Abstract
Mitotic index is an important component of histologic grade and has an etiologic role in breast tumorigenesis. Several small candidate gene studies have reported associations between variation in mitotic genes and breast cancer risk. We measured associations between 2156 single nucleotide polymorphisms (SNPs) from 194 mitotic genes and breast cancer risk, overall and by histologic grade, in the Breast Cancer Association Consortium (BCAC) iCOGS study (n = 39 067 cases; n = 42 106 controls). SNPs in TACC2 [rs17550038: odds ratio (OR) = 1.24, 95% confidence interval (CI) 1.16–1.33, P = 4.2 × 10−10) and EIF3H (rs799890: OR = 1.07, 95% CI 1.04–1.11, P = 8.7 × 10−6) were significantly associated with risk of low-grade breast cancer. The TACC2 signal was retained (rs17550038: OR = 1.15, 95% CI 1.07–1.23, P = 7.9 × 10−5) after adjustment for breast cancer risk SNPs in the nearby FGFR2 gene, suggesting that TACC2 is a novel, independent genome-wide significant genetic risk locus for low-grade breast cancer. While no SNPs were individually associated with high-grade disease, a pathway-level gene set analysis showed that variation across the 194 mitotic genes was associated with high-grade breast cancer risk (P = 2.1 × 10−3). These observations will provide insight into the contribution of mitotic defects to histological grade and the etiology of breast cancer.
INTRODUCTION
Inherited variation in genes encoding proteins involved in mitotic regulatory pathways, such as mitotic kinases and centrosome-related genes, has been associated with cancer risk in several small candidate gene studies. Common variants in mitotic genes have been associated with various cancer types such as prostate, lung, uterine, colorectal, and breast cancer (1–7). Specifically for breast cancer, genes involved in centrosome amplification, such as NIN, TACC3, GPSM2, CDC25C, NEK7 and MCPH1 and variation in mitotic regulators, including SART1, EIF3A, RRM2 and PSCD3 have been associated with breast cancer risk (8,9). SNP by SNP interactions for breast cancer risk have also been observed between SEPT4 and TEX14, both of which participate in the separation into daughter cells during cytokinesis (10). Finally, the mitotic kinases FYN, MAST2 and MAP2K4, identified through RNA interference-based functional screening of mitotic kinases in Drosophila (11), have been associated with breast cancer risk (12).
Multiple lines of evidence support an etiologic role for disruption of mitotic regulatory pathways in breast tumorigenesis. The disruption of chromosome segregation during mitosis is one mechanism of chromosomal instability, and ultimately aneuploidy, which has been found to occur early in breast tumor development (13,14), is found in ∼80% of all breast tumors and is thought to play a direct role in tumor progression (15,16). Further, somatic mutations in spindle assembly checkpoint genes have been identified in human breast tumors, and mutations in orthologous murine genes have been implicated in increased chromosomal instability and tumor development (13,14). Deregulation of mitosis is associated with the pathophysiology of breast cancer through the mitotic index, a component of the histologic grading system of breast tumors. Higher histologic grade is associated with increased aggressiveness and both high mitotic index and high grade are associated with poor prognosis (17). Given the relationship between histologic grade and mitotic index, we hypothesized that genetic variation in mitotic regulatory pathways is associated with high-grade breast cancer risk. Here we report on a comprehensive analysis of variation in mitotic genes in a study of nearly 80 000 subjects (n = 39 067 cases; n = 42 106 controls) with information on histopathologic grade from the Breast Cancer Association Consortium (BCAC). We evaluated 2135 single nucleotide polymorphisms (SNPs) in 194 genes involved in mitosis, encompassing those involved in mitotic entry and progression, the spindle assembly checkpoint and cytokinesis. Utilizing genotype data from a custom Illumina Infinium array (iCOGS) array (18), we investigated whether variation in these 194 mitotic genes influences the risk of breast cancer, both overall and with respect to histologic grade.
RESULTS
To determine whether variation in genes encoding mitotic regulatory proteins influences invasive breast cancer risk, we evaluated associations between 2156 SNPs in 194 mitotic genes (Supplementary Material, Table S1) and breast cancer risk among women of European ancestry using 39 067 breast cancer cases and 42 106 study-matched controls from BCAC (Supplementary Material, Table S2). Ten SNPs in three loci were significantly associated with overall breast cancer risk after Bonferroni correction (P < 2.3 × 10−5) (Table 1a). Six SNPs in the ITPR1 locus on chromosome 3, which has been previously reported as a breast cancer susceptibility locus by BCAC (18), were associated with overall breast cancer risk (Table 1a). Four of these SNPs achieved genome-wide significance with invasive breast cancer overall (rs6762644: odds ratio (OR) = 1.06, P = 1.1 × 10−8; rs6774180 OR = 1.06, P = 1.3 × 10−8; rs9867580 OR = 1.06, P = 4.2 × 10−8; rs13313995 OR = 1.06, P = 4.8 × 10−8) (Table 1a). Three SNPs in the TACC2 locus on chromosome 10 (rs17550038 OR = 1.15, P = 1.0 × 10−6; rs2461211 OR = 1.08, P = 1.8 × 10−6; rs2461210 OR = 1.08, P = 2.3 × 10−6) and one SNP in the EIF3H locus on chromosome 8 (rs799890 OR = 1.06, P = 1.4 × 10−5) were also significantly associated with overall breast cancer risk (Table 1a). Of these, the three TACC2 locus SNPs showed genome-wide significant associations with estrogen receptor (ER)-positive breast cancer but no significant associations with ER-negative breast cancer (Supplementary Material, Table S3a).
Table 1.
