Abstract
Background
Transcription factors of the RAV (RELATED TO ABI3 AND VP1) family are plant-specific and possess two DNA-binding domains. In Arabidopsis thaliana, the family comprises six members, including TEMPRANILLO 1 (TEM1) and TEM2. Arabidopsis RAV1 and TEM1 have been shown to bind bipartite DNA sequences, with the consensus motif C(A/C/G)ACA(N)2–8(C/A/T)ACCTG. Through direct binding to DNA, RAV proteins act as transcriptional repressors, probably in complexes with other co-repressors.
Scope and Conclusions
In this review, a summary is given of current knowledge of the regulation and function of RAV genes in diverse plant species, paying particular attention to their roles in the control of flowering in arabidopsis. TEM1 and TEM2 delay flowering by repressing the production of two florigenic molecules, FLOWERING LOCUS T (FT) and gibberellins. In this way, TEM1 and TEM2 prevent precocious flowering and postpone floral induction until the plant has accumulated enough reserves or has reached a growth stage that ensures survival of the progeny. Recent results indicate that TEM1 and TEM2 are regulated by genes acting in several flowering pathways, suggesting that TEMs may integrate information from diverse pathways. However, flowering is not the only process controlled by RAV proteins. Family members are involved in other aspects of plant development, such as bud outgrowth in trees and leaf senescence, and possibly in general growth regulation. In addition, they respond to pathogen infections and abiotic stresses, including cold, dehydration, high salinity and osmotic stress.
Keywords: RAV family, TEMPRANILLO genes, flowering, arabidopsis development, transcription factors, biotic/abiotic stress, photoperiod, gibberellins, flower development
INTRODUCTION
Flowering must occur at an appropriate time of the year to ensure offspring survival and species perpetuation. A delay in floral induction may lead to a robust plant, but be late for seed maturation. By contrast, a precocious flowering will result in a plant without enough energy for the development of fruits. Therefore, the time for floral induction is critical, and consequently both late induction and precocious flowering should be avoided. Plants respond to seasonal changes in daylength and temperature. In both inductive and non-inductive conditions flowering must be postponed until the plant obtains enough reserves for flower formation, and in unfavourable conditions it must be delayed to reach the appropriate time for seed-set. Arabidopsis thaliana is a good model to study this process. It is a facultative long day (LD) plant, i.e. it flowers rapidly when days are long, such as in spring, but it also eventually flowers in short days (SD). Several genetic pathways control flowering time in response to environmental or endogenous conditions. The major environmental effectors are daylength or photoperiod, seasonal and daily changes in temperature, and light intensity and quality (Thomas, 2006; Andrés and Coupland, 2012; Song et al., 2012, 2013). Among the endogenous factors are hormones such as gibberellins (GAs) and the age of the plant (Mutasa-Göttgens and Hedden, 2009; Huijser and Schmid, 2011). These pathways have been studied extensively in arabidopsis. The information provided by these genetic pathways is integrated in the activation of the expression of the so-called floral pathway integrators, FLOWERING LOCUS T (FT) and SUPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), which trigger flowering (Fornara et al., 2010; Wellmer and Riechmann, 2010). A major inducer of flowering in response to long days is CONSTANS (CO). CO transcript levels are high at the end of the light period under LD and its protein is stabilized only under light. If the expression coincides with the dark period, as in SD, the protein is immediately degraded. Therefore, CO is only active under LD (Suárez-López et al., 2001; Valverde et al., 2004; Jang et al., 2008; Liu et al., 2008).
Leaves perceive light and other environmental conditions, and CO is expressed in their vascular tissue, where it activates FT transcription (Takada and Goto, 2003; An et al., 2004). FT protein, identified as part of the florigen, travels to the shoot apical meristem (SAM), where flowers will be produced, to induce flowering (Corbesier et al., 2007; Jaeger and Wigge, 2007; Lin et al., 2007; Mathieu et al., 2007; Tamaki et al., 2007). In addition to FT, GAs are also mobile signals that travel from the leaves to the SAM to induce FT and SOC1 in order to trigger flowering (Eriksson et al., 2006). Different enzymatic activities give rise to the bioactive GA form GA4 (Mutasa-Göttgens and Hedden, 2009). As mentioned, these mobile inductive signals should be repressed for the correct timing of flowering. Several proteins have been identified as repressors and together prevent precocious flowering (Jarillo and Piñeiro, 2011). Two of these proteins are TEMPRANILLO1 (TEM1) and TEM2 (Castillejo and Pelaz, 2008; Osnato et al., 2012), which belong to the RAV (Related to ABI3/VP1) family of transcription factors.
