symbionts (algae) can be acquired to continue the beneficial symbiosis if environmental conditions change (Byler et al., 2013). More recently, significant photobiont flexibility has also been revealed in rock-inhabiting Xanthoparmelia species. Though no clear geographic patterns were identified, the local structure found with Trebouxia populations from sub-alpine meadows was significantly different from that of other niches (Leavitt et al., 2013b). We assume that the wider photobiont range of T. atra is a means to increase its ecological tolerance, the extent of its niche and hence its distribution range. Alternatively, the association with locally adapted photobionts (e.g. Trebouxia clade I–III or T. simplex) may have facilitated the persistence of the partnership in hostile environments and possible co-divergence of the symbionts. It is also noteworthy that the highly adapted photobionts of the six specific associations between Tephromela and Trebouxia (A–F) did not grow in cultures. So far, however, we cannot predict whether a low level of culturability is linked with specialization.
Plurality of photobionts in lichen thalli
The occurrence of multiple photobionts in individual lichen thalli was detected in foliose, epiphytic lichens (Piercey-Normore, 2006; del Campo et al., 2010; Mansournia et al., 2012; Nyati et al., 2013), as well as for crustose, epilithic lichens (Muggia et al., 2013). Recently, the application of microsatellite markers provided a detailed picture of algal plurality in lichen thalli (Dal Grande et al., 2014).
We repeatedly observed the algal plurality in T. atra specimens of different geographic origins, but the biological significance is not clear. Studies of the vegetatively propagating Ramalina farinacea from the Mediterranean regions showed consistent co-occurrence of Trebouxia ‘TR1’ and its sister taxon Trebouxia ‘TR9’ (del Campo et al., 2010). Experiments with these culture isolates demonstrated that they show different physiological responses to variable temperatures and irradiations, as well as tolerance to lead (Álvarez et al., 2012). Hence, it was hypothesized that photobiont plurality might contribute to symbiotic stability when environmental conditions are changeable (Casano et al., 2011). In sexually reproducing T. atra, where germinating fungal spores can develop symbiotic thalli only by resynthesis with suitable photobiont(s), an alternative hypothesis could be considered. It has been observed that lichen-forming fungi survive in association with algal strains until they encounter a suitable strain for thallus formation (Ott, 1987). We believe that sub-optimal strains might be maintained within the thallus at low frequency compared with the most abundant and preferred photobiont strain. The latter, dominating strain would be the one which creates the selectivity and specificity pattern found among our samples. This hypothesis requires further testing by separating and quantifying the different strains, e.g. with quantitative PCR. Potential primer bias may also be a problem in assessing differences in photobiont strain diversity among culture isolates and original thalli. We indeed experienced a very low success in amplifying some photobiont strains from our samples – only 143 individuals out of 300 could be sequenced. Obtaining sequences was straightforward for photobionts belonging to T. simpex, Trebouxia ‘TR1’, T. arboricola/T. decolorans and Trebouxia ‘sp.1’, while other were more difficult using PCR approaches. A similar primer bias effect has also been experienced recently in Antarctic lichens (Pérez-Ortega et al., 2012). Further, we found additional ITS alleles in individual cultures by sequencing SSCP bands. These corresponded neither to amplified products from the thallus nor to those from culture isolates. This phenomenon may hint either at inhomogeneity of the culture isolates or at an underestimated intragenomic diversity of the algal strains. This latter possibility can only be tested when Trebouxia genomes are available.
