Abstract
Genome-wide association studies (GWAS) in schizophrenia have focused on additive allelic effects to identify disease risk loci. In order to examine potential recessive effects, we applied a novel approach to identify regions of excess homozygosity in an ethnically homogenous cohort: 904 schizophrenia cases and 1640 controls drawn from the Ashkenazi Jewish (AJ) population. Genome-wide examination of runs of homozygosity identified an excess in cases localized to the major histocompatibility complex (MHC). To refine this signal, we used the recently developed GERMLINE algorithm to identify chromosomal segments shared identical-by-descent (IBD) and compared homozygosity at such segments in cases and controls. We found a significant excess of homozygosity in schizophrenia cases compared with controls in the MHC (P-value = 0.003). An independent replication cohort of 548 schizophrenia cases from Japan and 542 matched healthy controls demonstrated similar effects. The strongest case–control recessive effects (P = 8.81 × 10−8) were localized to a 53-kb region near HLA-A, in a segment encompassing three poorly annotated genes, TRIM10, TRIM15 and TRIM40. At the same time, an adjacent segment in the Class I MHC demonstrated clear additive effects on schizophrenia risk, demonstrating the complexity of association in the MHC and the ability of our IBD approach to refine localization of broad signals derived from conventional GWAS. In sum, homozygosity in the classical MHC region appears to convey significant risk for schizophrenia, consistent with the ecological literature suggesting that homozygosity at the MHC locus may be associated with vulnerability to disease.
INTRODUCTION
Schizophrenia (SCZ) is a severe and chronic psychiatric disorder that affects ∼1% of the global population. It is highly heritable (0.7–0.8) (1), but its complex pattern of inheritance has hindered efforts to identify susceptibility genes. With the advancement of high-throughput single-nucleotide genotyping technologies, genome-wide association studies (GWAS) have discovered the first robust and replicable risk loci for SCZ; however, these SNPs account for only a tiny proportion of the total genetic risk (2). Moreover, GWAS generally examine only common SNPs with additive allelic effects on disease susceptibility, but such effects contribute only about one-quarter of the variance in liability to SCZ (3). Therefore, complementary approaches are required to identify sources of the ‘missing heritability’ of this phenotype (4,5).
An alternative approach for investigating genetic association is homozygosity mapping (6). Although homozygosity mapping was originally developed to identify rare recessive effects in consanguineous families (7), long chromosomal segments of consecutive homozygous alleles can be found throughout the genome even in outbred individuals (8). These ‘runs of homozygosity’ (ROHs) represent a form of genetic variation that can be examined for associations with phenotypic variation, which are not additive and thus are poorly assessed by standard GWAS analysis. Such ROHs may harbor rare deleterious variants, and even in outbred populations, an excess of ROHs may produce negative phenotypic effects owing to a process similar to inbreeding depression (9,10). Recent evidence has shown that a genome-wide excess of ROHs may be associated with increased risk of SCZ (11,12), as well as Alzheimer's disease (13). Specific regions of excess homozygosity have further been implicated in risk for Alzheimer's disease (13), rheumatoid arthritis (14) and autism (15).
Recent studies of ROHs in SCZ and other complex disorders have been marked by several limitations. First, ROHs are sparsely distributed among individuals from outbred populations, imposing substantial limitations on power (12). Second, the relative presence of ROHs can be impacted by subtle differences in ancestry that are difficult to control for in heterogeneous populations, such as European-Americans (16,17). Third, commonly used methods for the identification of ROHs typically do not distinguish between different alleles that may be homozygous at a given locus (11). Finally, the presence of ROHs may be a function of common haplotypes and/or lack of recombination at a given chromosomal locus, rather than having any association with disease; studies to-date have not directly compared ROH prevalence against baseline levels of chromosomal sharing. Recently developed methods utilizing hidden Markov models to determine identity-by-descent (IBD) of chromosomal segments permit a more precise specification of true autozygosity (18–20).
The present study was designed to overcome the aforementioned limitations by examining ROHs in an SCZ case–control cohort drawn from a homogeneous Ashkenazi Jewish (AJ) founder population (21). Further, we utilized two complementary methods to examine ROHs, using both segment length and inferred IBD status to determine ROH thresholds. In our cohort of 904 AJ SCZ cases and 1640 AJ controls, genotyped for ∼1 million SNPs (Illumina Omni1-Quad platform), we compared the presence of ROHs between cases and controls using our previously published whole-genome homozygosity association (WGHA) method (11). We then further refined this signal using an IBD approach: we compared the presence of homozygous or heterozygous tracts of IBD both genome-wide as well as on a locus-by-locus basis. Finally, we sought to replicate our findings in an independent SCZ case–control cohort from a different ethnically homogeneous (though not founder) population from Japan.