Here we review the role of RAV genes in different species and show that they are involved in several plant processes such as flowering, bud outgrowth, leaf senescence, responses to hormones, stress and other environmental signals.
RAV FAMILY OF TRANSCRIPTION FACTORS
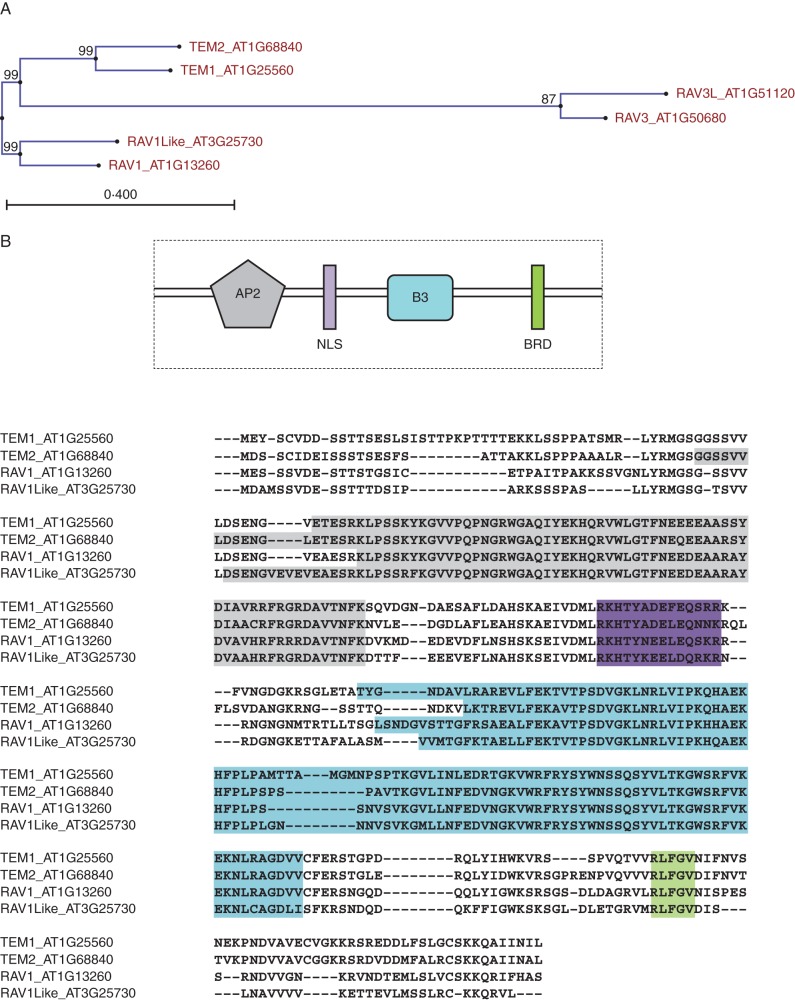
In arabidopsis there are six members of the RAV family of transcription factors: RAV1, RAV1-like, RAV2, RAV2-like, RAV3 and RAV3-like (Fig. 1A) (Riechmann et al., 2000). The first four have also been named ETHYLENE RESPONSE DNA BINDING FACTORS (EDF1–EDF4) (Alonso et al., 2003). Based on their function in flowering, RAV2-like and RAV2 were renamed TEM1 and TEM2, respectively (Castillejo and Pelaz, 2008). The main characteristic of RAV members is the presence of two different DNA-binding domains, a B3 and an AP2 domain (Fig. 1B). RAV family members have thus been classified as members of either the B3 super-family or the AP2/EREBP (APETALA2) family of transcription factors.
Fig. 1.
(A) Phylogenetic unrooted tree of the RAV family in Arabidopsis thaliana. Analysis was done using the CLC Genomics Workbench v6.5 program; bootstrap values are indicated. (B) Amino acid sequence of four RAV members, TEM1, TEM2, RAV1 and RAV1L, with the exact location of these domains. Analysis was done using Clustal v2.1 multiple amino acid alignment; substitution rate model = WAG. Main characteristic protein domains of RAV transcription factor family include: AP2 (grey) and B3 (blue) DNA-binding domains; nuclear localization signal (NLS) in purple; and B3 repression domain (BRD) in green.