Conclusions
Differential patterns of photobiont selectivity in the T. atra species complex may have consequences on the genetic structure of the fungal species. We observed that narrow photobiont selectivity correlates with six, well-resolved, monophyletic fungal clades which seem to be locally adapted, whereas broad selectivity corresponds to poor phylogenetic resolution of allelic richness in the widely distributed and ecologically plastic T. atra sensu stricto. Acceptance of a wide range of photobionts dramatically increases the population sizes of the fungal partners. As a consequence, genetic drift could be significantly retarded. Previously, slow genetic drift and the persistence of ancestral alleles have explained the genetic structure of a widely disjunct lichen species (Printzen et al., 2003). We also argue that association with a broad range of photobionts could also represent a bet-hedging strategy in anticipation of ecological changes (Beaumont et al., 2009). Germinating spores are left with potentially suitable photobionts encountered among a heterogeneous algal pool. Considering unpredictable ecological conditions, the mycobiont might therefore increase the likelihood of symbiotic success by temporarily associating with multiple photobionts (one of which might prove suitable for thallus formation). Problems with ecological changes have been observed in a common lichen species, Lecanora conizaeoides, which associates only with one algal lineage (Trebouxia simplex; Hauck et al., 2007). Even slight changes in abiotic parameters, such as raised substrate pH, have led to extensive and wide-ranging dieback of this ecologically restricted species (Hauck et al., 2011). We hypothesize that lichens with ample ecological niche and wide photobiont selectivity could better adapt to buffering ecological alterations by associating with alternative photobionts. The role of symbiont selectivity in adaptation merits further study in evolutionary biology. Habitat-adapted symbioses have not only been found in symbioses involving fungi, such as endophytes and lichens, but also in corals (Rowan, 2004), and may represent a general principle in evolution.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
L.M. and M.G. are grateful to the Austrian Science Foundation for financial support (L.M. for FWF Herta-Firnberg Project T481-B). S.P.O. is supported by the grant CTM2012-38222-467 C02-02 from the Spanish Ministry of Economy and Competitiveness. We thank colleague and friends for shipping specimens from their regions: Paul Kirika (Africa); Curtis Bjork and Bruce McCune (North America); Maria Ines Messuti (South America); Rebecca Yahr, Ave Suija, Mats Wedin, Jan Vondrak, Jaroslav Šoun, Philip Resl, Sergio Favero-Longo, Filipp Högnabba, Richard Tafner, Helmut Mayrhofer (Europe); Lidia Yakovchenko (Asian Russia); Yoshihito Ohmura (Japan); Jack Elix (Australia); and Gintaras Kantvilas (Tasmania). We thank Josef Hafellner for joint field work, Stefan Möstl for the help with the lab work, and Rebecca Yahr for revising the text.
LITERATURE CITED
- Ahmadjian V. A guide to the algae occurring as lichen symbionts: isolation, culture, cultural physiology, and identification. Phycologia. 1967;6:127–160. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez R, del Hoyo A, García-Breijo F, et al. Different strategies to achieve Pb-tolerance by the two Trebouxia algae coexisting in the lichen Ramalina farinacea. Journal of Plant Physiology. 2012;169:1797–806. doi: 10.1016/j.jplph.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Beaumont HJE, Gallie J, Kost C, Ferguson GC, Rainey PB. Experimental evolution of bet hedging. Nature Letters. 2009;462:90–93. doi: 10.1038/nature08504. [DOI] [PubMed] [Google Scholar]
- Beck A. Photobionts: diversity and selectivity in lichen symbioses. International Lichenological Newsletter. 2002;35:18–24. [Google Scholar]
- Beck A, Friedel T, Rambold G. Selectivity of photobiont choice in a defined lichen community: inference from cultural and molecular studies. New Phytologist. 1998;139:709–720. [Google Scholar]