Associations with overall and low-grade breast cancer risk
| SNP | Chr. | Position | Gene | Allelea | Controls | Cases | OR | P-value |
|---|---|---|---|---|---|---|---|---|
| (a) Overall breast cancer | ||||||||
| rs6762644 | 3 | 4717276 | ITPR1 | G | 42 100 | 39 055 | 1.06 (1.04–1.08) | 1.1 × 10−8 |
| rs6774180 | 3 | 4717779 | ITPR1 | A | 42 102 | 39 061 | 1.06 (1.04–1.08) | 1.3 × 10−8 |
| rs9867580 | 3 | 4722247 | ITPR1 | C | 42 101 | 39 058 | 1.06 (1.04–1.08) | 4.2 × 10−8 |
| rs13313995 | 3 | 4722360 | ITPR1 | A | 42 097 | 39 048 | 1.06 (1.04–1.08) | 4.8 × 10−8 |
| rs17550038 | 10 | 123780679 | TACC2 | C | 42 101 | 39 058 | 1.15 (1.09–1.22) | 1.0 × 10−6 |
| rs2461211 | 10 | 123783865 | TACC2 | A | 42 101 | 39 064 | 1.08 (1.05–1.12) | 1.8 × 10−6 |
| rs2461210 | 10 | 123784538 | TACC2 | A | 42 105 | 39 065 | 1.08 (1.05–1.12) | 2.3 × 10−6 |
| rs9830067 | 3 | 4731814 | ITPR1 | A | 42 104 | 39 062 | 1.05 (1.03–1.07) | 5.0 × 10−6 |
| rs2306881 | 3 | 4728712 | ITPR1 | G | 42 095 | 39 057 | 1.05 (1.03–1.07) | 6.1 × 10−6 |
| rs799890 | 8 | 117318782 | EIF3H | C | 42 102 | 39 063 | 1.06 (1.03–1.09) | 1.4 × 10−5 |
| (b) Low-grade breast cancer | ||||||||
| rs17550038 | 10 | 123780679 | TACC2 | C | 42 101 | 16 053 | 1.24 (1.16–1.33) | 4.2 × 10−10 |
| rs2461211 | 10 | 123783865 | TACC2 | A | 42 101 | 16 056 | 1.14 (1.09–1.19) | 4.8 × 10−10 |
| rs2461210 | 10 | 123784538 | TACC2 | A | 42 105 | 16 055 | 1.14 (1.09–1.18) | 7.1 × 10−10 |
| rs7898269 | 10 | 123784105 | TACC2 | A | 42 106 | 16 056 | 1.16 (1.09–1.22) | 2.1 × 10−7 |
| rs12146254 | 10 | 123793633 | TACC2 | A | 42 079 | 16 047 | 1.15 (1.08–1.21) | 1.3 × 10−6 |
| rs10887047 | 10 | 123770790 | TACC2 | A | 42 087 | 16 054 | 1.14 (1.08–1.21) | 2.2 × 10−6 |
| rs799890 | 8 | 117318782 | EIF3H | C | 42 102 | 16 055 | 1.07 (1.04–1.11) | 8.7 × 10−6 |
| rs799889 | 8 | 117320076 | EIF3H | C | 42 102 | 16 053 | 1.07 (1.04–1.10) | 1.8 × 10−5 |
| rs6762644 | 3 | 4717276 | ITPR1 | G | 42 100 | 16 052 | 1.06 (1.03–1.08) | 2.3 × 10−5 |
a Tested allele.
The 2156 mitotic SNPs were also assessed for associations with histologic grade of breast cancer, by comparing 19 475 low-grade breast cancers (Grades 1 and 2 combined) and 8780 high-grade (Grade 3) breast cancers to 42 106 controls in a polytomous logistic regression model. Similar to the overall breast cancer analysis, variants in the TACC2, EIF3H and ITPR1 loci were significantly associated with low-grade breast cancer risk (Table 1b). Three genotyped SNPs in the TACC2 locus showed genome-wide significant associations with risk of low-grade breast cancers (rs17550038 OR = 1.24, P = 4.2 × 10−10; rs2461211 OR = 1.14, P = 4.8 × 10−10; rs2461210 OR = 1.14, P = 7.1 × 10−10), and three others retained significance after Bonferroni correction (Table 1b). All six variants were located in intron 2 of TACC2 (Supplementary Material, Fig. S1). The levels of significance and the effect sizes for the associations with the six TACC2 SNPs were consistently greater in ER-positive than ER-negative low-grade breast cancers, although this may be due to reduced power for the ER-negative analysis (Supplementary Material, Table S3b). No SNPs in TACC2 were significantly associated with high-grade breast cancer risk (Supplementary Material, Table S4).
The TACC2 locus is located ∼390 kb downstream of FGFR2, a known breast cancer susceptibility locus (18–20), from which FGFR2 rs2981579 has been strongly associated with overall breast cancer risk in these data (OR = 1.32, P = 1.23 × 10−102) (Table 2a) (18). Although 1000 Genomes Project data showed little evidence of linkage disequilibrium (LD) between SNPs in the TACC2 and FGFR2 loci (Supplementary Material, Fig. S2), the proximity of the loci raised the possibility that associations between variants in TACC2 and low-grade breast cancer were accounted for by variation in the FGFR2 locus. To explore this in detail we investigated associations between 454 SNPs in the FGFR2 locus and low-grade breast cancer. By adjusting the top FGFR2 SNP, rs2981579, for each of the 453 remaining FGFR2 SNPs, rs78985527 was identified as an additional potentially independent FGFR2 signal for low-grade breast cancer (Table 2b). The analyses of the six significant TACC2 SNPs were then adjusted simultaneously for rs2981579 and rs78985527 (Table 2c). While the effect sizes and the significance of the findings were reduced, each of the six TACC2 SNPs remained strongly associated with low-grade breast cancer (Table 2c). In addition, there was no evidence for interaction between FGFR2 rs2981579, rs78985527, and any of the TACC2 SNPs (Supplementary Material, Table S5). For completeness, we also adjusted the top TACC2 SNP rs17550038 for each of the 454 FGFR2 SNPs, but did not find substantial evidence that FGFR2 SNPs account for the TACC2 signal (Supplementary Material, Fig. S3). These findings suggest that the TACC2 association is independent of previously described genetic associations at the FGFR2 locus. However, it will be necessary to take into account the potential for long-range transcriptional regulation in this region when exploring the exact functional mechanism underlying this signal.
Table 2.