RESULTS
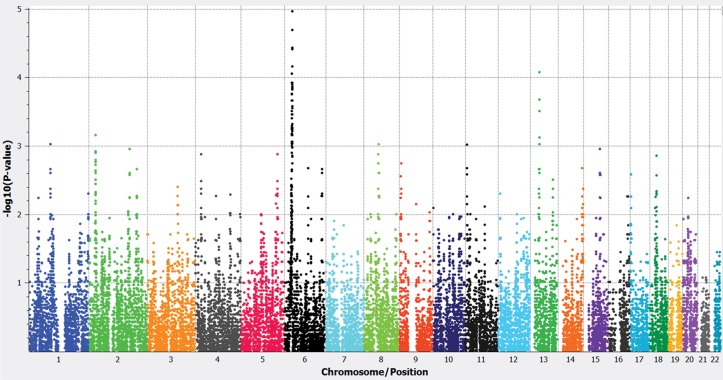
Principal component analysis (PCA) using ancestry informative markers (AIMs) that we have previously identified specifically for the AJ population (21) showed our AJ data set of 904 SCZ cases and 1640 controls to form a tight cluster (22); similar results are obtained with standard genome-wide PCA. After this QC, we performed WGHA in the AJ cohort. Specifically, we focused on large (>1.5 Mb in length with minimum 300 markers) ROHs for two reasons: (1) It has been shown that these are more likely to harbor rare, disease-associated variants (10,12) and (2) a comprehensive examination of ROHs across multiple worldwide populations has demonstrated that ∼1.5 Mb represents a reliable threshold for identifying ROHs that are due to true autozygosity (homozygosity by descent) (16). Comparing the tally of ROHs in cases versus controls at each locus, a genome-wide significant signal for excess ROHs within cases was observed at the MHC locus (P-value = 1.076 × 10−5, FDR = 0.0022) (Fig. 1). However, the observed peak was quite broad, extending ∼3 Mb (all P < 0.01 at hg18 coordinates chr6: 30053249–32848389).
Figure 1.
Manhattan plot for ROH analysis. Genome-wide significance threshold was set to P < 2.12 × 10−5 owing to non-independence across SNPs (mean ∼500 SNPs/ROH). The genome-wide significant signal was localized to the chr6 MHC region.
We next sought to refine localization of this signal using the GERMLINE algorithm (18). We also sought to determine whether these excess ROHs reflected specific haplotypes that are shared IBD between significantly larger numbers of case haplotype carriers compared with control haplotype carriers. We converted pairwise IBD segments into multi-sample haplotype clusters by using the DASH clustering algorithm (20). In our AJ data set, DASH analysis identified ∼1 million haplotype clusters genome wide. Each subject was then classified as a heterozygous carrier, homozygous carrier or non-carrier of each IBD segment, and the total number of heterozygous and homozygous IBD segments detected were counted for each subject.
In order to compare rates of homozygous IBD sharing in cases and controls, it was first necessary to control for baseline rates of IBD sharing, which can vary as a function of subtle sampling differences in ancestry (18). Our hypothesis therefore tests the rate of homozygous IBD sharing, controlling for overall rates of heterozygous IBD sharing using ANCOVA (Tables 1 and 2). Across the entire genome, we observed that the rate of carriage of homozygous IBD segments (controlling for the rate of heterozygous IBD sharing) was not significantly increased in SCZ cases compared with controls (Table 1) For the extended MHC region (Chr6: 25–35 Mb), however, we observed a significantly increased rate of homozygous IBD segments in cases relative to controls (P-value = 0.003; Table 1). Both MHC Class I and II regions show an excess of homozygous IBD segments in SCZ cases (Table 2), although the latter comparison only trends toward statistical significance owing to the lower overall rate of IBD sharing in that region. Effect sizes were small, but comparable, across each of the MHC segments (Cohen's d = 0.123 for the entire MHC, 0.118 for the Class I MHC and 0.082 for the Class II MHC).
Table 1.
Comparison of homozygous and heterozygous IBD segments across the whole genome and extended MHC region (chr6: 25–35 Mb) in AJ patients and controls
| Whole genome |
MHC locus chr6: 25–35 MB | |||
|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |
| Homozygous IBD segments | ||||
| Mean per subject (SD) | 30.97 (24.19) | 29.93 (21.13) | 10.23 (17.16) | 8.35 (13.08) |
| Heterozygous IBD segments | ||||
| Mean per subject (SD) | 7157.9 (913.0) | 7,258.6 (715.8) | 123.2 (43.4) | 125.14 (42.2) |
| ANCOVA (Hom controlling Het) | F1,2541 = 2.56 | P = 0.110 | F1,2541 = 8.72 | P = 0.003 |
Table 2.
