Abstract
Background
Rice (Oryza sativa) and Arabidopsis thaliana have been widely used as model systems to understand how plants control flowering time in response to photoperiod and cold exposure. Extensive research has resulted in the isolation of several regulatory genes involved in flowering and for them to be organized into a molecular network responsive to environmental cues. When plants are exposed to favourable conditions, the network activates expression of florigenic proteins that are transported to the shoot apical meristem where they drive developmental reprogramming of a population of meristematic cells. Several regulatory factors are evolutionarily conserved between rice and arabidopsis. However, other pathways have evolved independently and confer specific characteristics to flowering responses.
Scope
This review summarizes recent knowledge on the molecular mechanisms regulating daylength perception and flowering time control in arabidopsis and rice. Similarities and differences are discussed between the regulatory networks of the two species and they are compared with the regulatory networks of temperate cereals, which are evolutionarily more similar to rice but have evolved in regions where exposure to low temperatures is crucial to confer competence to flower. Finally, the role of flowering time genes in expansion of rice cultivation to Northern latitudes is discussed.
Conclusions
Understanding the mechanisms involved in photoperiodic flowering and comparing the regulatory networks of dicots and monocots has revealed how plants respond to environmental cues and adapt to seasonal changes. The molecular architecture of such regulation shows striking similarities across diverse species. However, integration of specific pathways on a basal scheme is essential for adaptation to different environments. Artificial manipulation of flowering time by means of natural genetic resources is essential for expanding the cultivation of cereals across different environments.
Keywords: Oryza sativa, rice, Arabidopsis thaliana, cereals, photoperiodic flowering, vernalization, florigen, flower development
INTRODUCTION
Floral initiation is a major physiological change that sets the switch from vegetative to reproductive development in most plant species. The transition from a vegetative (production of stem and leaves) to a reproductive stage (production of inflorescences and flowers) determines the time of flowering (or heading date in cereals) and is one of the most important developmental switches in the life cycle of plants. To maximize reproductive success and guarantee sufficient seed production for propagation of the species, flowering time should be tightly regulated through the integration of environmental inputs (daylength, temperature, light quality, water and nutrient availability) with endogenous cues (juvenility, stage of development). Depending on their requirement for daylength, plants can be classified into three categories. Long-day (LD) plants flower when the photoperiod exceeds a critical daylength, short-day (SD) plants flower when the photoperiod is shorter than a critical daylength and day-neutral plants flower regardless of daylength. The critical daylength for floral induction is specific to each species but often varies between accessions of the same species. In many plant species, flowering can also be stimulated by exposure to low non-freezing temperatures for several weeks. This process, known as vernalization, occurs in temperate zones during winter and prepares the plant to switch to reproductive growth only after the cold season, when temperatures become favourable again. Molecular genetic studies on model plants such as arabidopsis and rice (Oryza sativa) have allowed identification of genes controlling responses to environmental inputs and many of those have been shown to be conserved between the two model species, despite 150 million years of divergent evolution (Chaw et al., 2004). However, during evolution, other pathways have evolved and several factors have been recruited to novel functions, increasing the diversity of flowering behaviours and adapting the species to grow across broader areas. Although arabidopsis and rice have been crucial to establish the basal architecture of floral regulatory networks, studies conducted on other species, including temperate cereals (e.g. wheat and barley) greatly contributed to our understanding of the molecular mechanisms controlling flowering.
In this review we will focus on rice as a model for SD plants and discuss the pathways involved in daylength measurement and flowering, mainly in comparison with arabidopsis. We will discuss similarities of the core regulatory pathways controlling photoperiodic flowering but will also address the pathways that have evolved specifically in rice, which are not present in arabidopsis. Finally, we will contrast regulatory networks active in temperate cereals with those of arabidopsis and rice. We will take into account the spatial separation of functions in leaves and at the shoot apex.
Besides its importance as a model system, rice is a crop and staple food for most parts of the world. It was first domesticated in Southern China and has evolved to adapt to a range of geographical regions over time. Currently, rice is cultivated over a wide range of latitudes from 55°N to 36°S (Khush, 1997). This has been possible through the diversification of flowering time. In fact, flowering of rice is accelerated under SD conditions; however, artificial selection has led to successful cultivation even under LD conditions. Some varieties of rice with weak or no photoperiod sensitivity flower very early at Northern latitudes, particularly in Europe, Northern China and Japan. We will discuss the genetic factors that have allowed such expansion and that represent an increasing list of molecular tools that plant breeders can use to expand further rice cultivation worldwide.
THE PHOTOPERIODIC PATHWAY IN ARABIDOPSIS AND RICE
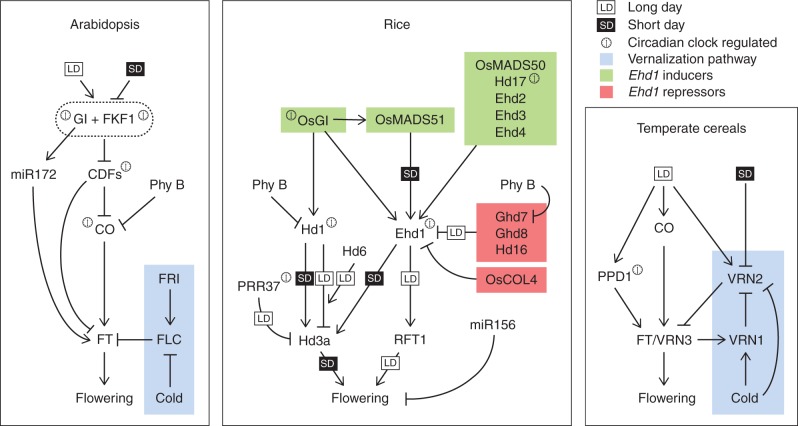
Facultative LD species, such as arabidopsis, accelerate flowering as days become longer, and several genes have been isolated that control responses to photoperiodic cues (Andrés and Coupland, 2012). The corresponding mutants flower later when exposed to long inductive days, typical of spring or early summer, but do not affect flowering time when plants are grown under SDs (Turck et al., 2008). Central to the photoperiod pathway in arabidopsis is CONSTANS (CO), a zinc finger transcription factor that integrates environmental and endogenous information to trigger flowering at the appropriate time of the year. In the vascular tissue of leaves, the CO protein directly activates expression of FLOWERING LOCUS T (FT), which encodes a florigenic protein promoting flowering (An et al., 2004; Tiwari et al., 2010). Expression of CO is regulated by the circadian clock that sets its rhythmic cycling to reach a peak at the end of the day under LDs. Cycling of CO mRNA depends on the activity of a protein complex formed by GIGANTEA (GI) and FLAVIN BINDING, KELCH REPEAT, F-BOX 1 (FKF1), a protein containing an F-box and blue light photoreceptor domains. The interaction between GI and FKF1 requires light, and the presence of GI protein is necessary to confer stability to the FKF1 protein (Sawa et al., 2008; Fornara et al., 2009). Upon interaction with GI, FKF1 targets a group of DOF transcription factors, collectively known as CYCLING DOF FACTOR (CDF) genes, for degradation (Fornara et al., 2009). CDF proteins directly bind to the promoters of CO and FT, to prevent their expression when plants are exposed to short photoperiods (Imaizumi et al., 2005; Y. H. Song et al., 2012). Degradation of the CDFs occurs at the DNA of target loci and results in de-repression of CO and FT, allowing flowering to occur (Fig. 1). Therefore, genotypes in which the activity of the CDF genes is strongly reduced are insensitive to daylength, and flower early under any photoperiodic condition (Fornara et al., 2009; Gómez-Ariza and Fornara, 2012).
Fig. 1.
Simplified regulatory networks controlling florigen production in leaves of different plant species. Networks of arabidopsis (left panel), rice (central panel) and temperate cereals (right panel) are compared. Small white and black boxes indicate regulatory connections occurring primarily under LDs and SDs, respectively. Arrows indicate transcriptional activation, whereas flat-ended arrows indicate transcriptional repression. The blue boxes in both the arabidopsis and the temperate cereal models include genes involved in vernalization responses. The green and red boxes in the rice model include positive and negative regulators of Ehd1 expression, respectively. A clock symbol close to a gene indicates that its transcription is controlled by the circadian clock.
CONSTANS is also tightly regulated at the post-transcriptional level. Despite its transcription being high during the night under both SDs and LDs, the protein does not accumulate in the dark because it is quickly directed to the proteasome through the activity of an E3 ubiquitin ligase encoded by CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) (Jang et al., 2008; Liu et al., 2008). Other proteins, including phytochromes and ubiquitin ligases, influence the stability of CO protein at different times of day, to ensure it accumulates only when the day is sufficiently long (Valverde et al., 2004; Pineiro et al., 2012; Y. H. Song et al., 2012).
Regulation of photoperiodic flowering through CO has features of the external coincidence model of photoperiodism, originally proposed by Pittendrigh and Minis (1964). According to this model, an endogenous oscillator sets the rhythmic phase of expression of target molecules. Coincidence of a particular phase of expression with an external factor, such as light, triggers a developmental response. Several steps of the photoperiodic cascade of arabidopsis represented in Fig. 1 require light at the appropriate time of a circadian cycle to induce flowering. The GI–FKF1 regulatory complex is expressed and stabilized only when the peak of mRNA expression of the two genes coincides with light under long daylengths. This allows the corresponding proteins to interact and the complex to be stabilized. Similarly, CO protein accumulation takes place only at the end of an LD, in the presence of light that stabilizes it. Only under these conditions can the CO protein accumulate, activate FT expression and induce flowering. Under SDs, GI and FKF1 proteins do not interact, preventing CO mRNA from increasing at the end of the day. Therefore, light is necessary at the appropriate time of day to activate the pathway. In agreement with this model, in mutants in which the diurnal waveform of CO mRNA is also displaced towards the light phase under SDs, or in which the protein is allowed to accumulate because of mutations in genes controlling its post-transcriptional stability, FT expression and flowering are activated regardless of daylength (Yanovsky and Kay, 2002; Valverde et al., 2004; Jang et al., 2008; Fornara et al., 2009). This mechanism incorporates endogenous and environmental information to synchronize flowering with seasons characterized by LDs.