Multivariable analysis of FGFR2 and TACC2 for low-grade breast cancer risk
| Gene | SNP | Adjustmentsa | OR (95% CI) | P-value |
|---|---|---|---|---|
| (a) Single SNP analysis | ||||
| TACC2 | rs17550038 | 1.24 (1.16–1.33) | 4.2 × 10−10 | |
| rs2461211 | 1.14 (1.09–1.19) | 4.8 × 10−10 | ||
| rs2461210 | 1.14 (1.09–1.18) | 7.1 × 10−10 | ||
| rs7898269 | 1.16 (1.09–1.22) | 2.1 × 10−7 | ||
| rs12146254 | 1.15 (1.08–1.21) | 1.3 × 10−6 | ||
| rs10887047 | 1.14 (1.08–1.21) | 2.2 × 10−6 | ||
| (b) FGFR2 2-SNP analysis | ||||
| rs2981579 | rs78985527 | 1.33 (1.30–1.37) | 8.3 × 10−106 | |
| rs78985527 | rs2981579 | 1.12 (1.06–1.18) | 5.7 × 10−5 | |
| (c) TACC2 + FGFR2 3-SNP analysis | ||||
| TACC2 | rs17550038 | rs2981579, rs78985527 | 1.14 (1.07–1.23) | 1.1 × 10−4 |
| rs2461211 | rs2981579, rs78985527 | 1.08 (1.04–1.13) | 2.1 × 10−4 | |
| rs2461210 | rs2981579, rs78985527 | 1.12 (1.04–1.12) | 2.6 × 10−4 | |
| rs7898269 | rs2981579, rs78985527 | 1.10 (1.04–1.16) | 1.3 × 10−3 | |
| rs12146254 | rs2981579, rs78985527 | 1.09 (1.03–1.15) | 3.2 × 10−3 | |
| rs10887047 | rs2981579, rs78985527 | 1.08 (1.03–1.15) | 4.2 × 10−3 | |
| FGFR2 | rs2981579 | rs17550038, rs78985527 | 1.32 (1.29–1.36) | 1.3 × 10−100 |
| rs2981579 | rs2461211, rs78985527 | 1.33 (1.29–1.36) | 1.7 × 10−100 | |
| rs2981579 | rs2461210, rs78985527 | 1.33 (1.29–1.36) | 1.2 × 10−100 | |
| rs2981579 | rs7898269, rs78985527 | 1.33 (1.29–1.36) | 2.8 × 10−102 | |
| rs2981579 | rs12146254, rs78985527 | 1.33 (1.30–1.37) | 8.8 × 10−103 | |
| rs2981579 | rs10887047, rs78985527 | 1.33 (1.30–1.37) | 7.3 × 10−103 | |
| FGFR2 | rs78985527 | rs17550038, rs2981579 | 1.11 (1.05–1.17) | 8.1 × 10−5 |
| rs78985527 | rs2461211, rs2981579 | 1.11 (1.05–1.17) | 1.4 × 10−4 | |
| rs78985527 | rs2461210, rs2981579 | 1.11 (1.05–1.17) | 1.5 × 10−4 | |
| rs78985527 | rs7898269, rs2981579 | 1.11 (1.05–1.17) | 8.2 × 10−5 | |
| rs78985527 | rs12146254, rs2981579 | 1.11 (1.06–1.18) | 5.8 × 10−5 | |
| rs78985527 | rs10887047, rs2981579 | 1.11 (1.06–1.17) | 7.4 × 10−5 | |
aIn addition to study and principal components.
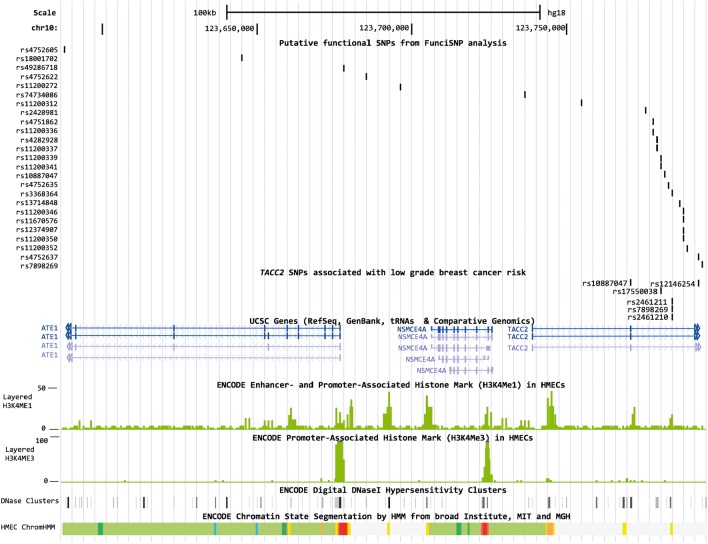
To identify putative functional SNPs in the TACC2 locus, we performed a FunciSNP analysis for rs17550038. A total of 27 SNPs in LD with rs17550038 (R2 ≥ 0.3), the majority of which were located in introns of TACC2 (n = 21) or ATE1 (n = 4), overlapped with at least one biofeature (Supplementary Material, Table S6, Fig. 1). Of these 27 SNPs, rs11200337 overlapped with biofeatures in three breast cell lines (HMEC, MCF7, T47D). rs11200337 is located 11.5 kb from the TACC2 index SNP (R2 = 0.53) in a methylated region in each of the cell lines and a DNaseI hypersensitivity (HS) site in HMEC and T47D cells. The SNP is also located in sites of histone modification and open chromatin in HMEC normal mammary epithelial cells. Three additional SNPs located in TACC2 introns overlapped with biofeatures in at least two of the cell lines (rs4282928, rs4752637, rs11200373).
Figure 1.
Overlap between putative functional SNPs and ENCODE tracks in HMECs. Figures were generated in the UCSC Genome Browser (http://genome.ucsc.edu, last accessed on 19 June 2014) using ENCODE and custom tracks. ChromHMM, Hidden Markov Model predicted chromatin state segmentation; bright red, active promoter; light red, weak promoter; purple, inactive promoter; orange, strong enhancer; yellow, weak enhancer; blue, insulator; dark green, transcriptional elongation; light green, weakly transcribed; dark gray, polycomb-repressed; light gray, repetitive/copy number variation.
We also performed an exploratory analysis of correlations between TACC2 expression and nearby SNPs, utilizing expression quantitative trait locus (eQTL) data available from 484 triple negative (TN) breast tumors from the Triple Negative Breast Cancer Consortium. Seven SNPs around TACC2 were associated with TACC2 expression at a 10% false discovery rate (FDR) threshold (P ≤ 5 × 10−5), although none of these SNPs were in LD with the risk-associated SNPs (Supplementary Material, Table S7). Similarly, an eQTL analysis using The Cancer Genome Atlas (TCGA) data identified an additional rare SNP, rs3752956 in intron 8 of TACC2, as an eQTL for TACC2 (P = 4.07 × 10−5) in ER-positive breast tumors (21). These data alone do not provide evidence that SNP-mediated deregulation of TACC2 underlies the breast cancer risk signal at this locus. Further functional analyses in low-grade breast tumors are necessary to understand the mechanistic basis of this association.
In addition to TACC2, two SNPs in the EIF3H locus (rs799890 OR = 1.07, P = 8.7 × 10−6, rs799889 OR = 1.07, P = 1.8 × 10−5) were associated with low-grade breast cancer risk (Table 1b). A total of 55 SNPs in the EIF3H locus were genotyped, with a single peak of association downstream of EIF3H (Supplementary Material, Fig. S4). Similar to the overall results, variants in EIF3H were associated with ER-positive low-grade breast cancer and marginally with ER-negative low-grade breast cancer, where the effect sizes were slightly larger but the association was less significant due to the small sample size (Supplementary Material, Table S3b). No SNPs in EIF3H were significantly associated with high-grade breast cancer risk (Supplementary Material, Table S8). We identified 19 putative functional SNPs correlated with rs799890 in the EIF3H locus, all of which were intergenic between the TRPS1 and EIF3H genes (Supplementary Material, Table S9). The only biofeatures associated with these SNPs were open chromatin states and sites of histone modification in HMEC cells.