Comparison of homozygous and heterozygous IBD segments for MHC Class I and II regions in AJ patients and controls
| MHC Class I region |
MHC Class II region |
|||
|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |
| Homozygous IBD segments | ||||
| Mean per subject (SD) | 4.36 (9.59) | 3.35 (7.38) | 0.60 (2.50) | 0.42 (1.86) |
| Heterozygous IBD segments | ||||
| Mean per subject (SD) | 57.12 (24.33) | 57.67 (23.16) | 17.22 (10.07) | 17.73 (9.94) |
| ANCOVA (Hom controlling Het) | F1,2541 = 8.498 | P = 0.004 | F1,2541 = 3.388 | P = 0.066 |
As a replication cohort, we used data from a Japanese GWAS of 548 patients with SCZ and 542 matched healthy controls with 265,558 markers after QC (23). All subjects were selected from the Tokai area of the Honshu Island of Japan. Previous studies have shown that this is an extremely homogeneous population, with slightly elevated rates of homozygosity compared with European cohorts (16). As presented in Table 3, we again observed higher homozygous IBD sharing in cases compared with controls across the extended MHC region, with comparable effect sizes as observed in the Ashkenazi cohort. It is important to note that the overall (genome wide) rate of homozygous IBD sharing in the outbred (though homogeneous) Japanese cohort was 30-fold lower than that in the Ashkenazi cohort, greatly reducing statistical power. However, meta-analysis revealed a robustly significant effect across the two cohorts (P = 0.0012).
Table 3.
Comparison of homozygous IBD segments across the extended MHC region (chr6: 25–35 Mb) in AJ and Japanese cohorts
| Homozygous IBD segments at MHC locus chr6: 25–35 Mb | ||||
|---|---|---|---|---|
| Cases | Controls | Effect sizea | P-value | |
| Ashkenazi Jewish Cohort | ||||
| Mean per subject (SD) | 10.23 (17.16) | 8.35 (13.08) | 0.123 | 0.003b |
| Japanese Cohort | ||||
| Mean per subject (SD) | 0.34 (1.66) | 0.23 (1.30) | 0.074 | 0.196b |
| Meta-analysis | 0.0012c | |||
aCohen's d comparison between means of cases and controls.
bP-value determined by ANCOVA, controlling for the rate of heterozygous segments.
cMeta-analytic P-value calculated using Stouffer's z trend procedure.
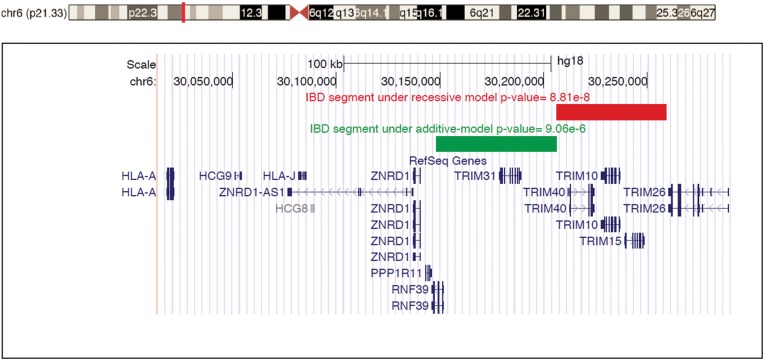
We next focused on individual homozygous IBD segments within the AJ SCZ cohort to pinpoint the source of the MHC signal. We performed association tests for IBD segments within the MHC, comparing additive, dominant and recessive models. Only two segments met the MHC-wide significance threshold (P < 1.09 × 10−4) for any model. As displayed in Table 4, one segment located at chr6: 30206368–30259349 (hg18) was strongly significant under the recessive model (chi-square = 28.62, df = 1, P = 8.81 × 10−8). Compared with the recessive model, the chi-square value for this same segment under the additive model was only half as large with P-value more than 3 orders of magnitude larger (chi-square = 14.47, df = 1, P = 1.42 × 10−4). This result is especially striking given that the additive model has enhanced power owing to measuring chromosomes (2n) rather than individuals (n). As displayed in Figure 2, this segment localized to the Class I MHC and encompassed three poorly annotated genes, TRIM10, TRIM15 and TRIM40. In contrast, a neighboring segment (chr6: 30148063–30206302) in the Class I MHC demonstrated evidence of MHC-wide significant additive effects (chi-square = 20.19, df = 1, P = 7.03 × 10−6), without evidence of strong recessive effects (Table 5); this segment contains two genes (RNF39 and TRIM31). No other IBD segments met the MHC-wide significance threshold under any model. Supplementary Material, Table S1, displays nine additional segments that approached significance (nominal P < 0.001 for any model); of these, seven segments had the strongest effects under the additive model, whereas two demonstrated strongest effects under the recessive model. These segments spanned both Class I and Class II MHC loci.
Table 4.
Association test for IBD region localized to chr6: 30206368–30259349
| Model | Allele count cases (carrier/non) | Allele count controls (carrier/non) | Chi-squared statistic | Degrees of freedom | P-value |
|---|---|---|---|---|---|
| Additive model | 352/1456 | 502/2778 | 14.47 | 1 | 1.42 × 10−4 |
| Dominant model | 306/598 | 478/1162 | 6.05 | 1 | 0.01394 |
| Recessive model | 46/858 | 24/1616 | 28.62 | 1 | 8.81 × 10−8 |
Figure 2.