Rice shares a similar photoperiodic pathway, mediating daylength responses. OsGI and Heading Date 1 (Hd1) have been cloned and shown to encode homologues of arabidopsis GI and CO, respectively (Yano et al., 2000; Hayama et al., 2003). Several homologues of FT are encoded in the rice genome and at least three of them can promote flowering when overexpressed, i.e. Hd3a, RICE FLOWERING LOCUS T 1 (RFT1) and FT-like 1 (FTL1) (Izawa et al., 2002; Kojima et al., 2002; Ogiso-Tanaka et al., 2013).
Similar to arabidopsis, control of Hd1 expression is crucial to confer a photoperiodic response (Brambilla and Fornara, 2013). Hd1 was originally identified as a major quantitative trait locus (QTL) and was later cloned by map-based approaches (Yano et al., 2000). Hd1 is highly homologous to CO and the Hd1 protein is thought to be involved in DNA binding. However, direct interaction of Hd1 with the Hd3a promoter has not been reported. Additionally, whereas CO promotes flowering under LDs, Hd1 promotes flowering under SDs but represses flowering under LDs. The bi-functionality of Hd1 became clear through the analysis of plants carrying hd1 loss-of-function alleles that cause late flowering under SDs and early flowering under LDs (Yano et al., 2000; Izawa et al., 2002). The effect on flowering is correlated with Hd3a mRNA levels that are reduced in hd1 mutants grown under SDs and increased when hd1 mutants are grown under LDs (Izawa et al., 2002). Therefore, Hd1 has opposite effects on Hd3a expression and flowering that switch depending on daylength.
OsGI is a positive regulator of Hd1 expression, under both SDs and LDs. In arabidopsis, overexpression of GI induces CO transcription and rapid flowering under any daylength. Also in rice, overexpression of OsGI in transgenic plants triggers higher Hd1 expression. However, because of the bi-functional features of Hd1, higher levels of OsGI and Hd1 lead to decreased Hd3a transcription and inhibition of flowering under both SDs and LDs (Hayama et al., 2003). The same inhibitory effect was observed when the Hd1 gene itself was overexpressed under SDs, abolishing its diurnal cycling and causing high levels of transcripts also to accumulate during the light phase. These data suggested that exposure of Hd1 protein to light converted it to a repressor of flowering, even if the length of the light phase was below the critical threshold necessary for flowering (Ishikawa et al., 2011). The molecular mechanisms responsible for switching Hd1 function are currently unknown. In arabidopsis, CO acts as floral activator only and, therefore, what is crucial in this species is the shape of CO protein accumulation during the day, which needs to be tightly controlled in order to reach a maximum at dusk under LDs. Conversely, accumulation of Hd1 protein occurs during both the light and dark phases, suggesting that a different mechanism is responsible for the bi-functional activity of Hd1 (Ishikawa et al., 2011).
OsGI transcription is controlled by the circadian clock and shows a rhythmic expression pattern which peaks at the end of the light period, similarly to arabidopsis (Hayama et al., 2003). Mutations in osgi affect a large set of rice genes, besides Hd1, and about 75 % of rice transcripts show altered levels in the mutant, during a diurnal time course (Izawa et al., 2011). Many of these genes encode proteins probably involved in circadian clock function, at least based on homology with known arabidopsis clock regulators, and OsGI can affect expression of LATE ELONGATED HYPOCOTYL (LHY), and of several PSEUDO RESPONSE REGULATOR (PRR) genes, including PRR1, PRR59 and PRR95. Interestingly, Hd2/PRR37 expression was not affected in the osgi mutant, possibly indicating independent control of heading date by these factors (Fig. 1). However, how OsGI protein activates expression of Hd1 has not been clarified yet. A homologue of FKF1 exists in rice and shows a diurnal expression pattern identical to that described in arabidopsis (Murakami et al., 2007; Higgins et al., 2010). DOF transcription factors are also present in the rice genome, and one of them, OsDOF12, is implicated in photoperiodic flowering (Li et al., 2008, 2009). Transcription of OsDOF12 is high in leaves and diurnally regulated with a trough during the night (Izawa et al., 2011). Transgenic rice overexpressing OsDOF12 flowers early compared with wild-type plants under LD, but not SD conditions, showing higher Hd3a expression compared with non-transgenic controls (Li et al., 2009). However, no altered levels of Hd1 mRNA were reported, suggesting a different genetic route for OsDOF12 action on flowering (Li et al., 2009).
Based on these observations, it was possible to conclude that the external coincidence model can be applied to rice, albeit with modifications from the scheme proposed for an LD plant (Hayama et al., 2003). Short-day plants measure the duration of the dark phase, during which expression of florigenic proteins starts. Hd1 is likely to act as a sensor of night length, being converted to a floral activator when exposed to darkness. Expression of Hd1 peaks during the night phase and, when this is sufficiently long, can promote enough Hd3a protein expression and initiate flowering. In this modified model, the coincidence of darkness with peak expression of Hd1 is crucial to trigger a developmental response.
THE EHD1–GHD7 PATHWAY IS UNIQUE TO RICE
Flowering in rice does not strictly require the OsGI–Hd1–Hd3a signalling pathway, and activation of florigens can be achieved through independent mechanisms. Natural variation between rice species and cultivars allowed cloning of EARLY HEADING DATE 1 (Ehd1) which encodes a B-type response regulator that does not have orthologues in arabidopsis and defines a unique floral activation pathway in rice (Doi et al., 2004). Ehd1 promotes flowering particularly under SDs, in parallel to Hd1, but can also promote flowering under LDs, when Hd1 acts as a repressor (Doi et al., 2004). The Ehd1 protein consists of a receiver domain at its N-terminus and a GARP DNA-binding motif (Riechmann et al., 2000). It induces flowering under SDs and LDs by upregulating Hd3a or RFT1 independently of Hd1, demonstrating the potential of Ehd1 and Hd1 to act redundantly on separate pathways (Doi et al., 2004).
Regulation of Ehd1 is crucial for correct flowering time in rice, and a large group of activators and repressors has been cloned and shown to modulate its expression (Fig. 1). Repressors of Ehd1 have a central role in the photoperiodic network and, among them, Grain number, plant height and heading date 7 (Ghd7) is particularly important to shape Ehd1 diurnal and seasonal transcription. Ghd7 encodes a CCT domain protein and is expressed at higher levels under LDs, correlating with limited expression and activity of Ehd1 under non-inductive photoperiods (Xue et al., 2008). Besides its effect on flowering, Ghd7 controls other traits in rice including grain number and plant height, indicating pleiotropic roles for the protein in other processes (Xue et al., 2008).
Analysis of Ghd7 and Ehd1 expression in response to light has defined a novel coincidence mechanism and a double gating system that sets critical daylength recognition for Hd3a expression under specific photoperiods (Itoh et al., 2010). Rice plants exposed to photoperiods shorter than 13·5 h induce Hd3a and Ehd1 expression, while reducing Ghd7 expression. Such interplay is achieved by a double regulatory mechanism dependent on OsGI and phytochromes. Induction of Ehd1 expression in the morning requires functional OsGI, which sets a gate (a sensitive phase set by the circadian clock in response to light) around dawn, which is independent of daylength, because it occurs at the same time under both LDs and SDs. The gate is sensitive to blue light in the morning and, when open, Ehd1 expression increases and Hd3a is activated. OsGI protein accumulation reaches its trough at dawn and therefore its effect on the gate is likely to be indirect. Under LDs, Ghd7 expression is high and its inducibility is gated at the same time as the OsGI and blue light-dependent gate. Under these conditions, induction of Ghd7 in the morning is sufficient to repress Ehd1 transcription and delay flowering. However, as daylength decreases under a critical threshold, maximum inducibility of Ghd7 is gated during the night, resulting in reduced expression the following morning and de-repression of Ehd1 (Itoh et al., 2010). Expression of Ghd7 requires functional phytochromes and it is abolished in the PHOTOPERIOD INSENSITIVITY 5 (SE5) mutant, which encodes a haem oxygenase very similar to LONG HYPOCOTYL 1 (HY1) of arabidopsis and is impaired in phytochrome chromophore biosynthesis (Izawa et al., 2000). Plants in which SE5 is mutated are insensitive to photoperiod and flower early under both SDs and LDs (Izawa et al., 2000). Expression of Ghd7 is also strongly reduced in plants where a long night is interrupted by a short red light pulse (a ‘night break’) that converts phytochromes to the inactive form. These data suggested that functional phytochromes are required for correct expression of Ghd7 and floral repression, and that red light signals are integrated in the photoperiodic flowering network through Ghd7. Rice has three phytochrome genes (PhyA–PhyC) but se5 single mutants lack all functional forms. Therefore, single and double phytochrome mutants have been useful tools to understand how correct Ghd7 expression is determined (Osugi et al., 2011). The results suggest that phytochromes are not required to set the light-sensitive phase for Ghd7 expression. However, PhyA homodimers and PhyB–PhyC heterodimers are independently sufficient to trigger Ghd7 transcription, whereas PhyB can repress it. The action of phytochromes on Ghd7 expression is therefore more complex than previously anticipated by the analysis of SE5.