The B3 domain was initially identified in the VIVIPAROUS1 (VP1) protein from Zea mays, and in the ABSCISIC ACID INSENSITIVE3 (ABI3), the VP1 orthologue from arabidopsis (Giraudat et al., 1992; Suzuki et al., 1997). B3 domains, consisting of a seven-stranded β-sheet arranged in an open barrel and two short α helices, generally share a common structural framework for DNA recognition (Yamasaki et al., 2004; Waltner et al., 2005). As mentioned, the RAV proteins are characterized by the presence of not only a C-terminal B3 domain that recognizes the consensus CACCTG sequence, but also an N-terminal AP2 domain that recognizes the consensus CAACA sequence (Kagaya et al., 1999). The AP2 domain is about 60 amino acids (aa) long (Okamuro et al., 1997; Riechmann and Meyerowitz, 1998; Riechmann et al., 2000; Sakuma et al., 2002; Magnani et al., 2004). This makes the RAV transcription factors unique, with two different DNA binding domains (Fig. 1B).
The contribution of transcriptional repressors may be of crucial importance in various plant biological processes. Around 10 % of arabidopsis transcription factors might be transcriptional repressors (Ikeda and Ohme-Takagi, 2009). Among the B3 super-family, it was found that many members had a repressive activity due to the existence of a 15-aa peptide (GNSKTLRLFGVNMEC), which has been named the B3 repression domain (BRD). Although replacement experiments pointed to the first leucine and/or the methionine residue (in bold) of the BRD (GNSKTLRLFGVNMEC) as crucial to maintain repressive activity, other amino acids of this domain are not always conserved. Deletion of the BRD of some B3 proteins revealed that only a short peptide of five amino acids, R/KLFGV, is essential as a repression domain. Four members of the RAV family, TEM1, TEM2, RAV1 and RAV1-like, share the core of the BRD (Ikeda and Ohme-Takagi, 2009). A quite similar sequence, MLFGV, is present in RAV3 and RAV3-like (Causier et al., 2012). The R/KLFGV sequence is also conserved in other RAV homologues from various plants such as rice (Ikeda and Ohme-Takagi, 2009). These results suggest strongly that RAV genes encoding RLFGV motifs could play roles as transcriptional repressors (Fig. 1B).
TEM GENES REPRESS FLOWERING IN TWO DIFFERENT PATHWAYS
As mentioned, FT plays a central role during the floral induction event (Turck et al., 2008) and is activated in response to CO (Kardailsky et al., 1999; Kobayashi et al., 1999; Samach et al., 2000). However, CO is already expressed in the phloem early in development (Takada and Goto, 2003), and changes in CO expression levels do not seem to account for the increase in FT accumulation for inducing flowering (Castillejo and Pelaz, 2008). Consequently, something else that accounts for this late FT accumulation must exist.
TEM genes affect the photoperiod pathway
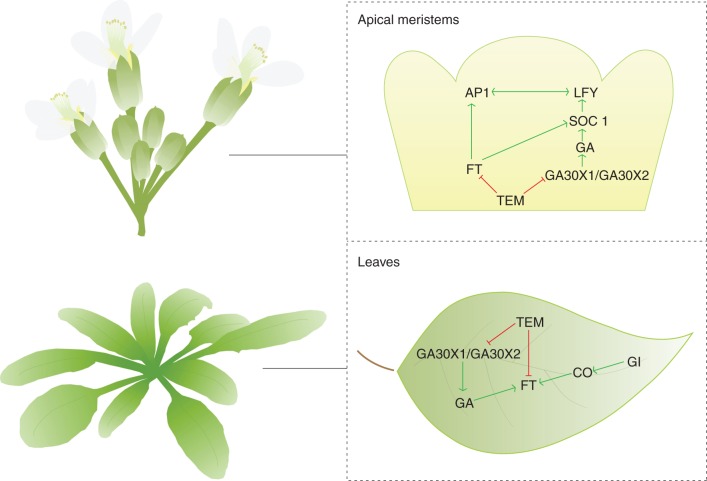
Regulation of flowering initiation in response to photoperiod is mediated by the interaction between external light signals and the circadian clock (Suárez-López et al., 2001; Yanovsky and Kay, 2002). In the photoperiod pathway, FT promotes flowering in response to LD. TEM1 and TEM2 were identified as repressors of flowering in the photoperiod pathway (Castillejo and Pelaz, 2008). Single loss-of-function alleles of TEM1, tem1–1, and TEM2, tem2–2, cause a slight early flowering phenotype in LD, and a double tem1–1 tem2–2 mutant shows enhanced early flowering compared with the single mutants under LD conditions. In this photoperiod, tem1–1 tem2–2 flowers as early as CO overexpressors (35S::CO). Supporting these results, it was found that both 35S::TEM1 and 35S::TEM2 plants show the opposite phenotype and flower extremely late under LD conditions (Castillejo and Pelaz, 2008; Osnato et al., 2012). Consequently, TEMs seem to play a pivotal role as repressors in floral induction (Fig. 2).