The most significant homozygous IBD segment under the recessive model (shown as red bar) that was associated with SCZ. The top signal was localized to TRIM10, TRIM15 and TRIM40. The green bar represents the IBD segment with the strongest P-value under the additive model, localized to RNF39 and TRIM31.
Table 5.
Association test for IBD region localized to chr6: 30148063–30206302
| Model | Allele count cases (carrier/non) | Allele count controls (carrier/non) | Chi-squared statistic | Degrees of freedom | P-value |
|---|---|---|---|---|---|
| Additive model | 273/1535 | 355/2925 | 19.7 | 1 | 9.06 × 10−6 |
| Dominant model | 249/655 | 340/1300 | 15.2 | 1 | 9.66 × 10−5 |
| Recessive model | 24/880 | 15/1625 | 11.69 | 1 | 6.28 × 10−4 |
DISCUSSION
The major histocompatibility complex (MHC) has emerged as a region of major interest in SCZ genetics. The first evidence supporting the MHC as a potential SCZ susceptibility locus dates back to 1974 (24). More recently, large-scale GWAS studies have converged to demonstrate that the MHC contains the strongest association signal (2,25–28). Importantly, prior GWAS studies have been unable to precisely localize the source of this signal, owing to the extensive long-range linkage disequilibrium throughout the MHC; different studies have identified top SNPs ranging across a nearly 10-Mb extent (coordinates ranging from 25 to 35 Mb on Chromosome 6). Additionally, prior association studies of the MHC have exclusively examined an additive model of risk, in which each additional allele at a given locus contributes to risk in a linear manner.
The present study has extended prior research on the MHC in SCZ in two ways. First, we identified a global increase in MHC homozygosity in SCZ cases compared with controls. Second, utilizing an IBD approach in our founder population, we were able to further localize this effect to at least one specific segment in the Class I MHC; the strongest case–control recessive effects were observed near the HLA-A gene, in a segment encompassing three poorly annotated genes, TRIM10, TRIM15 and TRIM40. A run of homozygosity in this region, while relatively rare (<2% frequency in AJ controls), conveyed a 3-fold increase in the risk for SCZ. At the same time, we observed that these recessive effects coexist side-by-side with other association signals within the MHC that are more consistent with an additive effect. In particular, a segment encompassing RNF39 and TRIM31 demonstrated significant additive effects in our Ashkenazi cohort. While these genes are also poorly annotated, it is noteworthy that a prior GWAS has detected a strong additive association with the levels of the MHC component molecule beta-2 microglobulin to an SNP in TRIM31 (29).
Prior GWAS implicating the MHC in SCZ have been interpreted in the context of growing evidence that immune system dysfunction might play an important role in the etiology of SCZ. For example, in epidemiologic studies, several MHC-linked autoimmune/inflammatory disorders are found to have significantly increased prevalence in patients with SCZ (and their relatives) (30–32); moreover, exposure to prenatal infections has repeatedly been found to enhance risk of developing SCZ (33–36). Additionally, numerous studies have detected elevated levels of inflammatory markers (e.g. IL-2 and TNF-α) associated with immune system activation in patients with SCZ (37–39). It is important to emphasize that the present report, as with all genetic association studies, is not able to determine the mechanism by which MHC homozygosity affects risk; further research is needed in this area.
To the extent that immune mechanisms might be mediating the observed increase in homozygosity, however, it may be relevant to consider the heterozygote advantage hypothesis of the MHC. This hypothesis proposes that individuals heterozygous at MHC loci are able to respond to a greater range of pathogen peptides than homozygous individuals, and consequently, are more likely to have higher relative fitness (40–42). However, other possible explanations cannot be ruled out: (1) the specific rare alleles identified in our study may increase susceptibility or (2) excess homozygosity may mark individuals with increased risk owing to maternal–fetal HLA matching (43). It should be noted that a negative result has been reported (44) by a recent study using a large, well-characterized trio cohort to determine whether non-random mating, parent-of-origin effects for HLA alleles and maternal–fetal genotype incompatibility in the HLA increased risk for SCZ. Because our samples were drawn from a case–control cohort, further studies would be required to test this hypothesis more directly.