Neither such a double gating mechanism nor the existence of homologues of Ehd1 and Ghd7 has been observed in arabidopsis. Expression of FT, as opposed to Hd3a expression, showed no critical daylength threshold in mathematical modelling experiments based on biological data (Salazar et al., 2009). Additionally, the flowering time of arabidopsis accessions grown under several SD and LD photoperiods indicated that most ecotypes could discriminate variations of 2 h under several SDs and LDs, and no critical photoperiod could be determined (Giakountis et al., 2010). Despite the fact that broad genetic variation could also be expected among rice varieties, these data point to a crucial difference in the way photoperiod is perceived in an LD and SD plant to promote flowering, probably reflecting the different adaptation to their environment.
Regulators of Ghd7
The double gating mechanism based on the interplay between Ehd1 and Ghd7 is crucial for daylength measurement, and proper regulation of Ghd7 expression and activity sets the sensitivity of the measure. Consistent with Ghd7 being central to the pathway, extensive natural genetic variation was reported at the Ghd7 locus and at loci regulating its activity. Such variation has allowed the cloning of a number of additional QTLs involved in flowering.
Map-based cloning of Heading date 16 (Hd16) revealed that this gene encodes a caseine kinase-I protein (Hori et al., 2013; Kwon et al., 2014). The Hd16 protein directly interacts with and phosphorylates Ghd7, thus converting it to an active repressor of flowering. Natural allelic variants of Hd16 showing reduced functionality de-repress expression of Ehd1 and the florigens, leading to accelerated flowering particularly under LD conditions. Natural variation at the Ehd3 locus allowed cloning of a repressor of Ghd7. Ehd3 encodes a nuclear protein with two PHD-finger motifs, and Matsubara et al. (2011) demonstrated that it is a repressor of Ghd7 and activator of Ehd1 expression particularly under LDs. The two functions can be genetically separated, indicating that activation of Ehd1 by Ehd3 can also be achieved independently of Ghd7 repression (Matsubara et al., 2011).
Finally, cloning of Hd17 revealed that it encodes a homologue of EARLY FLOWERING 3 (ELF3) which represses Ghd7 expression under SDs and LDs, resulting in increased levels of Ehd1. The effect on flowering is probably indirect and due to the influence of OsELF3 on circadian clock function (Saito et al., 2012; Zhao et al., 2012; Y. Yang et al., 2013).
Activators of Ehd1 expression
Several genes have been cloned whose activity eventually converges on Ehd1 transcriptional regulation.
Early heading date 2 (Ehd2) encodes a putative transcription factor with zinc finger motifs, which is an orthologue to INDETERMINATE1 (ID1) of maize (Matsubara et al., 2008a). Similarly to ID1, Ehd2 is expressed mainly in leaves (Colasanti et al., 2006). Plants mutated in Ehd2 prevent upregulation of Ehd1, show delayed flowering under SDs and cannot flower under LDs, indicating that Ehd2 is a fundamental gene for the LD promotion of flowering (Matsubara et al., 2008a; Wu et al., 2008).
In addition to Ehd2, OsMADS51, a type I MADS-box transcription factor, promotes flowering upstream of Ehd1 under SD conditions (Kim et al., 2007). Plants in which expression of OsMADS51 is artificially increased or decreased display altered heading dates that can be observed only under SD, but not under LD conditions. Expression of OsMADS51 depends on OsGI, and Kim et al. (2007) demonstrated that reduction of OsGI transcription by RNA interference (RNAi) is correlated with lower expression of OsMADS51 and Ehd1. This study showed how an OsGI-dependent signalling cascade can activate Hd3a under SDs, independently of Hd1, through Ehd1 and OsMADS51.
The Ehd4 locus encodes a nuclear-localized CCCH-type zinc finger protein unique to the Oryza genus (Gao et al., 2013). Loss-of-function mutants flower late under any condition, but particularly under LDs. Delayed flowering is associated with low levels of Ehd1 and the florigens, but not of Hd1, indicating that Ehd4 specifically targets the Ehd1 pathway (Fig. 1).
Repressors of Ehd1 expression
Several repressors of Ehd1 expression have been cloned that can function in a range of photoperiods. Hd5/DAYS TO HEADING 8 (DTH8)/Ghd8 (from here on Ghd8) encodes a putative HEME ACTIVATOR PROTEIN 3 (HAP3) subunit of the CCAAT-box-binding transcription factor complex. It acts as floral repressor under LD conditions and delays flowering by downregulating the expression of Ehd1, Hd3a and RFT1 (Wei et al., 2010). Under SDs, Ghd8 was reported to induce expression of these floral regulators, promoting flowering and showing some degree of bi-functionality, similarly to Hd1 (Yan et al., 2011). Expression of Ghd8 is not influenced by Ghd7 and Hd1, two major LD repressors, indicating a distinct genetic pathway for flowering time control (Wei et al., 2010). However, in arabidopsis, HAP3 and HAP5 proteins have been shown to interact physically with CO protein, forming a CCAAT-box-binding complex directly controlling FT expression (Wenkel et al., 2006; Cai et al., 2007; Kumimoto et al., 2010). If such a mechanism were operating in rice, HAP/Ghd8 proteins would act in the same genetic pathway as Hd1 to control Hd3a expression, perhaps directly. It will be interesting to determine whether Ghd8 and Hd1 proteins can physically interact to control expression of Ehd1 or the florigens and influence photoperiodic flowering responses. Genetic data indicate that when overexpressed in arabidopsis, Ghd8 triggers early flowering under LDs, and causes no alteration of flowering time under SDs, similarly to overexpression of other HAP subunits of arabidopsis, providing evidences that the function of this class of proteins is conserved between monocots and dicots (Kumimoto et al., 2010; Yan et al., 2011).
OsLFL1 (Oryza sativa LEC2 and FUSCA3 Like 1) is a B3 transcription factor that can delay flowering upon overexpression, by repressing Ehd1 and its downstream targets (Peng et al., 2008). Repression of Ehd1 mediated by OsLFL1 is probably direct, as demonstrated by chromatin immunoprecipitation and gel shift assays (Peng et al., 2007). Binding of OsLFL1 protein is mediated by RY motifs present in the promoter region of Ehd1. Such motifs can also mediate transactivation of a reporter gene in yeast when OsLFL1 protein is expressed from an effector plasmid. OsLFL1 is the only direct regulator of Ehd1 reported to date. Its transcriptional control is mediated by chromatin modifications that require O. sativa VERNALIZATION INSENSITIVE LIKE 2 (OsVIL2) and O. sativa EMBRYONIC FLOWER 2b (OsEMF2b), the former encoding a PHD finger histone-binding protein, and the latter encoding a component of Polycomb Repressor Complex 2 (PRC2) (J. Yang et al., 2013). A protein complex containing OsVIL2 and OsEMF2b can associate with the OsLFL1 promoter and enrich histones with H3K27me3 marks, leading to silencing of the locus. Consistently, osvil2 and osemf2b mutants are late flowering and show decreased expression of Ehd1 and the florigens (J. Yang et al., 2013). These mechanisms highlight the importance of epigenetic regulation of gene expression to fine-tune environmental responses. The fundamental nature of these processes also accounts for its occurrence across divergent plant groups (Sung et al., 2006; Oliver et al., 2009; J. Yang et al., 2013).
Fig. 2.
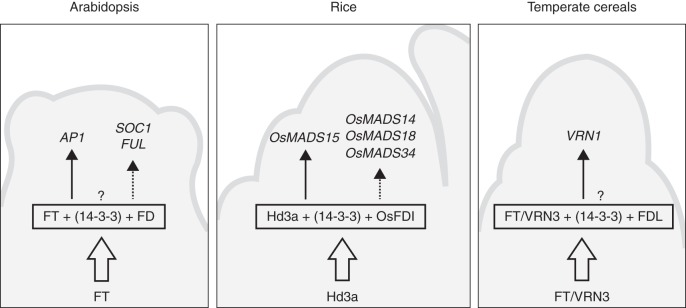
Molecular responses of the shoot apical meristem to florigenic proteins. Interaction of florigen with FD-like genes is required to promote expression of MADS-box transcription factors, one of the first molecular events occurring upon floral transition. The requirement for 14-3-3 proteins has not been demonstrated in arabidopsis and temperate cereals, and the linker protein is therefore indicated with a question mark on the top. Arrows indicate direct transcriptional activation. Dashed arrows indicate indirect transcriptional activation.
OsCO-Like 4 (OsCOL4) is a member of the CONSTANS-LIKE (COL) family in rice. It is a constitutive flowering repressor that functions under both SD and LD conditions (Lee et al., 2010). The OsCOL4 mutant plants showed early flowering under both SDs and LDs, while the overexpressing transgenic lines showed a late flowering phenotype (Lee et al., 2010). The expression of Ehd1 and Hd3a was higher in OsCOL4 mutants, suggesting that it functions upstream of these floral regulators. Neither the overexpressors nor the mutant plants had altered transcription levels of Hd1 or OsGI, indicating that OsCOL4 is specific to the Ehd1 pathway.