Similarly, a single SNP in the ITPR1 locus remained statistically significant among low-grade breast cancers (rs6762644 OR = 1.06, P = 2.3 × 10−5) (Table 1b). As with the TACC2 SNPs, the ITPR1 SNP was only associated with ER-positive low-grade breast cancers (Supplementary Material, Table S3b). Several SNPs in ITPR1, including rs6762644, were also marginally significantly associated with high-grade breast cancer (Table 1c), suggesting that the ITPR1 locus is associated with breast cancer risk regardless of histologic grade. SNPs in the ITPR1 locus that are associated with breast cancer risk have been previously annotated for effects on chromatin using Encyclopedia of DNA Elements (ENCODE) biofeatures identified in HMECs (22). Here we identified 14 SNPs correlated with rs6762644 that also overlap with DNaseI HS sites, Formaldehyde-assisted isolation of regulatory elements (FAIRE) open chromatin signals, and sites of histone modification in T47D and/or MCF7 cells located within introns of EGOT (Supplementary Material, Table S10).
No individual SNPs were significantly associated with high-grade breast cancer (Supplementary Material, Table S11). However, considering the original hypothesis that variation in mitotic genes is associated with high-grade breast cancer risk and the limited power to detect single SNP associations for high-grade breast cancer, we evaluated whether variation in the 194 mitotic genes influenced high-grade breast cancer risk when analyzed as a pathway. A two-step gene set analysis (PC-GM) was conducted, in which each of the 194 mitotic genes were summarized by principal component analysis and then combined into a single test statistic to evaluate whether the gene set was associated with risk (23). Based on this method, the mitotic pathway was significantly associated with overall breast cancer risk (P = 2.6 × 10−3). This association was maintained even after excluding SNPs in the TACC2, EIF3H and ITPR1 loci (filtered P = 2.5 × 10−3). In contrast to the findings with single SNPs, the pathway as a whole was associated with high-grade breast cancer (P = 2.1 × 10−3; filtered P = 2.6 × 10−3) rather than low-grade breast cancer risk (P = 0.065; filtered P = 0.063). This suggests that variation in mitotic genes is relevant to high-grade breast cancer risk; however these result are preliminary, and it is necessary to replicate this analysis in an independent population and to functionally validate the role of these genetic variants in high-grade breast cancer to confirm these findings.
DISCUSSION
In this analysis of 194 genes involved in mitotic regulation, we have shown that SNPs in TACC2, EIF3H and ITPR1 are associated with risk of low-grade but not high-grade breast cancer, with the greatest effects observed for ER-positive tumors. Several of the TACC2 SNPs remained associated with low-grade breast cancer risk after adjustment for the nearby FGFR2 breast cancer risk SNP rs2981579, suggesting that the TACC2 locus is a new genome-wide significant genetic risk factor for low-grade breast cancer. The association of SNPs in FGFR2 and TACC2 with breast cancer suggests a complex relationship between SNPs and genes in this region of chromosome 10. Indeed, it is possible that the underlying functional effect captured by this new signal in the TACC2 locus is related mechanistically to previously described associations in the FGFR2 locus, in that variants in the TACC2 locus may influence TACC2 and/or long-range transcriptional regulation of FGFR2. Analyses of common variants in these loci using ENCODE and eQTL data identified several candidate functional SNPs, which will need to be explored in future in vitro and in vivo studies to elucidate the underlying biological mechanisms at this locus that influence risk of low-grade breast cancer.
While we generally observed greater effects for ER-positive low-grade tumors, we had limited power to detect significant associations with the modest number of low-grade, ER-negative breast cancers genotyped (n = 1447) given the relatively small effect sizes for the TACC2, EIF3H and ITRP1 SNPs. Future studies by BCAC and other consortia that incorporate large numbers of ER-negative breast cancers with complete histologic grade data will be necessary to completely understand the relationship between these SNPs, grade and ER subtype. In contrast to single SNP effects, variation in the 194 mitotic genes was associated with high-grade breast cancer risk in a pathway-level analysis, although these findings require replication in an independent sample and functional validation. It is important to note that while the total sample size was large, the number of high-grade breast cancers was comparatively small and the statistical power to detect associations with SNPs with small effect sizes was limited. Additionally, due to the design of the iCOGS array, SNP coverage of the genes varied and some known mitotic genes were not represented at all. Nevertheless, we successfully identified biologically interesting genes that appear to influence breast tumor grade, and a series of candidate functional SNPs in these loci that warrant follow-up in future studies.
The TACC2 gene is a member of the transforming acidic coiled-coil-containing protein family and is located on chromosome 10q26 (24). TACC proteins are an essential component of the centrosome–spindle apparatus during mitosis, and TACC2 is strongly concentrated at centrosomes throughout the cell cycle (25). Interestingly, mutants lacking the Drosophila melanogaster TACC gene, d-tacc, experience high rates of chromosomal segregation defects (26). In a study of fresh frozen primary human breast cancer tissues, TACC2 expression was increased in high-grade compared with low-grade tumors and in tumors from patients with poor clinical outcomes including metastasis, recurrence, and breast cancer related death, reflected by a shorter disease-free survival for patients with high TACC2 expression (24). However, multiple other studies suggest that TACC2 can be up- or down-regulated in different types of cancer even in the same tissue, including breast (27–29).
Less is known about the exact role of EIF3H, located on chromosome 8q23, in cell cycle regulation. The EIF3H gene encodes the H subunit of the eukaryotic translation initiation factor 3 (eIF-3) complex, which is required for several steps in the initiation of protein synthesis including mRNA recruitment and disassembly of ribosomal complexes (30). Translational control is a crucial component of cancer development and progression (31), and EIF3H in particular is frequently amplified in breast and prostate cancers (32). Overexpression of eIF3 h in prostate cancers is also associated with increased grade as measured by the Gleason score (33). Two short interfering RNA (siRNA) screens in HeLa cells have identified EIF3H as essential for cell division, the disruption of which leads to cell cycle arrest and altered ploidy phenotype (34,35).
In summary, we have reported on a large-scale analysis of the relationship between common variation in mitotic genes and breast cancer grade in a study of ∼40 000 invasive breast cancer cases and study-matched controls with extensive histopathologic grade data. While the exact mechanism underlying the association between TACC2 and EIF3H and breast cancer grade are unclear, these results warrant follow-up in functional studies and larger studies of histopathologic subtypes of breast cancer.
MATERIALS AND METHODS
iCOGS genotyping
Subjects included in this analysis were a subset of those genotyped on the iCOGS array from the BCAC (18). Women with invasive breast cancer and study-matched controls from 40 studies (Supplementary Material, Table S2) with self-reported European ancestry and >95% subject call rate for genotyping (n = 39 067 cases; n = 42 106 controls) were included. These 40 studies have been described previously (18). The design of the iCOGS array (211 155 SNPs), genotyping methods, and quality control have been previously described (18). Samples were genotyped as part of the Collaborative Oncological Gene-environment Study (COGS) project using the iCOGS array at four genotyping centers. Genotype calling and quality-control analyses were conducted by a single analysis center at the University of Cambridge (18).