At the same time, many genes within the MHC are not primarily related to immune function and point to other possible neurodevelopmental mechanisms including synaptic plasticity, neurotransmitter signaling and brain myelination (45). For example, the class I HLA region includes two non-immune-related candidate genes for SCZ: MOG, which encodes myelin oligodendrocyte glycoprotein, a critical constituent of the myelin sheath which may be aberrant in SCZ (46), and GABBR1, which encodes a subunit of the metabotropic GABA(B) receptor, which is also implicated in SCZ pathophysiology (47). Moreover, HLA molecules themselves have recently been demonstrated to be directly expressed in the brain at varying levels during early development, even in the absence of infection or insult (48). In a recent study, the carriers of an SCZ-associated risk allele in the Class I MHC demonstrated larger ventricular volume (49), consistent with prior evidence of enlarged ventricles in Class I-deficient mice (50). It should be noted that the genes identified in our top recessive segment (TRIM10, TRIM15 and TRIM40) are only weakly expressed in mouse brain and are variably expressed in adult human brain according to the Allen Brain Atlas. Therefore, the limited knowledge on the biological function of TRIM10, TRIM15 and TRIM40 does not allow extensive speculation on the role of these genes in the etiology of the disease.
SUBJECTS AND METHOD
Participants
A total of 1156 SCZ cases from ethnically homogenous population (Ashkenazi Jews) have been collected and ascertained at the Hebrew University, Israel (Hebrew University Genetic Resource, HUGR, http://hugr.huji.ac.il). Cases were recruited from hospitalized inpatients at seven medical centers in Israel. All diagnoses were assigned after direct interview using the structured clinical interview, a questionnaire with inclusion and exclusion criteria, and cross-references to medical records. The inclusion criteria specified that subjects had to be diagnosed with SCZ or schizoaffective disorder by the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV), that all four grandparents of each subject were reported by the subject to be of AJ ethnic origin, and that each subject or the subject's legal representative has signed the informed consent form. The exclusion criteria eliminated subjects diagnosed with at least one of the following disorders: psychotic disorder owing to a general medical condition, substance-induced psychotic disorder or any Cluster A (schizotypal, schizoid or paranoid) personality disorder. Control samples (n = 2279) from healthy Ashkenazi individuals were collected from volunteers at the Israeli Blood Bank; these subjects were not psychiatrically screened but reported no chronic disease and were taking no medication at the time of blood draw. Corresponding institutional review boards and the National Genetic Committee of the Israeli Ministry of Health approved the studies. All samples were fully anonymized immediately after collection.
As a replication cohort, we used data from a Japanese SCZ GWAS of 575 patients with SCZ and 564 matched healthy controls as previously described (23). All subjects were unrelated, living in the Tokai area of the mainland of Japan, and self-identified as Japanese. Patients were included if they (1) met DSM-IV criteria for SCZ and (2) were physically healthy. Patients were excluded if they had a history of substance abuse, neurodevelopmental disorders, epilepsy or known mental retardation. Consensus diagnoses were made by at least two experienced psychiatrists according to DSM-IV criteria on the basis of unstructured interviews with patients, their families and review of medical records. Controls were selected from the general population with no history of mental disorders based upon self-report. After description of the study, written informed consent was obtained from each subject. This study was approved by the ethics committees of each participating university.
Genotyping and quality control
Ashkenazi Jewish samples were genotyped on 1 million genome-wide SNPs using Illumina HumanOmni1-Quad arrays according to manufacturers' specifications. SNP data for the Japanese SCZ cohort were obtained using the Affymetrix Human SNP array 5.0. To obtain high-quality data for further analysis, we performed QC steps for both data sets as follows: SNPs were filtered on call rate <98%, minor allele frequency <0.02 and Hardy–Weinberg exact test P < 0.000001 in controls. Samples were filtered based on genotype quality control filtration (sample call rate <97%, gender mismatch) and examined for cryptic identity and first- or second-degree relatedness using pairwise IBD estimation (PI_HAT) in PLINK (51) with 128,403 LD pruned (r2 > 0.2) genome-wide SNPs. Samples were excluded based on PI_Hat >0.125; the individual with lower call rate from each control/control or case/case pair was excluded, and controls were excluded from case–control pairs. After QC, there were 762,372 and 265,558 markers obtained for the AJ data set and Japan data set, respectively.
Population stratification
To obtain a homogenous population, the AJ samples after QC were further examined for underlying population stratification using PCA with AIMs specific for the AJ population (21). Samples with PCA results suggestive of one or more non-AJ grandparents were identified as outliers based on first principal component score of >0.01 and were excluded from further analysis (n = 607). We then compared the remaining AJ samples with HapMap CEU, JPT and CHB and YRI population. Following sample quality control filters and PCA-based population homogeneity, a total of 904 AJ SCZ cases and 1640 AJ controls as well as 548 Japanese SCZ cases and 542 Japanese controls were included in our IBD analysis. All PCA plots were obtained using the SVS7 (Golden Helix, Inc.) software package.