- Blaha J, Baloch E, Grube M. High photobiont diversity in symbioses of the euryoecious lichen Lecanora rupicola (Lecanoraceae, Ascomycota) Biological Journal of the Linnean Society. 2006;88:283–293. [Google Scholar]
- Bold HC. The morphology of Chlamydomonas chlamydogama sp. nov. Bulletin of the Torrey Botanical Club. 1949;76:101–108. [Google Scholar]
- Bischoff HW, Bold HC. Phycological studies IV. Some soil algae from Enchanted Rock and related algal species. Univeristy of Texas Publication; 1963. pp. 1–95. 6318. [Google Scholar]
- Byler KA, Carmi-Veal M, Fine M, Goulet TL. Multiple symbiony acquisition strategies as an adaptive mechanism in the coral Stylophora pistillata. PLoS One. 2013;8:e59596. doi: 10.1371/journal.pone.0059596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campo EM, Grimeno J, De Nova JPG, et al. South European populations of Ramalina farinacea (L.) Ach. share different Trebouxia algae. In: Nash TH III, Geiser L, McCune B, Triebel D, Tomescu AMF, Sanders WB, editors. Biology of lichens – symbiosis, ecology, environmental monitoring, systematics and cyber applications. . Stuttgart: J. Cramer in der Gebrüder Borntraeger Verlagsbuchhandlung; 2010. pp. 247–256. Bibliotheca Lichenologica No. 105. [Google Scholar]
- del Campo EM, Cabalá S, Gimeno J, et al. The genetic structure of the cosmopolitan three-partner lichen Ramalina farinacea evidences the concerted diversification of symbionts. FEMS Microbiology Ecology. 2013;83:310–323. doi: 10.1111/j.1574-6941.2012.01474.x. [DOI] [PubMed] [Google Scholar]
- Casano LM, del Campo EM, García-Breijo FJ, et al. Two Trebouxia algae with different physiological performances are ever-present in lichen thalli of Ramalina farinacea. Coexistence versus competition? Environmental Microbiology. 2011;13:806–818. doi: 10.1111/j.1462-2920.2010.02386.x. [DOI] [PubMed] [Google Scholar]
- Crespo A, Kauff F, Divakar P, et al. Phylogenetic generic classification of parmelioid lichens (Parmeliaceae, Ascomycota) based on molecular, morphological and chemical evidence. Taxon. 2010;59:1735–1753. [Google Scholar]
- Cubero OF, Crespo A, Fatehi J, Bridge PD. DNA extraction and PCR amplification method suitable for fresh, herbarium stored and lichenized fungi. Plant Systematics and Evolution. 1999;217:243–249. [Google Scholar]
- Dal Grande F, Alors D, Divakar PK, Balint M, Crespo A, Schmitt I. Insights into intrathalline genetic diversity of the cosmopolitan lichen symbiotic green algae Trebouxia decolorans Ahmadjian using microsatellites markers. Molecular Phylogenetics and Evolution. 2014;72:54–60. doi: 10.1016/j.ympev.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Domaschke S, Fernández-Mendoza F, García MA, Martín MP, Printzen C. Low genetic diversity in Antarctic populations of the lichen-forming ascomycete Cetraria aculeata and its photobionts. Polar Research. 2012;31:17353. [Google Scholar]
- Fernandez-Mendoza F, Domaschke S, Garcia MA, Jordan P, Martin MP, Printzen C. Population structure of mycobionts and photobionts of the widespread lichen Cetraria aculeata. Molecular Ecology. 2011;20:1208–1232. doi: 10.1111/j.1365-294X.2010.04993.x. [DOI] [PubMed] [Google Scholar]
- Fox LR, Morrow PA. Specialization: species property or local phenomenon? Science. 1981;211:887–893. doi: 10.1126/science.211.4485.887. [DOI] [PubMed] [Google Scholar]
- Friedl T. Systematik und Biologie von Trebouxia (Microthamniales, Chlorophyta) als Phycobiont der Parmeliaceae (lichenisierte Ascomyceten) Bayreuth: Universität Bayreuth; 1989. [Google Scholar]
- Grube M, Kroken S. Molecular approaches and the concept of species and species complexes in lichenized fungi. Mycological Research. 2000;104:1284–1294. [Google Scholar]
- Grube M, Lindblom L, Mayrhofer H. Contribution to the lichen flora of Crete: a compilation of references and some new records. Studia Geobotanica. 2001;20:41–59. [Google Scholar]
- Guzow-Krzemińska B. Photobiont flexibility in the lichen Protoparmeliopsis muralis as revealed by ITS rDNA analysis. The Lichenologist. 2006;38:469–476. [Google Scholar]
- Hall TA. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symposium Series. 1999;41:95–98. [Google Scholar]