DIFFERENTIAL REGULATION OF FLORIGEN EXPRESSION UNDER LONG AND SHORT DAYS
Plants exposed to inductive daylengths activate expression of florigenic proteins in the vasculature of leaves. In arabidopsis, FT and TWIN SISTER OF FT (TSF) proteins are expressed in the phloem of leaves and act as mobile, long-distance signals to trigger developmental reprogramming at the shoot apical meristem (SAM) (An et al., 2004; Yamaguchi et al., 2005; Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007). Hd3 and RFT1 encode two rice florigens and, similarly to their arabidopsis orthologues, are expressed in the rice phloem and move to the SAM (Tamaki et al., 2007; Komiya et al., 2009). Plants suppressing both Hd3a and RFT1 mRNA expression by RNAi did not flower after 300 d under SDs. RNAi suppression of Hd3a only delayed flowering under SDs, whereas suppression of RFT1 expression delayed flowering under LDs but not under SDs, indicating that the two florigens have distinct effects on floral promotion depending on the photoperiod. Additionally, RFT1 is redundant to Hd3a under SDs (Komiya et al., 2008, 2009). Critical daylength recognition does not affect RFT1 expression as severely as Hd3a expression, and RFT1 is essentially less influenced by LD repression as compared with Hd3a (Itoh et al., 2010). These data have contributed to build a model by which RFT1 and Hd3a encode LD- and SD-specific florigens, respectively. The diverse impact of these two florigens on photoperiodic flowering raises interesting observations, as they are very similar in structure and located physically close to each other (11·5 kb), suggesting a common origin through tandem duplication, but also indicating the existence of distinct mechanisms of transcriptional regulation (Komiya et al., 2008). Accumulating evidence indicates that changes in the chromatin state can influence florigen expression, in both rice and arabidopsis, and possibly cause differential regulation of florigens in rice (Komiya et al., 2008; Adrian et al., 2010; Gu et al., 2013). Periodic deacetylation of histones at the FT locus in arabidopsis are associated with transcriptional repression of FT. Components of a histone deacetylase complex (HDAC) associate with the FT locus to limit its expression at the end of the day (Gu et al., 2013). Plants mutated in such components show enrichment of acetylated histones at FT, increase FT expression and cause earlier flowering compared with wild-type controls. Interestingly, in such mutants, FT induction still requires functional CO, suggesting that chromatin modifications leading to de-repression of FT still need the presence of a transcriptional activator. Although acetylation of histones at the FT locus has been associated with increased transcription of FT, these chromatin modifications were reported to be a consequence of FT activation, rather than its cause, suggesting that the timing of these changes needs to be carefully monitored in order to reach a conclusion about a causal relationship (Adrian et al., 2010). In rice, accumulation of RFT1 mRNA in Hd3a RNAi-suppressed plants was associated with increased H3K9 acetylation at the RFT1 locus, indicating a possible relationship between histone modifications and transcriptional activity at florigenic loci (Komiya et al., 2008). Whether such modifications also involve the Hd3a locus and precede transcriptional activation remains to be determined. A mutant defective in a histone methyltransferase, SDG724, demonstrated delayed flowering and reduced levels of Hd3a and RFT1 (Sun et al., 2012). Interestingly, the OsMADS50 and RFT1 loci, but not the Hd3a locus, showed enrichment of H3K36me2/me3 chromatin marks, associated with transcriptionally active chromatin. These data indicate differential regulation of the LD flowering pathway by histone methylation and provide an example of how RFT1 and Hd3a could be differentially controlled by the dynamics of chromatin states.
Recent cloning of Hd2 showed that the gene underlying the QTL is encoded by OsPRR37, a homologue of PRR7 of arabidopsis and PPD1 of wheat and barley (Koo et al., 2013). Flowering of plants carrying loss-of-function alleles of OsPRR37 is accelerated under any daylength, but is particularly enhanced under LDs. Under such conditions, OsPRR37 suppresses Hd3a but not RFT1 expression, indicating differential sensitivity of the florigens to the presence of this floral regulator. The molecular mechanisms that allow OsPRR37 to discriminate between Hd3a and RFT1 are unclear, but provide another layer of control that fine-tunes photoperiodic responses (Fig. 1).
A GENE NETWORK AT THE SHOOT APICAL MERISTEM INTEGRATES ENVIRONMENTAL CUES
In previous sections, we have described how daylength affects flowering and how light duration is monitored through a regulatory network. The products of such a network are florigenic proteins which are highly expressed in response to inductive photoperiods and encode long-distance transmissible signals. Florigens have been isolated from several plant species and shown to control floral induction. In arabidopsis and rice, FT, Hd3a and RFT1 proteins are produced in leaves and transported to the SAM upon perception of the appropriate photoperiods, initiating panicle development (Corbesier et al., 2007; Tamaki et al., 2007). A complex network of regulatory proteins controls perception of florigenic signals at the apex and drives downstream developmental events.
Proteins that interact with Hd3a at the shoot apical meristem
Florigens cannot directly function as transcriptional regulators in meristematic cells and thus interaction with other transcription factors is essential for their functioning. In several plant species, basic leucine zipper (bZIP) transcription factors have been described as FT-interacting proteins required for florigen activity at the apical meristem (Abe et al., 2005; Wigge et al., 2005; Muszynski et al., 2006; Li and Dubcovsky 2008; Taoka et al., 2011; Dong et al., 2012). Arabidopsis FD and rice OsFD1 encode bZIP transcription factors required for florigen function. According to current models, the heterodimer formed by FT and FD in arabidopsis is a molecular hub integrating environmental cues and spatial information at the apical meristem (Abe et al., 2005; Wigge et al., 2005; Jaeger et al., 2013). The rice homologue of FD, OsFD1, interacts with Hd3a and the dimer fulfils similar roles to the FT–FD unit. Direct interaction between OsFD1 and Hd3a could not be demonstrated, but contact between the two proteins was shown to be mediated by 14-3-3 proteins, now considered to be receptors of florigens (Taoka et al., 2011, 2013). Initial studies performed in different species and aimed at identifying interactors of florigens were performed in yeast that contains proteins probably mediating the interaction between FD homologues and florigens (Pnueli et al., 2001; Abe et al., 2005; Wigge et al., 2005; Li and Dubcovsky, 2008). The use of yeast as a heterologous system to test protein–protein interactions might have thus hidden the nature of the florigen receptor complex (or FAC, florigen activation complex). Structural and in vivo analyses in rice have demonstrated that the FAC unit is actually a heterohexamer formed by two molecules each of Hd3a, OsFD1 and a 14-3-3 protein that bridges the interaction between the florigen and the bZIP transcription factor. A similar structure for the FAC might apply to other plant species (Fig. 2). Mutagenesis of key residues at the interaction surface between Hd3a and 14-3-3 and between 14-3-3 and OsFD1 has uncovered several features of the complex and contributed to build the current model of florigen action at the SAM (Taoka et al., 2013). Florigen is first received by a 14-3-3 protein in the cytosol of a meristematic cell, from which it is translocated to the nucleus where it binds OsFD1. Phosphorylation of OsFD1 is required for binding to 14-3-3 receptors, and to activate target genes (Taoka et al., 2011). The presence of the bZIP transcription factor is a prerequisite for contacting DNA at the promoters of target genes, because florigens or 14-3-3 proteins have no DNA binding property. The full extent of potential targets of the FAC is currently unknown; however, it is becoming clear that assembly of the FAC is to a certain extent combinatorial, and bZIPs homologous to OsFD1 can replace it to generate complexes controlling processes other than flowering (Tsuji et al., 2013). The OsFD2 transcription factor is one such example of a bZIP protein capable of forming a FAC, but controlling leaf development rather than floral transition. Plant architecture is altered when OsFD2 is overexpressed, but not when a mutated version unable to bind to 14-3-3 receptors is overexpressed. Since 14-3-3 proteins are ubiquitously expressed and florigens are detected in the entire meristem, bZIP proteins are probably restricting different FAC complexes to different cell types. The broad extent of florigen–bZIP interactions and the potential role of FACs in rice development are still to be fully explored. Additionally, the role of RFT1 in FAC formation has not been addressed yet, which might suggest additional combinatorial possibilities. These aspects are opening up novel possibilities for dissecting the full range of florigen functions in rice.
Molecular events occurring at the shoot apical meristem in response to photoperiodic induction
Inductive photoperiods trigger florigen expression and movement to the apical meristem, affecting the regulation of genes that are involved in inflorescence formation. In arabidopsis apices the FT–FD complex is recruited to the promoter of APETALA1 (AP1), encoding a MADS-box transcription factor necessary for flower development, and triggers its activation (Wigge et al., 2005). Early events occurring during the floral transition also include upregulation of other related MADS-box transcription factors, including SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) and FRUITFULL (FUL), which are required for the promotion of flowering by FT (Fig. 2). In rice protoplasts, co-expression of Hd3a and OsFD1 proteins is required to induce expression of OsMADS15, a homologue of AP1 (Taoka et al., 2011). Mutagenized variants of Hd3a unable to bind to 14-3-3 proteins cannot activate OsMADS15, and an RNAi mutant simultaneously silencing four isoforms of 14-3-3 proteins also fails to induce OsMADS15 to the levels observed in wild-type plants (Taoka et al., 2011). A homologue of SOC1, encoded by OsMADS50, has been isolated in rice and shown to be required for flowering (Lee et al., 2004). It is unclear if OsMADS50 participates in the network regulating flowering at the SAM, but its transcription can be detected at the apex, suggesting that it shares features of its arabidopsis homologue (Kobayashi et al., 2012). Other MADS-box genes, including OsMADS14, OsMADS18 and OsMADS34, are upregulated at the apex in response to reproductive transition and are necessary for correct inflorescence development (Kobayashi et al., 2012). These data indicate the existence of a conserved mechanism for floral induction at the apical meristem of plants, whereby florigens, interacting with FD-like transcription factors, activate expression of a set of MADS-box genes at the early stages of inflorescence development (Fig. 2). Further regulatory layers are connected with this basic developmental plan to fine-tune and stabilize floral transition from environmental noise (Fornara et al., 2009; Jaeger et al., 2013). Additionally, partial or complete redundancy between FD-like or MADS-box genes probably contributes to co-ordinate and stabilize inflorescence meristem specification downstream of florigenic signals (Kobayashi et al., 2012; Torti and Fornara, 2012; Jaeger et al., 2013).