Runs of homozygosity
We performed WGHA using the SVS7 software package (GoldenHelix, Inc.) following methods described previously (11). Briefly, SNP data from each chromosome of each subject were interrogated for ROHs, which are long series of consecutive SNPs that are homozygous. Based on prior literature (10,12), we were interested in long, rare ROHs, so minimum run length was set to 1500 kb (with minimum 300 SNPs and no gap >100 kb to exclude centromeres and other anomalies). To permit some tolerance for genotyping error, up to one heterozygote call and up to five missing genotypes were permitted within a called ROH. After the identification of ROHs, each subject's SNP data were then converted to binary calls (0 or 1) at each position indicating whether that SNP is a member of a ROH for that individual. Next, at each position, data from all subjects were examined to determine how many cases and controls share a ROH call at a given position. There were 708,433 SNP positions, which were part of an ROH for >1 individual in the cohort; frequency of ROH in cases versus controls was compared at each such position using chi-square tests. Given that at least 300 consecutive SNPs were to comprise a ROH call, a threshold of P < 0.05/(708433/300) or P < 2.12 × 10−5 was required for genome-wide significance.
Identity-by-descent and homozygosity detection
As a complementary approach to identifying homozygosity, we utilized the GERMLINE algorithm (18), which uses a dynamically adjusted sliding window to detect, across all pairs of samples, genomic segments shared IBD from a recent ancestor. Genotype data were first phased using Beagle (version 3.3.2) (52,53) with default parameters. The phased genotype data were interrogated for pairwise shared IBD segments using GERMLINE (18). We ran GERMLINE with parameters tuned to identify IBD segments of 3 cM or greater, with a window size of 128 sites and 3 mismatching homozygous sites allowed. We next executed DASH (20) on the IBD segments to identify the specific cases and controls sharing each particular IBD segment. Using in-house perl scripts, we categorized each segment in each subject as homozygous or heterozygous and compared rates of homozygous IBD segments in cases versus controls, controlling for rates of heterozygous sharing, using ANCOVA (implemented in SPSS 22).
After conducting global tests of homozygous versus heterozygous IBD segments, we sought to localize the source of signal within the MHC by performing association tests in PLINK. For each of 457 IBD segments tested in the MHC region, we compared allelic, dominant and recessive models. We sought to identify segments that attained significance below a Bonferroni-corrected threshold (0.05/457 = 1.09 × 10−4) for any of these models. For each significant segment, we further sought to determine the best fitting model.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by grants from the NIH to S.M. (K99 MH101255), T.L. (RC2 MH089964; R01 MH084098) and A.M. (P50 MH080173). S.M. has received compensation for consultation from Genomind, Inc.
Supplementary Material
REFERENCES
- 1.Sullivan P.F., Kendler K.S., Neale M.C. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Ripke S., Sanders A.R., Kendler K.S., Levinson D.F., Sklar P., Holmans P.A., Lin D.Y., Duan J., Ophoff R.A., Andreassen O.A., et al. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S.H., Decandia T.R., Ripke S., Yang J., Sullivan P.F., Goddard M.E., Keller M.C., Visscher P.M., Wray N.R. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat. Genet. 2012;44:831. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mowry B.J., Gratten J. The emerging spectrum of allelic variation in schizophrenia: current evidence and strategies for the identification and functional characterization of common and rare variants. Mol. Psychiatry. 2013;18:38–52. doi: 10.1038/mp.2012.34. [DOI] [PubMed] [Google Scholar]
- 5.Owen M.J., Craddock N., O'Donovan M.C. Suggestion of roles for both common and rare risk variants in genome-wide studies of schizophrenia. Arch. Gen. Psychiatry. 2010;67:667–673. doi: 10.1001/archgenpsychiatry.2010.69. [DOI] [PubMed] [Google Scholar]
- 6.Morrow E.M., Yoo S.Y., Flavell S.W., Kim T.K., Lin Y., Hill R.S., Mukaddes N.M., Balkhy S., Gascon G., Hashmi A., et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander E.S., Botstein D. Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science. 1987;236:1567–1570. doi: 10.1126/science.2884728. [DOI] [PubMed] [Google Scholar]
- 8.Gibson J., Morton N.E., Collins A. Extended tracts of homozygosity in outbred human populations. Hum. Mol. Genet. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]
- 9.Ku C.S., Naidoo N., Teo S.M., Pawitan Y. Regions of homozygosity and their impact on complex diseases and traits. Hum. Genet. 2011;129:1–15. doi: 10.1007/s00439-010-0920-6. [DOI] [PubMed] [Google Scholar]