- Hauck M, Helms G, Friedl T. Photobiont selectivity in the epiphytic lichens Hypogymnia physodes and Lecanora conizaeoides. The Lichenologist. 2007;39:195–204. [Google Scholar]
- Hauck M, Otto PI, Dittrich S, et al. Small increase in sub-stratum pH causes the dieback of one of Europe's most common lichens, Lecanora conizaeoides. Annals of Botany. 2011;108:359–366. doi: 10.1093/aob/mcr136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger R. The symbiotic phenotype of lichen-forming ascomycetes and their endo- and epibionts. In: Esser K, editor. The Mycota – a comprehensive treatise on fungi as experimental system for basic and applied research. Fungal association IX. 2nd edn. Berlin: Springer; 2012. pp. 288–339. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Kalb K, Hafellner J. Bemerkenswerte Flechten und lichenicole Pilze von der Insel Madeira. Herzogia. 1992;9:45–102. [Google Scholar]
- Kroken S, Taylor JW. Phylogenetic species, reproductive mode, and specificity of the green alga Trebouxia forming lichens with the fungal genus Letharia. The Bryologist. 2000;103:645–660. [Google Scholar]
- Leavitt SD, Fernández-Mendoza F, Pérez-Ortega S, et al. Local representation of global diversity in a cosmopolitan lichen-forming fungal species complex (Rhizoplaca, Ascomycota) Journal of Biogeography. 2013a;40:1792–1806. [Google Scholar]
- Leavitt SD, Nelsen MP, Lumbsch HT, Johnson LA, St. Clair LL. Symbiont flexibility in subalpine rock shield lichen communities in the Southwestern USA. The Bryologist. 2013b;116:149–161. [Google Scholar]
- Mansournia MR, Wu B, Matsushita M, Hogetsu T. Genotypic analysis of the foliose lichen Parmotrema tinctorum using microsatellite markers: association of mycobiont and photobiont, and their reproductive modes. The Lichenologist. 2012;44:419–440. [Google Scholar]
- Miadlikowska J, Kauff F, Hofstetter V, et al. New insights into classification and evolution of the Lecanoromycetes (Pezizomycotina, Ascomycota) from phylogenetic analyses of three ribosomal RNA and two protein-coding genes. Mycologia. 2006;98:1088–1103. [PubMed] [Google Scholar]
- Muggia L, Grube M. Fungal composition of lichen thalli assessed by single strand conformation polymorphism. The Lichenologist. 2010;42:461–473. [Google Scholar]
- Muggia L, Grube M, Tretiach M. Genetic diversity and photobiont associations in selected taxa of the Tephromela atra group (Lecanorales, lichenised Ascomycota) Mycological Progress. 2008;7:147–160. [Google Scholar]
- Muggia L, Zellnig G, Rabensteiner J, Grube M. Morphological and phylogenetic study of algal partners associated with the lichen-forming fungus Tephromela atra from the Mediterranean region. Symbiosis. 2010;51:149–160. [Google Scholar]
- Muggia L, Vancurova L, Skaloud P, Peksa O, Wedin M, Grube M. The symbiotic playground of lichen thalli – a highly flexible photobiont association in rock-inhabiting lichens. FEMS Microbiology Ecology. 2013;85:313–323. doi: 10.1111/1574-6941.12120. [DOI] [PubMed] [Google Scholar]
- Muggia L, Pérez-Ortega S, Fryday A, Spribille T, Grube M. Global assessment of genetic variation and phenotypic plasticity in the lichen-forming species Tephromela atra. Fungal Diversity. 2014;64:233–251. [Google Scholar]
- Nash TH., III . Lichen biology. 2nd edn. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- Nimis PL, Martellos S. A second checklist of the lichens of Italy with a thesaurus of synonyms. Monografie del Museo Regionale di Scienze Naturali. 4th edn. Valle d'Aosta, Italy: Museo Regionale di Scienze Naturali Saint-Pierre; 2003. [Google Scholar]
- Nozaki H, Ito M, Sano R, Uchida H, Watanabe MM, Kuroiwa T. Phylogenetic relationships within the colonial Volvocales (Chlorophyta) inferred from rbcL gene sequence data. Journal of Phycology. 1995;31:970–979. [Google Scholar]
- Nyati S, Werth S, Honegger R. Genetic diversity of sterile cultured Trebouxia photobionts associated with the lichen-forming fungus Xanthoria parietina visualized with RAPD-PCR fingerprinting techniques. The Lichenologist. 2013;45:825–840. [Google Scholar]
- O'Brien H. A preliminary look at host association patterns in Trebouxia. Photobiont Diversity. 2013. wordpress.com : http://dx.doi.org/10.6084/m9.figshare.711786 .