Gene and SNP selection
The iCOGS array included SNPs from 194 genes encoding proteins implicated in normal control of mitotic entry, spindle assembly checkpoint and cytokinesis (GO: http://www.geneontology.org; KEGG http://www.genome.jp/kegg/) (Supplementary Material, Table S1). All 2351 SNPs on the iCOGS array within each of the 194 genes and within a 50 kb window from the beginning and end of the longest transcript were selected. A total of 2156 SNPs had a call rate >95% and were included in the analysis.
Pathology
The collection of pathology and tumor marker information for BCAC has been described previously (36). Briefly, studies provided information on ER status and grade of differentiation. The most common source of data for ER status was medical records, followed by inmunohistochemistry performed on tumor tissue microarrays or whole section tumor slides. ER-negative status was defined as < 10% of the tumor cells stained for a number of participating studies, where patients were recruited from Europe (n = 30), Australia (n = 3), Canada (n = 2) and the USA (n = 5) from 1972–2011 (median recruitment year = 2004). Histologic grade was reported using the Nottingham combined grading system. For the purpose of this analysis, Grades 1 and 2 were jointly considered ‘low grade’ while Grade 3 was considered ‘high grade’.
Statistical analyses
Single SNP analyses were conducted in PLINK (37), and polytomous logistic regression was implemented in R (http://cran.us.r-project.org/, last accessed on 19 June 2014) when comparing histopathologic subtypes to a common set of controls. SNP associations were tested in a log-additive model and were adjusted for study and European ancestry-specific principal components as described by Michailidou et al. (18). Consideration of age, assessed by both the exclusion of studies for which the age of controls was not known and the adjustment for age in 5-year categories and as a continuous covariate, made no substantial difference to the results.
The two-step gene set pathway analysis (PC-GM) has been previously described (23). Briefly, we first performed principal component (PC) analysis for each of the 194 mitotic genes. The PCs that captured at least 80% of variation in each gene were used to assess the significance of the associations between each gene and breast cancer risk in a logistic regression model. Following determination of these gene-level associations for each of the 194 genes, the P-values were summarized using the gamma method (23) to obtain a pathway-level test statistic based on observed data. Empirical gene set association P-values and pathway-level test statistics were determined from 1000 permutations, where the response variable (case–control status) was permuted while keeping genotype and covariate data fixed. The final pathway P-value was determined as the proportion of permutations in which the empirical pathway-level test statistic was greater than the observed pathway-level test statistic.
FunciSNP annotation
The FunciSNP package (38) was implemented in R using default parameters with a search window of ±500 kb. Analyses were run separately for each of three index SNPs: rs17550038 (TACC2), rs799890 (EIF3H) and rs6762644 (ITPR1). The FunciSNP tool identified all SNPs from the 1000 genomes project (http://www.1000genomes.org/, last accessed on 19 June 2014) within 500 kb of the index SNP that overlapped with at least one biofeature. The biofeatures included in this analysis were (1) built-in consensus promoter regions, ENCODE DNaseI HS and CTCF sites from the getFSNPs function and (2) HS sites, FAIRE signals and histone modification ChIP-seq data (H3K4me1, me2, me3, H3K9Ac and H3K27Ac) downloaded as bed files from ENCODE Build 37 production data (http://genome.ucsc.edu/ENCODE/, last accessed on 19 June 2014) for HMEC normal mammary epithelial cells, and the MCF7 and T47D breast cancer cell lines when available (Supplementary Material, Table S12). Recognizing that observed SNP associations may capture functional SNPs even at relatively low levels of LD, we defined LD with the index SNP at R2 ≥ 0.3.
Triple negative breast cancer expression quantitative trait loci (eQTL) analyses
Expression profiles were generated for 596 triple negative (TN) breast tumors (Supplementary Material, Table S13) using the Illumina Whole Genome cDNA-mediated Annealing, Selection, extension and Ligation (DASL) v4.0 assay. Study sites have been described previously (39,40). Whole formalin fixed paraffin embedded tumor sections were macrodissected for enrichment of tumor cells, guided by a pathologist-read hematoxylin and eosin-stained slide. RNA was extracted using the Roche High Pure RNA Isolation Kit (Indianapolis, USA). DASL expression profiling was performed by the Mayo Clinic Medical Genome Facility Gene Expression Core (Rochester, MN). After log2-transformation of raw intensity values, a per-sample quality (stress) measure was calculated (41). Log2-transformed intensity values were median-quantile normalized. Probes with a P-value of detection >0.05 in all samples were excluded (n = 713) yielding 28 664 high-quality probes. Samples were median-centered by 96-well plate to correct for batch effects. Of the 596 TN tumors with high-quality expression data, germline genotype data from the Illumina 660-Quad, HumanHap 500k DUO, CNV370DUO, or iCOGS custom genotyping array (18,40), were available for 516 of the same individuals. cis-eQTLs for TACC2 were defined as associations between ILMN_2315780, ILMN_1686442, ILMN_2363165 probe expression and SNPs within 1 MB of these probes in a robust linear regression model. An FDR was generated using 100 permutations of the genome-wide analysis (cis associations between 8 969 066 SNPs and 28 504 probes), and cis-eQTLs were excluded at a 10% FDR threshold (equivalent to P ≤ 5.0 × 10−5).