Fig. 2.
Floral transition model in Arabidopsis thaliana. TEM1 and TEM2 genes play a central role in regulating the flowering process by repressing at least the photoperiod and gibberellin pathways under inductive and non-inductive daylengths, in leaves and apical meristems.
TEM1 transcript levels follow a diurnal oscillation, such that TEM1 abundance is low during the daytime and peaks at dusk. Similar developmental and circadian regulations were observed for TEM1 and TEM2, supporting the proposed redundant role of both genes (Castillejo and Pelaz, 2008; Osnato et al., 2012). Moreover, TEM1 mRNA abundance is very high during early stages of seedling development but a pronounced decline takes place just before floral transition. CO expression remains almost unaltered throughout development, although a subtle increase occurs during the transition to flowering (Castillejo and Pelaz, 2008).
In addition in wild-type plants, FT mRNA remains at basal levels until the transition to flowering, at days 10–12, when there is a pronounced increase in FT accumulation. However, FT expression increases from day 6 in the tem1–1 tem2–2 double mutant, when plants had only formed the first two true leaves (Osnato et al., 2012). The significant increase of FT expression responsible for floral induction is abolished in the 35S::TEM1 seedlings (Castillejo and Pelaz, 2008). Therefore, TEM1 represses FT expression at early developmental stages.
The identical precocious flowering phenotypes of 35S::CO and tem1 tem2 plants suggested strongly that only when TEM levels drop drastically can CO activate FT to reach the threshold level necessary to trigger the floral transition under inductive photoperiods (Castillejo and Pelaz, 2008). When both CO and TEM levels are elevated, in 35S::CO 35S::TEM1 plants, the balance between the activator and the repressor is restored and consequently these plants flower after producing a wild-type number of leaves. The late-flowering phenotype of 35S::TEM1 plants is completely suppressed by the constitutive expression of FT, which is consistent with FT acting downstream of TEM1. The combination of tem1–1 and ft mutants confirmed the epistatic relationship between both genes, as the double mutant tem1–1 ft-101 flowers at the same time as ft-101 alone (Castillejo and Pelaz, 2008). These results also suggest that FT is the primary downstream target of TEM1 to repress flowering.
TEM1 expression is detected in all vegetative tissues (Castillejo and Pelaz, 2008). It has been proposed that TEM could act in the vascular bundles of leaves, together with CO, to tightly control FT accumulation; however, TEM1 is expressed throughout the leaf as well as in the SAM and the hypocotyl. An artificial micro RNA (amiRNA) targeted against TEM1 and TEM2 genes was expressed under control of the KNAT1 promoter to drive their silencing only in the SAM and hypocotyls. An early flowering phenotype of pKNAT1::amiR-TEM lines was associated with an up-regulation of FT expression. All this indicated that TEM has a role in controlling flowering, at least in the SAM (Osnato et al., 2012).
RAV binding motifs (Kagaya et al., 1999) were found in the 5′ untranslated region (UTR) of the FT gene. In vitro and in vivo interactions of TEM1 protein with the FT 5′UTR were confirmed by gel-shift and chromatin immunoprecipitation (ChIP) assays, respectively. Interestingly, the RAV binding site in FT is located just next to the CO binding site found 43 bp upstream of the ATG (Wenkel et al., 2006; Castillejo and Pelaz, 2008). Therefore, precise control of flowering time could be explained if the CO and TEM proteins compete for their respective binding sites to directly regulate FT accumulation. Consequently, FT levels are the result of a quantitative balance between the respective promoter and repressive activities of CO and TEM (Castillejo and Pelaz, 2008).