- Orita M, Iwahan H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proceeding of the National Academy of Sciences, USA. 1989;86:2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott S. Sexual reproduction and developmental adaptations in Xanthoria parietina. Nordic Journal of Botany. 1987;7:219–228. [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pérez-Ortega S, Ortiz-Álvarez S, Allan Green TG, de los Ríos A. Lichen myco- and photobiont diversity and their relationships at the edge of life (McMurdo Dry Valleys, Antarctica) FEMS Microbiology Ecology. 2012;82:429–448. doi: 10.1111/j.1574-6941.2012.01422.x. [DOI] [PubMed] [Google Scholar]
- Piercey-Normore MD. The lichen-forming ascomycete Evernia mesomorpha associates with multiple genotypes of Trebouxia jamesii. New Phytologist. 2006;169:331–344. doi: 10.1111/j.1469-8137.2005.01576.x. [DOI] [PubMed] [Google Scholar]
- Piercey-Normore MD, DePriest PT. Algal switching among lichen symbioses. American Journal of Botany. 2001;88:1490–1498. [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest – testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Printzen C, Ekman S, Tonsberg T. Phylogeography of Cavernularia hultenii: evidence of slow genetic drift in a widely disjunct lichen. Molecular Ecology. 2003;12:1473–1486. doi: 10.1046/j.1365-294x.2003.01812.x. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. Tracer. 2007. Available from: beast.bio.ed.ac.uk/Tracer . [Google Scholar]
- Rodriguez RJ, Henson J, Van Volkenburgh E, et al. Stress tolerance in plants via habitat-adapted symbiosis. ISME Journal. 2008;2:404–416. doi: 10.1038/ismej.2007.106. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP, Van der Mark P. 2005. MrBayes 3·1 Manual. http://mrbayes.csit.fsu.edu/mb3.1_manual.pdf . [Google Scholar]
- Rowan R. Coral bleaching: thermal adaptation in reef coral symbionts. Nature. 2004;430:742. doi: 10.1038/430742a. [DOI] [PubMed] [Google Scholar]
- Ruprecht U, Brunauer G, Printzen C. Genetic diversity of photobionts in Antarctic lecideoid lichens from an ecological view point. The Lichenologist. 2012;44:661–678. [Google Scholar]
- Schwieger F, Tebbe CC. A new approach to utilize PCR-single-strand conformation polymorphism for 16S rRNA gene-based microbial community analysis. Applied and Environmental Microbiology. 1998;64:4870–4876. doi: 10.1128/aem.64.12.4870-4876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-iii: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Vargas Castillo R, Beck A. Photobiont selectivity and specificity in Caloplaca species in a fog-induced community in the Atacama Desert, northern Chile. Fungal Biology. 2012;116:665–676. doi: 10.1016/j.funbio.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Werth S. Fungal–algal interactions in Ramalina menziesii and its associated epiphytic lichen community. The Lichenologist. 2012;44:543–560. [Google Scholar]
- Yahr R, Vilgalys R, DePriest PT. Strong fungal specificity and selectivity for algae symbionts in Florida scrub Cladonia lichens. Molecular Ecology. 2004;13:3367–3378. doi: 10.1111/j.1365-294X.2004.02350.x. [DOI] [PubMed] [Google Scholar]
- Yahr R, Vilgalys R, DePriest PT. Geographic variation in algal partners of Cladonia subtenuis (Cladoniaceae) highlights the dynamic nature of a lichen symbiosis. New Phytologist. 2006;171:847–860. doi: 10.1111/j.1469-8137.2006.01792.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.