SUPPLEMENTARY MATERIAL
FUNDING
Part of this work was supported by the National Institutes of Health (NIH) grant (CA128978) and a Specialized Program of Research Excellence (SPORE) in Breast Cancer to Mayo Clinic (P50 CA116201), the Breast Cancer Research Foundation, the Department of Defence (W81XWH-10-1-0341) and the European Community’s Seventh Framework Programme under grant agreement number 223175 (grant number HEALTH-F2-2009-223175) (COGS). Funding for the iCOGS infrastructure came from: the European Community's Seventh Framework Programme under grant agreement no. 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692), the National Institutes of Health Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112—the GAME-ON initiative), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure and the Ovarian Cancer Research Fund. The ABCFS, NC-BCFR and OFBCR work was supported by the National Cancer Institute, National Institutes of Health under RFA-CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario (U01 CA69467), Northern California Cancer Center (U01 CA69417), University of Melbourne (U01 CA69638). Samples from the NC-BCFR were processed and distributed by the Coriell Institute for Medical Research. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Australia Fellow and a Victorian Breast Cancer Research Consortium Group Leader. M.C.S. is a NHMRC Senior Research Fellow and a Victorian Breast Cancer Research Consortium Group Leader. The ABCS study was supported by the Dutch Cancer Society (grants NKI 2007-3839; 2009 4363) and BBMRI-NL, which is a Research Infrastructure financed by the Dutch government (NWO 184.021.007) The Australian Breast Cancer Tissue Bank is generously supported by the National Health and Medical Research Council of Australia, The Cancer Institute NSW and the National Breast Cancer Foundation. The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. The BBCS is funded by Cancer Research UK and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). The BCAC is funded by CR-UK (C1287/A10118 and C1287/A12014). Meetings of the BCAC have been funded by the European Union COST programme (BM0606). D.F.E. is a Principal Research Fellow of CR-UK. E.S. is supported by NIHR Comprehensive Biomedical Research Centre, Guy's and St. Thomas' NHS Foundation Trust in partnership with King's College London, UK I.T. is supported by the Oxford Biomedical Research Centre. The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ) The CECILE study was funded by Fondation de France, Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Ligue contre le Cancer Grand Ouest, Agence Nationale de Sécurité Sanitaire (ANSES), Agence Nationale de la Recherche (ANR) The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council and Herlev Hospital The CNIO-BCS was supported by the Genome Spain Foundation, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI081120). The Human Genotyping-CEGEN Unit (CNIO) is supported by the Instituto de Salud Carlos III. The CTS was initially supported by the California Breast Cancer Act of 1993 and the California Breast Cancer Research Fund (contract 97-10500) and is currently funded through the National Institutes of Health (R01 CA77398). Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. HAC receives support from the Lon V Smith Foundation (LVS39420). The work of Demokritos has been co-financed by the European Union (European Social Fund—ESF) and Greek national funds through the Operational Program ‘Education and Lifelong Learning’ of the National Strategic Reference Framework (NSRF)—Research Funding Program: Thales. Investing in knowledge society through the European Social Fund. The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The GC-HBOC was supported by Deutsche Krebshilfe (107 352) The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The HEBCS was financilly supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society, The Nordic Cancer Union and the Sigrid Juselius Foundation. The HMBCS was supported by a grant from the Friends of Hannover Medical School and by the Rudolf Bartling Foundation. Financial support for KARBAC was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, The Swedish Cancer Society, the Stockholm Cancer Foundation, and Bert von Kantzow's foundation. The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, the Academy of Finland and by the strategic funding of the University of Eastern Finland kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia and the Cancer Foundation of Western Australia. R.B. was a Cancer Institute NSW Fellow. LMBC is supported by the ‘Stichting tegen Kanker’ (232-2008 and 196-2010). D.L. is supported by the FWO and the KULPFV/10/016-SymBioSysII. The MARIE study was supported by the Deutsche Krebshilfe e.V. (70-2892-BR I), the Hamburg Cancer Society, the German Cancer Research Center and the genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany (01KH0402). MBCSG is supported by grants from the Italian Association for Cancer Research (AIRC) and by funds from the Italian citizens who allocated the 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects ‘5 × 1000’). The MCBCS was supported by the National Institutes of Health grant (CA128978), an National Institutes of Health Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), the Breast Cancer Research Foundation and the Komen Race for the Cure and a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation. MCCS cohort recruitment in was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. The MEC was support by National Institutes of Health grants CA63464, CA54281, CA098758 and CA132839. The Canadian Breast Cancer Research Initiative supported the initial case–control study. This work was supported by the Quebec Breast Cancer Foundation, the Canadian Institutes of Health Research for the ‘CIHR Team in Familial Risks of Breast Cancer’ program—grant # CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade—grant # PSR-SIIRI-701. The NBCS was supported by grants from the Norwegian Research council, 155218/V40, 175240/S10 to ALBD, FUGE-NFR 181600/V11 to VNK and a Swizz Bridge Award to ALBD. The NBHS was supported by National Institutes of Health grant R01CA100374. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. The OBCS was supported by research grants from the Finnish Cancer Foundation, the Academy of Finland, the University of Oulu and the Oulu University Hospital. This work for OFBCR was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the BCFR. The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The pKARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer. The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009-4318). The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health and the Susan G. Komen Breast Cancer Foundation. The SBCS was supported by Yorkshire Cancer Research S295, S299, S305PA SEARCH is funded by a programme grant from Cancer Research UK (C490/A10124) and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. SKKDKFZS is supported by the DKFZ. The SZBCS was supported by Grant PBZ_KBN_122/P05/2004. The TNBCC was supported by a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), grants from the Komen Foundation for the Cure and the Breast Cancer Research Foundation and a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation. The UKBGS is funded by Breakthrough Breast Cancer and the Institute of Cancer Research (ICR). ICR acknowledges NHS funding to the NIHR Biomedical Research Centre.
Supplementary Material
Acknowledgements