- 10.Szpiech Z.A., Xu J., Pemberton T.J., Peng W., Zollner S., Rosenberg N.A., Li J.Z. Long runs of homozygosity are enriched for deleterious variation. Am. J. Hum. Genet. 2013;93:90–102. doi: 10.1016/j.ajhg.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lencz T., Lambert C., DeRosse P., Burdick K.E., Morgan T.V., Kane J.M., Kucherlapati R., Malhotra A.K. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc. Natl. Acad. Sci. USA. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keller M.C., Simonson M.A., Ripke S., Neale B.M., Gejman P.V., Howrigan D.P., Lee S.H., Lencz T., Levinson D.F., Sullivan P.F. Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet. 2012;8:e1002656. doi: 10.1371/journal.pgen.1002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghani M., Sato C., Lee J.H., Reitz C., Moreno D., Mayeux R., St George-Hyslop P., Rogaeva E. Evidence of recessive Alzheimer disease loci in a Caribbean Hispanic data set: genome-wide survey of runs of homozygosity. JAMA Neurol. 2013;70:1261–1267. doi: 10.1001/jamaneurol.2013.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H.C., Chang L.C., Liang Y.J., Lin C.H., Wang P.L. A genome-wide homozygosity association study identifies runs of homozygosity associated with rheumatoid arthritis in the human major histocompatibility complex. PloS one. 2012;7:e34840. doi: 10.1371/journal.pone.0034840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casey J.P., Magalhaes T., Conroy J.M., Regan R., Shah N., Anney R., Shields D.C., Abrahams B.S., Almeida J., Bacchelli E., et al. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum. Genet. 2012;131:565–579. doi: 10.1007/s00439-011-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pemberton T.J., Absher D., Feldman M.W., Myers R.M., Rosenberg N.A., Li J.Z. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirin M., McQuillan R., Franklin C.S., Campbell H., McKeigue P.M., Wilson J.F. Genomic runs of homozygosity record population history and consanguinity. PloS one. 2010;5:e13996. doi: 10.1371/journal.pone.0013996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gusev A., Lowe J.K., Stoffel M., Daly M.J., Altshuler D., Breslow J.L., Friedman J.M., Pe'er I. Whole population, genome-wide mapping of hidden relatedness. Genom. Res. 2009;19:318–326. doi: 10.1101/gr.081398.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Browning B.L., Browning S.R. A fast, powerful method for detecting identity by descent. Am. J. Hum. Genet. 2011;88:173–182. doi: 10.1016/j.ajhg.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gusev A., Kenny E.E., Lowe J.K., Salit J., Saxena R., Kathiresan S., Altshuler D.M., Friedman J.M., Breslow J.L., Pe'er I. DASH: a method for identical-by-descent haplotype mapping uncovers association with recent variation. Am. J. Hum. Genet. 2011;88:706–717. doi: 10.1016/j.ajhg.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guha S., Rosenfeld J.A., Malhotra A.K., Lee A.T., Gregersen P.K., Kane J.M., Pe'er I., Darvasi A., Lencz T. Implications for health and disease in the genetic signature of the Ashkenazi Jewish population. Genom. Biol. 2012;13:R2. doi: 10.1186/gb-2012-13-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lencz T., Guha S., Liu C., Rosenfeld J., Mukherjee S., DeRosse P., John M., Cheng L., Zhang C., Badner J.A., et al. Genome-wide association study implicates NDST3 in schizophrenia and bipolar disorder. Nat. Commun. 2013;4:2739. doi: 10.1038/ncomms3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda M., Aleksic B., Kinoshita Y., Okochi T., Kawashima K., Kushima I., Ito Y., Nakamura Y., Kishi T., Okumura T., et al. Genome-wide association study of schizophrenia in a Japanese population. Biol. Psychiat. 2011;69:472–478. doi: 10.1016/j.biopsych.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Cazzullo C.L., Smeraldi E., Penati G. The leucocyte antigenic system HL-A as a possible genetic marker of schizophrenia. Br. J. Psychiat. 1974;125:25–27. doi: 10.1192/bjp.125.1.25. [DOI] [PubMed] [Google Scholar]
- 25.Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi J., Levinson D.F., Duan J., Sanders A.R., Zheng Y., Pe'er I., Dudbridge F., Holmans P.A., Whittemore A.S., Mowry B.J., et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stefansson H., Ophoff R.A., Steinberg S., Andreassen O.A., Cichon S., Rujescu D., Werge T., Pietilainen O.P., Mors O., Mortensen P.B., et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinberg S., de Jong S., Andreassen O.A., Werge T., Borglum A.D., Mors O., Mortensen P.B., Gustafsson O., Costas J., Pietilainen O.P., et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum. Mol. Genet. 2011;20:4076–4081. doi: 10.1093/hmg/ddr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tin A., Astor B.C., Boerwinkle E., Hoogeveen R.C., Coresh J., Kao W.H. Genome-wide association study identified the human leukocyte antigen region as a novel locus for plasma beta-2 microglobulin. Hum. Genet. 2013;132:619–627. doi: 10.1007/s00439-013-1274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benros M.E., Nielsen P.R., Nordentoft M., Eaton W.W., Dalton S.O., Mortensen P.B. Autoimmune diseases and severe infections as risk factors for schizophrenia: a 30-year population-based register study. Am. J. Psychiat. 2011;168:1303–1310. doi: 10.1176/appi.ajp.2011.11030516. [DOI] [PubMed] [Google Scholar]