In arabidopsis, the vegetative to reproductive phase change is largely controlled by microRNAs, including miR156 and miR172. Expression of miR156 decreases as plants age, and this pattern is complementary to that shown by miR172, whose expression increases during development. Overexpression of miR156 delays the floral transition in arabidopsis by limiting expression of SQUAMOSA PROMOTER BINDING LIKE (SPL) transcription factors (Wu and Poethig, 2006). Conversely, miR172 accelerates flowering, by promoting FT expression in leaves and by targerting six related members of the APETALA2 (AP2) clade, that repress flowering (Jung et al., 2007; Yant et al., 2010). The transcriptional dynamics of miR172 depend on some of the SPLs targeted by miR156, generating a molecular loop that allows progression of phase change as plants age (Wang et al., 2009; Wu et al., 2009). Heading date in rice is only partially dependent on these microRNAs. Rice plants overexpressing miR172 showed altered flower development, including homeotic convertions of floral organs and loss of determinacy (Zhu et al., 2009). However, no altered heading date could be observed, unlike arabidopsis plants overexpressing mir172 that showed extremely early flowering (Yant et al., 2010). Overexpression of rice miR156 influenced heading date, delaying flowering by several days and was associated with a strong increase in tiller number and dwarfism (Xie et al., 2006). Whether the levels of mir156 and mir172 are reciprocally regulated in rice remains to be established.
SEASONAL FLOWERING RESPONSES IN TEMPERATE CEREALS
Flowering time pathways share a high degree of conservation between monocots and dicots. However, homologous genes could also be recruited to different functions during evolution, and novel pathways could evolve in specific lineages, to allow adaptation of species to different environments.
Most plants adapted to temperate climates, including arabidopsis and temperate cereals such as wheat (Triticum spp.) and barley (Hordeum vulgare), require prolonged exposure to cold temperatures before flowering. This process is known as vernalization. Since rice was domesticated in tropical regions it does not show vernalization responses. However, this is a crucial adaptation for species and varieties adapted to higher latitudes, because it prevents flowering when temperature is unfavourable, thus protecting the delicate inflorescence meristem from cold damage. Arabidopsis has been used extensively to understand the genetic and molecular bases of vernalization responses, and two genes, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC), play a major role in preventing flowering before cold exposure (Ream et al., 2012). Plants experiencing low temperatures repress FLC expression and maintain its repression also when returned to warm temperatures. Stable downregulation of FLC expression is associated with epigenetic silencing of FLC chromatin that is converted from active to inactive (J. Song et al., 2012). Until recently it was believed that no homologue of FLC existed in monocots, but a recent report suggested that this is not the case. Ruelens et al. (2013) showed that tandem arrangements of MADS-box genes, including FLC, are evolutionarily conserved across Angiosperms, and FLC homologues can also be traced in monocot genomes (Ruelens et al., 2013). These studies also suggest that OsMADS51 and OsMADS37 are the closest homologues of FLC in rice. Interestingly, OsMADS51 has been shown to control heading date as an activator of Ehd1 expression, indicating an opposite function in regulation of florigenic proteins to that performed by FLC in arabidopsis (Kim et al., 2007).
In temperate cereals, vernalization responses are controlled by VERNALIZATION (VRN) loci (Ream et al., 2012). In wheat and barley, broad genetic variation in the vernalization responses has been reported, and many varieties are known to have strict or no vernalization requirement. Varieties that need to be exposed to cold are planted before winter and flower only during the subsequent spring, whereas vernalization-insensitive accessions can be planted after winter. Such variation has been instrumental in isolating genes controlling the vernalization process and in establishing regulatory connections between vernalization genes (Fig. 1) (Yan et al., 2003, 2004; Trevaskis et al., 2003; Karsai et al., 2005; Hemming et al., 2008; Ream et al., 2012). Temperate cereal varieties showing vernalization requirements express VRN2 at high levels before vernalization. The VRN2 locus encodes a CCT-domain protein showing sequence similarity to Ghd7 of rice, and acts as a potent floral repressor that has to be downregulated during floral transition. Mutations in VRN2 cause insensitivity to vernalization and confer a spring habit (Yan et al., 2004). Exposure to low temperatures increases expression of VRN1, a floral promoter homologue of FUL and AP1 of arabidopsis, and reduces expression of VRN2 (Trevaskis et al., 2006). Dominant allelic variants of VRN1 carrying mutations in its regulatory regions express VRN1 independently of exposure to low temperatures, and confer a spring growth habit (Loukoianov et al., 2005). High levels of VRN1 expression are associated with repression of VRN2 transcription, which supported the idea that VRN1 acts as repressor of VRN2. However, by loss-of-function vrn1 mutants, it became clear that VRN1 induction is not necessary to initiate repression of VRN2 during vernalization, but is required to maintain its repression after exposure to cold (Chen and Dubcovsky, 2012). These data indicate that cold signals co-ordinately repress VRN2 and activate VRN1 expression during vernalization, whereas after vernalization VRN1 maintains the repressed state at the VRN2 locus (Fig. 1).
In barley, transcriptional dynamics of VRN1 mRNA are probably caused by changes in the chromatin state of the VRN1 locus, in which cold promotes an active chromatin state that is later maintained after plants are exposed to warm temperatures (Oliver et al., 2009). Changes in the chromatin state were not observed at the VRN2 locus, suggesting that VRN1 is the primary target of chromatin remodelling complexes during vernalization (Oliver et al., 2009).
As VRN2 levels decrease, the vernalization requirement is satisfied and if plants are exposed to LDs, VRN3, a homologue of Hd3a and FT, is transcribed and moves to the apical meristem where it promotes flowering during spring (Yan et al., 2006; Hemming et al., 2008). The molecular mechanisms through which VRN3 promotes flowering at the apex are conserved (Fig. 2). In wheat, VRN3/TaFT protein can interact with TaFDL transcriptional regulators, homologues of FD and OsFD1, and promote expression of VRN1 (Li and Dubcovsky, 2008). Expression of VRN1 is directly controlled by the TaFT–TaFDL heterodimer as at least one TaFDL protein can bind the promoter of VRN1. Whether 14-3-3 proteins mediate the interaction between TaFT and TaFDL is currently unclear (Fig. 2).
As in arabidopsis, temperate cereals flower earlier if exposed to LDs, whereas flowering is delayed under SDs. Photoperiodic control of flowering becomes relevant only after the vernalization requirement has been satisfied and plants become competent to respond to daylength. The major gene controlling photoperiod sensitivity in both wheat and barley is PHOTOPERIOD 1 (Ppd1), encoding a pseudoresponse regulator similar to OsPRR37 of rice and PRR7 of arabidopsis (Turner et al., 2005; Beales et al., 2007). Photoperiod-insensitive wheat plants carry Ppd-D1a, a semi-dominant allele that bears a 2 kb deletion in its upstream regulatory region and induces early flowering under both LDs and SDs (Beales et al., 2007). The Ppd-D1a deletion causes increased expression of Ppd-D1a also during the night under SDs, when transcription of the wild-type gene is normally repressed. Misexpression induces TaFT constitutive activation and flowering, irrespective of daylength. In barley, the recessive ppd-H1 loss-of-function allele cannot induce high expression of HvFT when plants are exposed to LD conditions. This results in delayed flowering and is advantageous for spring-sown varieties that can prolong vegetative growth, producing more biomass and eventually seeds (Turner et al., 2005).
Interestingly, the effect of homologues of OsPRR37 on flowering depends on the species. In LD plants such as arabidopsis and temperate cereals, functional alleles promote flowering through transcriptional induction of florigens; conversely, in SD species such as rice and sorghum, PRR genes repress expression and flowering of florigens under LDs (Murphy et al., 2011; Koo et al., 2013). The molecular mechanisms underlying PRR function will provide important clues to understand how information on daylength is elaborated and the causes FT activation.
Homologues of CO and Hd1 have been cloned from temperate cereals; however, they seem not to be crucial to confer a photoperiodic response (Nemoto et al., 2003; Shimada et al., 2009). Overexpression of HvCO1 in a ppd-H1-deficient accession accelerates flowering under both LD and SD conditions, indicating that HvCO is independent of the PRR pathway. However, plants overexpressing HvCO retained responsiveness to photoperiod, flowering later under SDs and indicating the existence of additional factors with major effects on flowering. Indeed, allelic variation at Ppd-H1 was shown to be a major determinant of HvFT1 expression and flowering time, and acted independently of HvCO1 (Campoli et al., 2012). Similar conclusions were suggested from studies in wheat (Shaw et al., 2012) and point to a model whereby florigen expression is the convergence point of independent pathways with limited cross-talk (Fig. 1).
In temperate cereals the responses to daylength and vernalization are integrated. After a period of growth under LDs, exposure to SDs accelerates flowering in wheat, and can largely substitute for vernalization treatments (Dubcovsky et al., 2006). Flowering is due to downregulation of VRN2 expression under SDs, an effect also observed in barley and Brachypodium (Trevaskis et al., 2006; Ream et al., 2012). Thus, VRN2 is a convergence point integrating photoperiodic and temperature information, and its correct expression is key for flowering at the most appropriate time of year.
RICE ADAPTATION TO LONG-DAY CONDITIONS INVOLVED ALLELIC CHANGES AT HEADING DATE LOCI
Rice domestication started about 10 000–13 000 years ago in the surroundings of the Pearl River in Southern China (Huang et al., 2012). During this process, the founder ecotypes, probably belonging to the O. rufipogon progenitor, split into the five groups of cultivated rice known to date. Over the centuries, the area of rice cultivation expanded, first within tropical and sub-tropical Asia and then to other regions of the world, reaching temperate areas at higher latitudes. The success of rice adaptation depended on the acquisition of cold tolerance traits and the loss of photoperiod sensitivity. In temperate areas, seasonal variations in temperatures limit the period of rice cultivation from late spring to early autumn. Thus, rice flowering occurs during summer days, which are warm but long, and varieties adapted to temperate climates show reduced sensitivity to changes in daylength, flowering under conditions normally non-inductive.