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. This study would not have been possible without the contributions of the following: Andrew Berchuck (OCAC), Rosalind A. Eeles, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL), Antonis Antoniou, Lesley McGuffog, Ken Offit (CIMBA), Joe Dennis, Alison M. Dunning, Andrew Lee and Ed Dicks, Craig Luccarini and the staff of the Centre for Genetic Epidemiology Laboratory, Anna Gonzalez-Neira and the staff of the CNIO genotyping unit, Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière and Frederic Robidoux and the staff of the McGill University and Génome Québec Innovation Centre, the staff of the Copenhagen DNA laboratory, and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer and the staff of Mayo Clinic Genotyping Core Facility. ABCFS: Maggie Angelakos, Judi Maskiell, Gillian Dite. ABCS: Sanquin Research. ABCTB Investigators: Christine Clarke,Rosemary Balleine, Robert Baxter,Stephen Braye, Jane Carpenter, Jane Dahlstrom, John Forbes, Soon Lee, Debbie Marsh, Adrienne Morey, Nirmala Pathmanathan, Rodney Scott, Allan Spigelman, Nicholas Wilcken, Desmond Yip. Samples are made available to researchers on a non-exclusive basis. Silke Landrith, Sonja Oeser, Matthias Rübner. BBCS: Eileen Williams, Elaine Ryder-Mills, Kara Sargus. BIGSS: Niall McInerney, Gabrielle Colleran, Andrew Rowan, Angela Jones. BSUCH: Peter Bugert, Medical Faculty Mannheim. CGPS: Staff and participants of the Copenhagen General Population Study. For the excellent technical assistance: Dorthe Uldall Andersen, Maria Birna Arnadottir, Anne Bank, Dorthe Kjeldgård Hansen. The Danish Breast Cancer Group (DBCG) is acknowledged for the tumor information. CNIO-BCS: Guillermo Pita, Charo Alonso, Daniel Herrero, Nuria Álvarez, Pilar Zamora, Primitiva Menendez, the Human Genotyping-CEGEN Unit (CNIO). The CTS Steering Committee includes Leslie Bernstein, James Lacey, Sophia Wang, Huiyan Ma, Yani Lu, and Jessica Clague DeHart at the Beckman Research Institute of the City of Hope, Dennis Deapen, Rich Pinder, Eunjung Lee and Fred Schumacher at the University of Southern California, Pam Horn-Ross, Peggy Reynolds, and David Nelson at the Cancer Prevention Institute of California, and Hannah Park at the University of California Irvine. ESTHER: Hartwig Ziegler, Sonja Wolf, Volker Hermann. GC-HBOC: Heide Hellebrand, Stefanie Engert and GC-HBOC. The GENICA Network: Dr Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany; [HB, Wing-Yee Lo, Christina Justenhoven], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [YDK, Christian Baisch], Institute of Pathology, University of Bonn, Germany [Hans-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [Ute Hamann], Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, Germany [TB, Beate Pesch, Sylvia Rabstein, Anne Lotz]; Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany [Volker Harth]. HEBCS: Taru A. Muranen, Kirsimari Aaltonen, Karl von Smitten, Sofia Khan, Tuomas Heikkinen, Irja Erkkilä. HMBCS: Peter Hillemanns, Hans Christiansen, Johann H. Karstens, and Natalia Antonenkova. KBCP: Eija Myöhänen, Helena Kemiläinen. kConFab/AOCS: We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow-up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab. LMBC: Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel and Kathleen Corthouts. MARIE would like to thank Alina Vrieling, Katharina Buck, Ursula Eilber, Muhabbet Celik and Sabine Behrens. MBCSG: Siranoush Manoukian, Bernard Peissel, Daniela Zaffaroni, Giulietta Scuvera and Jacopo Azzolini of the Fondazione IRCCS Istituto Nazionale dei Tumori (INT); Bernardo Bonanni, Irene Feroce, Angela Maniscalco, Alessandra Rossi of the Istituto Europeo di Oncologia (IEO) and Loris Bernard and the personnel of the Cogentech Cancer Genetic Test Laboratory. MTLGEBCS: The authors gratefully acknowledge the assistance of Lesley Richardson and Marie-Claire Goulet in conducting the study. We would like to Martine Tranchant (Cancer Genomics Laboratory, CHU de Québec Research Center), Marie-France Valois, Annie Turgeon and Lea Heguy (McGill University Health Center, Royal Victoria Hospital; McGill University) for DNA extraction, sample management and skillful technical assistance. J.S. is Chairholder of the Canada Research Chair in Oncogenetics. NBHS: We thank study partcipants and research staff for their contributions and commitment to this study. OBCS: Mervi Grip, Meeri Otsukka, Kari Mononen. OFBCR: Teresa Selander, Nayana Weerasooriya. ORIGO: We thank E. Krol-Warmerdam and J. Blom for patient accrual, administering questionnaires, and managing clinical information. PBCS: Louise Brinton, Mark Sherman, Stephen Chanock, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, Michael Stagner. pKARMA: The Swedish Medical Research Counsel. RBCS: Petra Bos, Jannet Blom, Ellen Crepin, Anja Nieuwlaat, Annette Heemskerk, the Erasmus MC Family Cancer Clinic. SASBAC: The Swedish Medical Research Counsel. SBCS: Sue Higham, Helen Cramp, Ian Brock, Sabapathy Balasubramanian and Dan Connley. SEARCH: The SEARCH and EPIC teams. SKKDKFZS: We thank all study participants, clinicians, family doctors, researchers and technicians for their contributions and commitment to this study. UKBGS: We thank Breakthrough Breast Cancer and the Institute of Cancer Research for support and funding of the Breakthrough Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study.
Conflicts of Interest statement: None declared.
REFERENCES
- 1.Hawkins G.A., Mychaleckyj J.C., Zheng S.L., Faith D.A., Kelly B., Isaacs S.D., Wiley K.E., Chang B.L., Ewing C.M., Bujnovszky P., et al. Germline sequence variants of the LZTS1 gene are associated with prostate cancer risk. Cancer Genet. Cytogenet. 2002;137:1–7. doi: 10.1016/s0165-4608(02)00549-6. [DOI] [PubMed] [Google Scholar]
- 2.Guo Y., Zhang X., Yang M., Miao X., Shi Y., Yao J., Tan W., Sun T., Zhao D., Yu D., et al. Functional evaluation of missense variations in the human MAD1L1 and MAD2L1 genes and their impact on susceptibility to lung cancer. J. Med. Genet. 2010;47:616–622. doi: 10.1136/jmg.2009.074252. [DOI] [PubMed] [Google Scholar]
- 3.Milam M.R., Gu J., Yang H., Celestino J., Wu W., Horwitz I.B., Lacour R.A., Westin S.N., Gershenson D.M., Wu X., et al. STK15 F31I polymorphism is associated with increased uterine cancer risk: a pilot study. Gynecol. Oncol. 2007;107:71–74. doi: 10.1016/j.ygyno.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed S., Thomas G., Ghoussaini M., Healey C.S., Humphreys M.K., Platte R., Morrison J., Maranian M., Pooley K.A., Luben R., et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ewart-Toland A., Dai Q., Gao Y.T., Nagase H., Dunlop M.G., Farrington S.M., Barnetson R.A., Anton-Culver H., Peel D., Ziogas A., et al. Aurora-A/STK15 T+91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types. Carcinogenesis. 2005;26:1368–1373. doi: 10.1093/carcin/bgi085. [DOI] [PubMed] [Google Scholar]
- 6.Jang J.S., Kim K.M., Kang K.H., Choi J.E., Lee W.K., Kim C.H., Kang Y.M., Kam S., Kim I.S., Jun J.E., et al. Polymorphisms in the survivin gene and the risk of lung cancer. Lung Cancer. 2008;60:31–39. doi: 10.1016/j.lungcan.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Gazouli M., Tzanakis N., Rallis G., Theodoropoulos G., Papaconstantinou I., Kostakis A., Anagnou N.P., Nikiteas N. Survivin -31G/C promoter polymorphism and sporadic colorectal cancer. Int. J. Colorectal Dis. 2009;24:145–150. doi: 10.1007/s00384-008-0601-2. [DOI] [PubMed] [Google Scholar]