- 31.Chen S.J., Chao Y.L., Chen C.Y., Chang C.M., Wu E.C., Wu C.S., Yeh H.H., Chen C.H., Tsai H.J. Prevalence of autoimmune diseases in in-patients with schizophrenia: nationwide population-based study. Br. J. Psychiat. 2012;200:374–380. doi: 10.1192/bjp.bp.111.092098. [DOI] [PubMed] [Google Scholar]
- 32.Eaton W.W., Byrne M., Ewald H., Mors O., Chen C.Y., Agerbo E., Mortensen P.B. Association of schizophrenia and autoimmune diseases: linkage of Danish national registers. Am. J. Psychiat. 2006;163:521–528. doi: 10.1176/appi.ajp.163.3.521. [DOI] [PubMed] [Google Scholar]
- 33.Brown A.S., Derkits E.J. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. Am. J. Psychiat. 2013;167:261–280. doi: 10.1176/appi.ajp.2009.09030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khandaker G.M., Zimbron J., Lewis G., Jones P.B. Prenatal maternal infection, neurodevelopment and adult schizophrenia: a systematic review of population-based studies. Psychol. Med. 2013;43:239–257. doi: 10.1017/S0033291712000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown A.S., Patterson P.H. Maternal infection and schizophrenia: implications for prevention. Schizop. Bull. 2011;37:284–290. doi: 10.1093/schbul/sbq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sorensen H.J., Mortensen E.L., Reinisch J.M., Mednick S.A. Association between prenatal exposure to bacterial infection and risk of schizophrenia. Schizop. Bull. 2009;35:631–637. doi: 10.1093/schbul/sbn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller N., Schwarz M.J. Immune system and schizophrenia. Curr. Immun. Rev. 2010;6:213–220. doi: 10.2174/157339510791823673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saetre P., Emilsson L., Axelsson E., Kreuger J., Lindholm E., Jazin E. Inflammation-related genes up-regulated in schizophrenia brains. BMC Psychiatry. 2007;7:46. doi: 10.1186/1471-244X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soderlund J., Schroder J., Nordin C., Samuelsson M., Walther-Jallow L., Karlsson H., Erhardt S., Engberg G. Activation of brain interleukin-1beta in schizophrenia. Mol. Psychiat. 2009;14:1069–1071. doi: 10.1038/mp.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doherty P., Zinkernagel R. A biological role for the major histocompatibility antigens. Lancet. 1975;1:1406–1409. doi: 10.1016/s0140-6736(75)92610-0. [DOI] [PubMed] [Google Scholar]
- 41.Hughes A.L., Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 42.Spurgin L.G., Richardson D.S. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B Biol. Sci. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer C.G., Hsieh H.J., Reed E.F., Lonnqvist J., Peltonen L., Woodward J.A., Sinsheimer J.S. HLA-B maternal-fetal genotype matching increases risk of schizophrenia. Am. J. Hum. Genet. 2006;79:710–715. doi: 10.1086/507829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y., Ripke S., Kirov G., Sklar P., Purcell S.M., Owen M.J., O'Donovan M.C., Sullivan P.F. Non-random mating, parent-of-origin, and maternal-fetal incompatibility effects in schizophrenia. Schizop. Res. 2013;143:11–17. doi: 10.1016/j.schres.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Debnath M., Cannon D.M., Venkatasubramanian G. Variation in the major histocompatibility complex [MHC] gene family in schizophrenia: Associations and functional implications. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2013;42:49–62. doi: 10.1016/j.pnpbp.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Cannon D.M., Walshe M., Dempster E., Collier D.A., Marshall N., Bramon E., Murray R.M., McDonald C. The association of white matter volume in psychotic disorders with genotypic variation in NRG1, MOG and CNP: a voxel-based analysis in affected individuals and their unaffected relatives. Transl. Psychiat. 2012;2:e167. doi: 10.1038/tp.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X., Davis R.L. The GABAergic anterior paired lateral neuron suppresses and is suppressed by olfactory learning. Nat. Neurosci. 2009;12:53–59. doi: 10.1038/nn.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sterner K.N., Weckle A., Chugani H.T., Tarca A.L., Sherwood C.C., Hof P.R., Kuzawa C.W., Boddy A.M., Abbas A., Raaum R.L., et al. Dynamic gene expression in the human cerebral cortex distinguishes children from adults. PloS one. 2012;7:e37714. doi: 10.1371/journal.pone.0037714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agartz I., Brown A.A., Rimol L.M., Hartberg C.B., Dale A.M., Melle I., Djurovic S., Andreassen O.A. Common sequence variants in the major histocompatibility complex region associate with cerebral ventricular size in schizophrenia. Biol. Psychiat. 2011;70:696–698. doi: 10.1016/j.biopsych.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 50.Huh P.W., Belayev L., Zhao W., Koch S., Busto R., Ginsberg M.D. Comparative neuroprotective efficacy of prolonged moderate intraischemic and postischemic hypothermia in focal cerebral ischemia. J. Neurosurg. 2000;92:91–99. doi: 10.3171/jns.2000.92.1.0091. [DOI] [PubMed] [Google Scholar]
- 51.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Browning B.L., Browning S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.