Several genetic studies have been carried out in order to identify the molecular mechanisms that allow rice to flower at high latitudes. Five major QTLs controlling heading date in response to photoperiod were identified and described in detail in previous sections (Yamamoto et al., 1998, 2000). All genes underlying these major QTLs have been cloned and, interestingly, four of them (Hd1, Hd2/PRR37, Hd4/Ghd7 and Hd5/DTH8/Ghd8) were demonstrated to be repressors of flowering under LD conditions, whereas Hd3a encodes the major florigen normally targeted by the Hd1–Hd4 repressors (Yano et al., 2000; Kojima et al., 2002; Xue et al., 2008; Wei et al., 2010; Yan et al., 2011; Fujino et al., 2012; Koo et al., 2013). This genetic architecture probably reflects the tropical origin of the species, and indicates that floral repression is a default state that needs to be overcome in order for flowering to occur. However, it also provides the substrate for artificial selection of varieties better adapted to regions where daylength is not permissive.
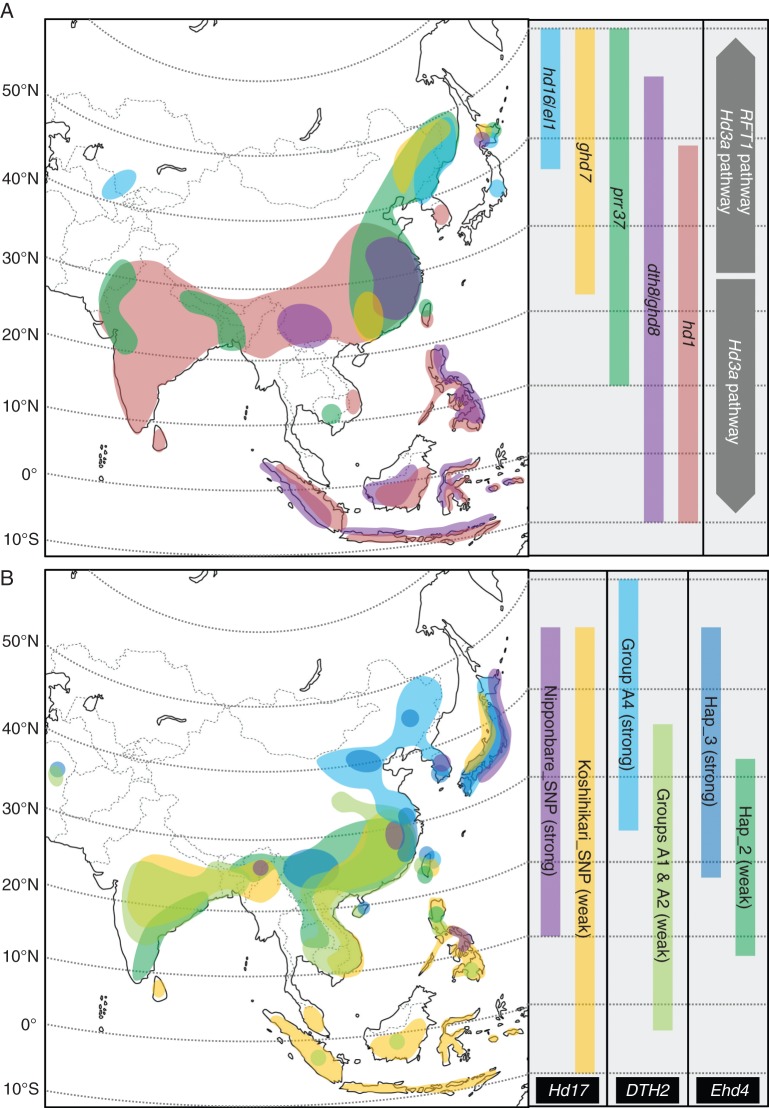
Polymorphisms at loci encoding florigens exist and can partly account for flowering diversity. In particular, variations at the Hd3a promoter regions contribute to diversification of flowering time of a rice core collection (Takahashi et al., 2009). A recent study has demonstrated how single nucleotide polymorphisms (SNPs) in the regulatory genomic region and an amino acid substitution in the protein sequence of RFT1 provide flowering time divergence under LD conditions (Ogiso-Tanaka et al., 2013). Rice varieties growing under natural LD conditions (where daylength is longer than 13 h and latitude over 23·6 °N) use both the RFT1- and Hd3a-dependent pathways to promote flowering, whereas rice varieties growing at southern latitudes mostly use the Hd3a pathway (Fig. 3A) (Ogiso-Tanaka et al., 2013). However, since florigens are highly conserved across rice varieties and species, flowering diversification has mainly resulted from the regulation of florigen expression levels that are highly correlated with flowering time.
Fig. 3.
Distribution of alleles influencing heading date in rice. The maps show the distribution of loss-of-function alleles of floral repressors, including hd16, hd1, prr37, ghd7 and dth8/ghd8 (A) or allelic variants of floral promoters associated with weak or strong activity, which include Hd17, DTH2 and Ehd4 (B). The colour of the distribution matches that of the corresponding gene, which is indicated on the right-hand side of the map. The length of the coloured bars on the right covers the latitudinal range across which varieties bearing the allelic variant are grown. Grey arrows (represented only in A) indicate the requirement for Hd3a expression at southern latitudes and for Hd3a and RFT1 expression at higher latitudes. The maps are based on data reported in Xue et al. (2008), Takahashi et al. (2009), Wei et al. (2010), Fujino et al. (2012), Matsubara et al. (2012), Gao et al. (2013), Koo et al. (2013), Kwon et al. (2013) and Wu et al. (2013).
Repressors (or suppressors) of flowering play a crucial role in reducing florigen gene expression under LD conditions, leading to a strong delay in heading date. Non-functional alleles of repressors (or suppressors) of LD-dependent flowering have been associated with loss of sensitivity to photoperiod. Loss-of-function alleles of such genes cause an increase in florigen gene expression to promote flowering under LDs. Thus, defective alleles of repressors can be used by breeders to introduce variations in flowering time in rice varieties that grow under LD conditions. These alleles have been useful tools to introduce tropical varieties into temperate areas and to increase the northern limit of rice cultivation.
Non-functional alleles at Hd1, PRR37, Ghd7 and Ghd8 loci are generated by SNPs, insertions or deletions, leading to dramatic changes in florigen expression and heading dates. Special attention is required for Hd1, which acts as a repressor under LDs but as an inducer under SDs. A high occurrence of natural polymorphisms in Hd1 has been correlated with variation in flowering time and Hd3a mRNA levels (Takahashi et al., 2009). Rice cultivars with functional Hd1 alleles showed higher Hd3a expression levels and earlier flowering times under SD conditions, whereas those with non-functional Hd1 alleles showed lower Hd3a expression levels and later flowering times (Takahashi et al., 2009). Since the presence of non-functional alleles of Hd1 influences flowering in opposite ways depending on daylength, rice varieties carrying natural hd1 mutants have been found in a wide range of latitudes (Fig. 3A). Varieties bearing non-functional Hd1 alleles grown under SDs will delay flowering time, which could be important to elongate the vegetative phase in order to increase grain production. The effect of non-functional Hd1 alleles under SDs can be reinforced by the presence of non-functional Ehd1 alleles, as observed in some Taiwanese rice varieties (Doi et al., 2004). Conversely, non-functional Hd1 alleles in varieties grown under LDs will anticipate heading, contributing to cultivation at high latitudes (Izawa, 2007). A recent study has revealed that Hd2/PRR37 downregulates Hd3a expression under LD conditions and has demonstrated that natural variation at PRR37 in many Asian rice cultivars has contributed to the expansion of rice cultivation to temperate areas, similar to previous reports in sorghum (Murphy et al., 2011; Koo et al., 2013). Non-functional PRR37 alleles (Fig. 3A) were detected in a wide range of latitudes, including the northern limit of rice cultivation (Koo et al., 2013). Genetic analysis revealed that the effect of PRR37 on heading date is additive to that of Ghd7, and rice varieties carrying non-functional PRR37 and Ghd7 showed extremely early flowering under LDs (Koo et al., 2013). Ghd7 has been previously described as a key component in the adaptation of rice to northern latitudes because it downregulates the expression of Ehd1 and, consequently, that of Hd3a and RFT1 under LDs. Natural ghd7 mutants (Fig. 3A) were found in early flowering rice varieties grown in central and southern China and in varieties from the Heilongjiang Province of North-eastern China, the latter being characterized by cool summers and a short growing season (Xue et al., 2008). Japonica cultivars with both Ghd7 and PRR37 mutations were also found at high-latitude regions of North-eastern Asia, including Northern Japan. This suggests that naturally occurring mutations in PRR37 and Ghd7 play an important role in rice adaptation from low to high latitudes (Koo et al., 2013). However, Ghd7 acts on a separate genetic pathway to that of PRR37 (Xue et al., 2008; Fujino et al., 2012; Koo et al., 2013). This might indicate that pyramiding of non-functional alleles in cultivated varieties has probably allowed further expansion of the cultivation area, and artificial construction of early flowering genotypes has been particularly successful when independent repressor pathways were targeted (Ebana et al., 2011). Polymorphisms in the DTH8/Ghd8 sequence that create non-functional alleles have been related to loss of photoperiod sensitivity. Natural ghd8 mutants were found in several provinces of China, the Philippines, Indonesia and Northern Japan (Wei et al., 2010; Fujino et al., 2012) (Fig. 3A). The mutant allele has been used in breeding programmes outside of Japan, its country of origin, and spread to Europe, where it probably conferred an agronomic advantage over functional alleles (Wei et al., 2010; Fujino et al., 2012). Ghd8 expression does not affect Ghd7 or Hd1 expression, suggesting that multiple targeting of repressor pathways has the potential to accelerate flowering strongly. Combinations of defective alleles generate stronger phenotypes, as demonstrated with prr37 ghd7 cultivars under LDs (Koo et al., 2013), suggesting that accumulation of additional Hd mutant alleles could contribute to further reduction of photoperiodic sensitivity and crop cycle.