- 8.Olson J.E., Wang X., Pankratz V.S., Fredericksen Z.S., Vachon C.M., Vierkant R.A., Cerhan J.R., Couch F.J. Centrosome-related genes, genetic variation, and risk of breast cancer. Breast Cancer Res. Treat. 2011;125:221–228. doi: 10.1007/s10549-010-0950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson J.E., Wang X., Goode E.L., Pankratz V.S., Fredericksen Z.S., Vierkant R.A., Pharoah P.D., Cerhan J.R., Couch F.J. Variation in genes required for normal mitosis and risk of breast cancer. Breast Cancer Res. Treat. 2010;119:423–430. doi: 10.1007/s10549-009-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelemen L.E., Wang X., Fredericksen Z.S., Pankratz V.S., Pharoah P.D., Ahmed S., Dunning A.M., Easton D.F., Vierkant R.A., Cerhan J.R., et al. Genetic variation in the chromosome 17q23 amplicon and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2009;18:1864–1868. doi: 10.1158/1055-9965.EPI-08-0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bettencourt-Dias M., Giet R., Sinka R., Mazumdar A., Lock W.G., Balloux F., Zafiropoulos P.J., Yamaguchi S., Winter S., Carthew R.W., et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- 12.Wang X., Fredericksen Z.S., Vierkant R.A., Kosel M.L., Pankratz V.S., Cerhan J.R., Justenhoven C., Brauch H., Olson J.E., Couch F.J. Association of genetic variation in mitotic kinases with breast cancer risk. Breast Cancer Res. Treat. 2010;119:453–462. doi: 10.1007/s10549-009-0404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson S.L., Bakhoum S.F., Compton D.A. Mechanisms of chromosomal instability. Curr. Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo Y.L., Yu J.C., Chen S.T., Yang H.C., Fann C.S., Mau Y.C., Shen C.Y. Breast cancer risk associated with genotypic polymorphism of the mitosis-regulating gene Aurora-A/STK15/BTAK. Int. J. Cancer. 2005;115:276–283. doi: 10.1002/ijc.20855. [DOI] [PubMed] [Google Scholar]
- 15.Tirkkonen M., Tanner M., Karhu R., Kallioniemi A., Isola J., Kallioniemi O.P. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- 16.Mendelin J., Grayson M., Wallis T., Visscher D.W. Analysis of chromosome aneuploidy in breast carcinoma progression by using fluorescence in situ hybridization. Lab. Invest. 1999;79:387–393. [PubMed] [Google Scholar]
- 17.Ignatiadis M., Sotiriou C. Understanding the molecular basis of histologic grade. Pathobiology. 2008;75:104–111. doi: 10.1159/000123848. [DOI] [PubMed] [Google Scholar]
- 18.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K., et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q., Seo J.H., Stranger B., McKenna A., Pe'er I., Laframboise T., Brown M., Tyekucheva S., Freedman M.L. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell. 2013;152:633–641. doi: 10.1016/j.cell.2012.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhie S.K., Coetzee S.G., Noushmehr H., Yan C., Kim J.M., Haiman C.A., Coetzee G.A. Comprehensive functional annotation of seventy-one breast cancer risk Loci. PLoS One. 2013;8:e63925. doi: 10.1371/journal.pone.0063925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fridley B.L., Jenkins G.D., Tsai Y.Y., Song H., Bolton K.L., Fenstermacher D., Tyrer J., Ramus S.J., Cunningham J.M., Vierkant R.A., et al. Gene set analysis of survival following ovarian cancer implicates macrolide binding and intracellular signaling genes. Cancer Epidemiol. Biomarkers Prev. 2012;21:529–536. doi: 10.1158/1055-9965.EPI-11-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng S., Douglas-Jones A., Yang X., Mansel R.E., Jiang W.G. Transforming acidic coiled-coil-containing protein 2 (TACC2) in human breast cancer, expression pattern and clinical/prognostic relevance. Cancer Genomics Proteomics. 2010;7:67–73. [PubMed] [Google Scholar]
- 25.Gergely F., Karlsson C., Still I., Cowell J., Kilmartin J., Raff J.W. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. U.S.A. 2000;97:14352–14357. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M.J., Gergely F., Jeffers K., Peak-Chew S.Y., Raff J.W. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- 27.Conte N., Delaval B., Ginestier C., Ferrand A., Isnardon D., Larroque C., Prigent C., Seraphin B., Jacquemier J., Birnbaum D. TACC1-chTOG-Aurora A protein complex in breast cancer. Oncogene. 2003;22:8102–8116. doi: 10.1038/sj.onc.1206972. [DOI] [PubMed] [Google Scholar]
- 28.Schuendeln M.M., Piekorz R.P., Wichmann C., Lee Y., McKinnon P.J., Boyd K., Takahashi Y., Ihle J.N. The centrosomal, putative tumor suppressor protein TACC2 is dispensable for normal development, and deficiency does not lead to cancer. Mol. Cell. Biol. 2004;24:6403–6409. doi: 10.1128/MCB.24.14.6403-6409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauffart B., Gangisetty O., Still I.H. Molecular cloning, genomic structure and interactions of the putative breast tumor suppressor TACC2. Genomics. 2003;81:192–201. doi: 10.1016/s0888-7543(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 30.Marchione R., Leibovitch S.A., Lenormand J.L. The translational factor eIF3f: the ambivalent eIF3 subunit. Cell. Mol. Life Sci. 2013;70:3603–3616. doi: 10.1007/s00018-013-1263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silvera D., Formenti S.C., Schneider R.J. Translational control in cancer. Nat. Rev. Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- 32.Nupponen N.N., Porkka K., Kakkola L., Tanner M., Persson K., Borg A., Isola J., Visakorpi T. Amplification and overexpression of p40 subunit of eukaryotic translation initiation factor 3 in breast and prostate cancer. Am. J. Pathol. 1999;154:1777–1783. doi: 10.1016/S0002-9440(10)65433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saramaki O., Willi N., Bratt O., Gasser T.C., Koivisto P., Nupponen N.N., Bubendorf L., Visakorpi T. Amplification of EIF3S3 gene is associated with advanced stage in prostate cancer. Am. J. Pathol. 2001;159:2089–2094. doi: 10.1016/S0002-9440(10)63060-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kittler R., Putz G., Pelletier L., Poser I., Heninger A.K., Drechsel D., Fischer S., Konstantinova I., Habermann B., Grabner H., et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 35.Kittler R., Pelletier L., Heninger A.K., Slabicki M., Theis M., Miroslaw L., Poser I., Lawo S., Grabner H., Kozak K., et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 2007;9:1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- 36.Yang X.R., Chang-Claude J., Goode E.L., Couch F.J., Nevanlinna H., Milne R.L., Gaudet M., Schmidt M.K., Broeks A., Cox A., et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J. Natl. Cancer Inst. 2011;103:250–263. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coetzee S.G., Rhie S.K., Berman B.P., Coetzee G.A., Noushmehr H. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 2012;40:e139. doi: 10.1093/nar/gks542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens K.N., Vachon C.M., Lee A.M., Slager S., Lesnick T., Olswold C., Fasching P.A., Miron P., Eccles D., Carpenter J.E., et al. Common breast cancer susceptibility loci are associated with triple-negative breast cancer. Cancer Res. 2011;71:6240–6249. doi: 10.1158/0008-5472.CAN-11-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia-Closas M., Couch F.J., Lindstrom S., Michailidou K., Schmidt M.K., Brook M.N., Orr N., Rhie S.K., Riboli E., Feigelson H.S., et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet. 2013;45:392–398. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahoney D.W., Therneau T.M., Anderson S.K., Jen J., Kocher J.P., Reinholz M.M., Perez E.A., Eckel-Passow J.E. Quality assessment metrics for whole genome gene expression profiling of paraffin embedded samples. BMC Res. Notes. 2013;6:33. doi: 10.1186/1756-0500-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.