In addition to these major loci, polymorphisms in the DNA sequence of other alleles have also contributed to the northern adaptation of rice. From additional QTL analyses, Hd6, a minor heading date allele, was detected (Yamamoto et al., 2000). Hd6 enhances the repressive activity of Hd1 and is defective in some japonica cultivars (Takahashi et al., 2001; Ogiso et al., 2010; Ebana et al., 2011). This reduces (but does not abolish) Hd1-mediated repression under LDs, further contributing to diversification of flowering time. Matsubara et al. (2008b) identified new QTLs related to photoperiodic flowering. Among them, recent studies have shown how naturally occurring variants of EL1/Hd16 alleles in japonica cultivars influence Ghd7 activity (Matsubara et al., 2012; Hori et al., 2013; Kwon et al., 2014). Hd16 acts as a suppressor of LD-dependent flowering by phosphorylating Ghd7 (Hori et al., 2013). Cultivars carrying non-functional EL1/Hd16 variants (Fig. 3A) are closely associated with high latitudes, whereas the cultivars carrying functional EL1/Hd16 variants are randomly distributed independently of latitude (Kwon et al., 2014).
Natural variation at loci encoding floral activators has recently been shown to have an important role in adaptation to northern latitudes. In contrast to all the genes described above, allelic variants of Hd17, encoding an OsELF3-like protein, DTH2, which encodes a CONSTANS-like protein, and Ehd4 do not create loss-of-function alleles but rather genetic variants showing a gradient of activity (Matsubara et al., 2012; Gao et al., 2013; Wu et al., 2013). Genetic studies demonstrated that the Nipponbare_SNP of Hd17, allele 4 (A4) of DTH2 and haplotype 3 of Ehd4 have been fixed during the domestication of rice at high latitudes (Fig. 3B). These showed a stronger effect as floral promoters under natural LD conditions in comparison with other alleles (Matsubara et al., 2012; Gao et al., 2013; Wu et al., 2013).
CONCLUSIONS AND PERSPECTIVES
Decades of research on flowering control have greatly expanded our understanding of the molecular mechanisms that initiate and drive reproductive phase transitions in different species. Molecular control networks are becoming increasingly complex as novel genes and regulatory mechanisms are described. Research that takes advantage of arabidopsis as a model organism often leads the way and opens up the possibility of exploring the function of orthologues from other species. However, arabidopsis is not representative of all plant species, and several examples discussed in this review indicate that several monocot-specific (or even Oryza-specific) genes do not have functional equivalents in dicots (Doi et al., 2004; Yan et al., 2004; Xue et al., 2008; Matsubara et al., 2011; Wang et al., 2013; Wu et al., 2013). Exploring genomes by DNA sequencing, QTL and association mapping, transcriptome profiling or mutant screens will keep providing new exciting insights into the way genes allow plants to interface with the environment. Mining genetic variation in crop species and their progenitors, and coupling it with the enormous potential of next-generation sequencing, will reach the dual objective of identifying novel regulators, perhaps difficult to pinpoint with other tools, and to exploit diversity to accelerate breeding programmes (Huang et al., 2011; Zhao et al., 2011).
ACKNOWLEDGEMENTS
This work was supported by an ERC Starting Grant (#260963) to F.F.
LITERATURE CITED
- Abe M, Kobayashi Y, Yamamoto S, et al. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Adrian J, Farrona S, Reimer JJ, Albani MC, Coupland G, Turck F. Cis-regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in arabidopsis. The Plant Cell. 2010;22:1425–1440. doi: 10.1105/tpc.110.074682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theoretical and Applied Genetics. 2007;115:721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- Brambilla V, Fornara F. Molecular control of flowering in response to day length in rice. Journal of Integrative Plant Biology. 2013;55:410–418. doi: 10.1111/jipb.12033. [DOI] [PubMed] [Google Scholar]
- Cai X, Ballif J, Endo S, et al. A putative CCAAT-binding transcription factor is a regulator of flowering timing in arabidopsis. Plant Physiology. 2007;145:98–105. doi: 10.1104/pp.107.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Drosse B, Searle I, Coupland G, von Korff M. Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS. The Plant Journal. 2012;69:868–880. doi: 10.1111/j.1365-313X.2011.04839.x. [DOI] [PubMed] [Google Scholar]
- Chaw S-M, Chang C-C, Chen H-L, Li W-H. Dating the monocot–dicot divergence and the origin of core eudicots using whole chloroplast genomes. Journal of Molecular Evolution. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- Chen A, Dubcovsky J. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genetics. 2012;8 doi: 10.1371/journal.pgen.1003134. e1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Tremblay R, Wong AYM, Coneva V, Kozaki A, Mable BK. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics. 2006;7:158. doi: 10.1186/1471-2164-7-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. FT protein movement contributes to long-distance signaling in floral induction of arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, et al. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes and Development. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Danilevskaya O, Abadie T, Messina C, Coles N, Cooper M. A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043450. e43450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Loukoianov A, Fu D, Valarik M, Sanchez A, Yan L. Effect of photoperiod on the regulation of wheat vernalization genes VRN1 and VRN2. Plant Molecular Biology. 2006;60:469–480. doi: 10.1007/s11103-005-4814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebana K, Shibaya T, Wu J, et al. Uncovering of major genetic factors generating naturally occurring variation in heading date among Asian rice cultivars. Theoretical and Applied Genetics. 2011;122:1199–1210. doi: 10.1007/s00122-010-1524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KCS, Gissot L, et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Developmental Cell. 2009;17:75–86. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- Fujino K, Yano M, Yamanouchi U. Roles of the Hd5 gene controlling heading date for adaptation to the northern limits of rice cultivation. Theoretical and Applied Genetics. 2012;126:611–618. doi: 10.1007/s00122-012-2005-5. [DOI] [PubMed] [Google Scholar]
- Gao H, Zheng X-M, Fei G, et al. Ehd4 encodes a novel and Oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genetics. 2013;9 doi: 10.1371/journal.pgen.1003281. e1003281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giakountis A, Cremer F, Sim S, Reymond M, Schmitt J, Coupland G. Distinct patterns of genetic variation alter flowering responses of arabidopsis accessions to different daylengths. Plant Physiology. 2010;152:177–191. doi: 10.1104/pp.109.140772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Ariza J, Fornara F. Photoperiodic control of flowering in arabidopsis. Acta Horticulturae. 2012;967:17–27. [Google Scholar]
- Gu X, Wang Y, He Y. Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biology. 2013;11 doi: 10.1371/journal.pbio.1001649. e1001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- Hemming MN, Peacock WJ, Dennis ES, Trevaskis B. Low-temperature and daylength cues are integrated to regulate FLOWERING LOCUS T in barley. Plant Physiology. 2008;147:355–366. doi: 10.1104/pp.108.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JA, Bailey PC, Laurie DA. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PloS One. 2010;5 doi: 10.1371/journal.pone.0010065. e10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Ogiso-Tanaka E, Matsubara K, Yamanouchi U, Ebana K, Yano M. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. The Plant Journal. 2013;76:36–46. doi: 10.1111/tpj.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Han B, Fujiyama A, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012;490:497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhao Y, Wei X, et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nature Genetics. 2011;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Aoki M, Kurotani K-I, et al. Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Molecular Genetics and Genomics. 2011;285:461–470. doi: 10.1007/s00438-011-0621-4. [DOI] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nature Genetics. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- Izawa T. Adaptation of flowering-time by natural and artificial selection in arabidopsis and rice. Journal of Experimental Botany. 2007;58:3091–3097. doi: 10.1093/jxb/erm159. [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant) The Plant Journal. 2000;22:391–399. doi: 10.1046/j.1365-313x.2000.00753.x. [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Sugiyama N, Tanisaka T, Yano M, Shimamoto K. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes and Development. 2002;16:2006–2020. doi: 10.1101/gad.999202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Mihara M, Suzuki Y, et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. The Plant Cell. 2011;23:1741–1755. doi: 10.1105/tpc.111.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. FT protein acts as a long-range signal in arabidopsis. Current Biology. 2007;17:1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in arabidopsis. The Plant Cell. 2013;25:820–833. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KCS, et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO Journal. 2008;27:1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, et al. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in arabidopsis. The Plant Cell. 2007;19:2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai I, Szucs P, Mészáros K, et al. The Vrn-H2 locus is a major determinant of flowering time in a facultative×winter growth habit barley (Hordeum vulgare L.) mapping population. Theoretical and Applied Genetics. 2005;110:1458–1466. doi: 10.1007/s00122-005-1979-7. [DOI] [PubMed] [Google Scholar]
- Khush GS. Origin, dispersal, cultivation and variation of rice. Plant Molecular Biology. 1997;35:25–34. [PubMed] [Google Scholar]
- Kim SL, Lee S, Kim HJ, Nam HG, An G. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiology. 2007;145:1484–1494. doi: 10.1104/pp.107.103291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Yasuno N, Sato Y, et al. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. The Plant Cell. 2012;24:1848–1859. doi: 10.1105/tpc.112.097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Takahashi Y, Kobayashi Y, et al. Hd3a, a rice ortholog of the arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant and Cell Physiology. 2002;43:1096–1105. doi: 10.1093/pcp/pcf156. [DOI] [PubMed] [Google Scholar]
- Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135:767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- Koo BH, Yoo SC, Park JW, et al. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Molecular Plant. 2013;6:1877–1888. doi: 10.1093/mp/sst088. [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Zhang Y, Siefers N, Holt BF., 3rd NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. The Plant Journal. 2010;63:379–391. doi: 10.1111/j.1365-313X.2010.04247.x. [DOI] [PubMed] [Google Scholar]
- Kwon CT, Yoo SC, Koo BH, et al. Natural variation in Early flowering1 contributes to early flowering in japonica rice under long days. Plant, Cell and Environment. 2014;37:101–112. doi: 10.1111/pce.12134. [DOI] [PubMed] [Google Scholar]
- Lee S, Kim J, Han JJ, Han MJ, An G. Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. The Plant Journal. 2004;38:754–764. doi: 10.1111/j.1365-313X.2004.02082.x. [DOI] [PubMed] [Google Scholar]
- Lee YS, Jeong DH, Lee DY, et al. OsCOL4 is a constitutive flowering repressor upstream of Ehd1 and downstream of OsphyB. The Plant Journal. 2010;63:18–30. doi: 10.1111/j.1365-313X.2010.04226.x. [DOI] [PubMed] [Google Scholar]
- Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. The Plant Journal. 2008;55:543–554. doi: 10.1111/j.1365-313X.2008.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, et al. A repressor complex governs the integration of flowering signals in arabidopsis. Developmental Cell. 2008;15:110–120. doi: 10.1016/j.devcel.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Li D, Yang C, Li X, Gan Q, Zhao X, Zhu L. Functional characterization of rice OsDof12. Planta. 2009;229:1159–1169. doi: 10.1007/s00425-009-0893-7. [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, et al. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in arabidopsis. The Plant Cell. 2008;20:292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loukoianov A, Yan L, Blechl A, Sanchez A, Dubcovsky J. Regulation of VRN-1 vernalization genes in normal and transgenic polyploid wheat. Plant Physiology. 2005;138:2364–2373. doi: 10.1104/pp.105.064287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in arabidopsis. Current Biology. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Wang Z-X, Minobe Y, Izawa T, Yano M. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiology. 2008a;148:1425–1435. doi: 10.1104/pp.108.125542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Kono I, Hori K, et al. Novel QTLs for photoperiodic flowering revealed by using reciprocal backcross inbred lines from crosses between japonica rice cultivars. Theoretical and Applied Genetics. 2008b;117:935–945. doi: 10.1007/s00122-008-0833-0. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Nonoue Y, et al. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. The Plant Journal. 2011;66:603–612. doi: 10.1111/j.1365-313X.2011.04517.x. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, Yano M. Natural variation in Hd17, a homolog of arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant and Cell Physiology. 2012;53:709–716. doi: 10.1093/pcp/pcs028. [DOI] [PubMed] [Google Scholar]
- Murakami M, Tago Y, Yamashino T, Mizuno T. Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant and Cell Physiology. 2007;48:110–121. doi: 10.1093/pcp/pcl043. [DOI] [PubMed] [Google Scholar]
- Murphy RL, Klein RR, Morishige DT, et al. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proceedings of the National Academy of Sciences, USA. 2011;108:16469–16474. doi: 10.1073/pnas.1106212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, et al. Delayed Flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiology. 2006;142:1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto Y, Kisaka M, Fuse T, Yano M, Ogihara Y. Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. The Plant Journal. 2003;36:82–93. doi: 10.1046/j.1365-313x.2003.01859.x. [DOI] [PubMed] [Google Scholar]
- Ogiso E, Takahashi Y, Sasaki T, Yano M, Izawa T. The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiology. 2010;152:808–820. doi: 10.1104/pp.109.148908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso-Tanaka E, Matsubara K, Yamamoto S-I, et al. Natural variation of the RICE FLOWERING LOCUS T 1 contributes to flowering time divergence in rice. PloS One. 2013;8 doi: 10.1371/journal.pone.0075959. pe75959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SN, Finnegan EJ, Dennis ES, Peacock WJ, Trevaskis B. Vernalization-induced flowering in cereals is associated with changes in histone methylation at the VERNALIZATION1 gene. Proceedings of the National Academy of Sciences, USA. 2009;106:8386–8391. doi: 10.1073/pnas.0903566106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osugi A, Itoh H, Ikeda-Kawakatsu K, Takano M, Izawa T. Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiology. 2011;157:1128–1137. doi: 10.1104/pp.111.181792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL. Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochemical and Biophysical Research Communications. 2007;360:251–256. doi: 10.1016/j.bbrc.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL. Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. Journal of Plant Physiology. 2008;165:876–885. doi: 10.1016/j.jplph.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Pineiro M, Jarillo JA, Valverde F, Lazaro A. The arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. The Plant Cell. 2012;24:982–999. doi: 10.1105/tpc.110.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, Minis DH. The entrainment of circadian oscillations by light and their role as photoperiodic clocks. American Naturalist. 1964;98:261–294. [Google Scholar]
- Pnueli L, Gutfinger T, Hareven D, et al. Tomato SP-interacting proteins define a conserved signaling system that regulates shoot architecture and flowering. The Plant Cell. 2001;13:2687–2702. doi: 10.1105/tpc.010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream TS, Woods DP, Amasino RM. The molecular basis of vernalization in different plant groups. Cold Spring Harbor Symposia on Quantitative Biology. 2012;77:105–115. doi: 10.1101/sqb.2013.77.014449. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Ruelens P, de Maagd RA Proost S, Theißen G, Geuten K, Kaufmann K. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nature Communications. 2013;4:2280. doi: 10.1038/ncomms3280. [DOI] [PubMed] [Google Scholar]
- Saito H, Ogiso-Tanaka E, Okumoto Y, et al. Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant and Cell Physiology. 2012;53:717–728. doi: 10.1093/pcp/pcs029. [DOI] [PubMed] [Google Scholar]
- Salazar JD, Saithong T, Brown PE, et al. Prediction of photoperiodic regulators from quantitative gene circuit models. Cell. 2009;139:1170–1179. doi: 10.1016/j.cell.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinov DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Turner AS, Laurie DA. The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum) The Plant Journal. 2012;71:71–84. doi: 10.1111/j.1365-313X.2012.04971.x. [DOI] [PubMed] [Google Scholar]
- Shimada S, Ogawa T, Kitagawa S, et al. A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. The Plant Journal. 2009;58:668–681. doi: 10.1111/j.1365-313X.2009.03806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Angel A, Howard M, Dean C. Vernalization – a cold-induced epigenetic switch. Journal of Cell Science. 2012;125:3723–3731. doi: 10.1242/jcs.084764. [DOI] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;25:1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Fang J, Zhao T, et al. The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. The Plant Cell. 2012;24:3235–3247. doi: 10.1105/tpc.112.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM. A PHD finger protein involved in both the vernalization and photoperiod pathways in arabidopsis. Genes and Development. 2006;20:3244–3248. doi: 10.1101/gad.1493306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Shomura A, Sasaki T, Yano M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proceedings of the National Academy of Sciences, USA. 2001;98:7922–7927. doi: 10.1073/pnas.111136798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proceedings of the National Academy of Sciences, USA. 2009;106:4555–4560. doi: 10.1073/pnas.0812092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476:332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- Taoka K, Ohki I, Tsuji H, Kojima C, Shimamoto K. Structure and function of florigen and the receptor complex. Trends in Plant Science. 2013;18:287–294. doi: 10.1016/j.tplants.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang H-C, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytologist. 2010;187:57–66. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- Torti S, Fornara F. AGL24 acts in concert with SOC1 and FUL during arabidopsis floral transition. Plant Signaling and Behavior. 2012;7:1251–1254. doi: 10.4161/psb.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES. MADS box genes control vernalization-induced flowering in cereals. Proceedings of the National Academy of Sciences, USA. 2003;100:13099–13104. doi: 10.1073/pnas.1635053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES. HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiology. 2006;140:1397–1405. doi: 10.1104/pp.105.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K. Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Current Opinion in Plant Biology. 2013;16:228–235. doi: 10.1016/j.pbi.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Wei X, Xu J, Guo H, et al. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiology. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of arabidopsis. The Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, et al. Integration of spatial and temporal information during floral induction in arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Wu C, You C, Li C, et al. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proceedings of the National Academy of Sciences, USA. 2008;105:12915–12920. doi: 10.1073/pnas.0806019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Zheng X-M, Lu G, et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proceedings of the National Academy of Sciences, USA. 2013;110:2775–2780. doi: 10.1073/pnas.1213962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiology. 2006;142:280–293. doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant and Cell Physiology. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Kuboki Y, Lin SY, Sasaki T, Yano M. Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single Mendelian factors. Theoretical and Applied Genetics. 1998;97:37–44. [Google Scholar]
- Yamamoto T, Lin H, Sasaki T, Yano M. Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny. Genetics. 2000;154:885–891. doi: 10.1093/genetics/154.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. Positional cloning of the wheat vernalization gene VRN1. Proceedings of the National Academy of Sciences, USA. 2003;100:6263–6268. doi: 10.1073/pnas.0937399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Blechl A, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303:1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, et al. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proceedings of the National Academy of Sciences, USA. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W-H, Wang P, Chen H-X, et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant. 2011;4:319–330. doi: 10.1093/mp/ssq070. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee S, Hang R, et al. OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. The Plant Journal. 2013;73:566–578. doi: 10.1111/tpj.12057. [DOI] [PubMed] [Google Scholar]
- Yang Y, Peng Q, Chen G-X, Li X-H, Wu C-Y. OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Molecular Plant. 2013;6:202–215. doi: 10.1093/mp/sss062. [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. The Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, et al. Orchestration of the floral transition and floral development in arabidopsis by the bifunctional transcription factor APETALA2. The Plant Cell. 2010;22:2156–2170. doi: 10.1105/tpc.110.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Huang X, Ouyang X, et al. OsELF3-1, an ortholog of arabidopsis early flowering 3, regulates rice circadian rhythm and photoperiodic flowering. PloS One. 2012;7 doi: 10.1371/journal.pone.0043705. pe43705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nature Communications. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa) BMC Plant Biology. 2009;9:149. doi: 10.1186/1471-2229-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]