GIGANTEA (GI), a circadian clock regulator, plays a role in floral induction through regulation of the timing and amplitude of CO expression (Fowler et al., 1999; Park et al., 1999; Mizoguchi et al., 2005; Sawa et al., 2007). GI and FLAVIN-BINDING, KELCH REPEAT, F BOX protein 1 (FKF1) form a protein complex that mediates the degradation of CYCLING DOF FACTOR 1 (CDF1), a key CO repressor. Under LDs, GI and FKF1 expression peak at the same time, at the end of the day, leading to the optimal formation of the GI–FKF1 complex. However, under SDs, the expression of GI peaks a few hours before the peak of FKF1 expression, resulting in low levels of the GI–FKF1 complex and maintenance of the repressor CDF1 (Sawa and Kay, 2011).
CO and FT are mainly expressed in vascular tissue, whereas (and similarly to TEM genes) GI is expressed in various tissues including vascular bundles, mesophyll, SAM and root (Takada and Goto, 2003; An et al., 2004; Winter et al., 2007). In fact, GI expression in either mesophyll and/or vascular tissue rescues the late-flowering phenotype of the gi-2 mutant under both SD and LD conditions (Sawa and Kay, 2011). It was observed that the GI N-terminal region was able to interact with TEM1 and TEM2 through yeast-two-hybrid (Y2H) assays. Moreover, the in vivo physical interactions of these proteins were found to take place in the nucleus but not in the cytosol (Sawa and Kay, 2011). These authors also showed that GI activates FT expression independently of CO through direct binding to FT promoter regions (alone or in a complex with another protein). A possible explanation is that GI could neutralize the TEM1 and TEM2 repressors by interfering with their access to the FT promoter or their activity and/or stability.
TEM genes also regulate the GA pathway
By contrast, under SD conditions, in which CO is inactive, flowering is induced in the SAM by GAs through activation of the floral integrator SOC1, and the floral meristem identity gene LEAFY (LFY) (Blazquez and Weigel, 2000; Moon et al., 2003; Mutasa-Göttgens and Hedden, 2009). Under SDs, tem1–1 tem2–2 double mutants still flower much earlier than wild-type plants. When expression levels of SOC1 and LFY are analysed in wild-type and tem1–1 tem2–2 mutant plants under SDs, a significant enhancement of SOC1 and LFY expression is observed in tem1–1 tem2–2, indicating an additional role of TEM in flowering-time regulation under SD conditions (Osnato et al., 2012). By contrast, 35S::TEM1 plants flower extremely late under SDs, most of them remaining at the vegetative phase and producing leaves indefinitely. In this photoperiod, TEM mRNA levels are low during the light period, start to increase at dusk and peak early in the night in wild-type plants. TEM1 and TEM2 expression patterns are similar, except for an extra TEM2 peak late at night (Osnato et al., 2012).
pKNAT1::amiR-TEM plants have elongated hypocotyls both in LD and in SD conditions, while 35S::TEM1 plants show, apart from the extremely late flowering, a dwarf phenotype, loss of apical dominance and shorter hypocotyls (Osnato et al., 2012). These are phenotypes typical of GA-deficient mutants, such as ga3ox1–3 and the double mutant ga3ox1 ga3ox2 (Eriksson et al., 2006; Mitchum et al., 2006). When GA is sprayed onto the 35S::TEM1 plants the apical dominance and flowering phenotypes are rescued (Osnato et al., 2012), suggesting that TEM genes play a major role in the GA pathway.
Furthermore, a significant down-regulation of GA20OX2, GA3OX1 and GA3OX2 expression is found in 35S::TEM1, whereas an up-regulation of GA3OX1 and GA3OX2 is observed in tem1–1 and tem1–1 tem2–2 in comparison with the wild type (Osnato et al., 2012). 35S::TEM1 produces a down-regulation of GUS expression in plants carrying a GA3OX1::GUS reporter construct (Mitchum et al., 2006), specifically in the SAM of young plants and in leaves of older plants (Osnato et al., 2012). These results indicate a clear effect of TEM on the enzymes that catalyse the last step of GA4 biosynthesis. In addition, ChIP assays show that TEM1 is a direct in vivo regulator of the GA4 biosynthetic genes GA3OX1 and GA3OX2 by binding an RAV binding site positioned in the first exon in both cases (Osnato et al., 2012). These data therefore corroborate that TEM directly represses GA3OX genes, which may result in a reduction of bioactive GA4. tem1 tem2 ga3ox1 triple mutant plants flower later than tem1 tem2 plants but still earlier than the wild type and ga3ox1 single mutant, indicating that the early flowering phenotype of tem1 tem2 double mutants in LD is due at least partially to the GA3OX1 up-regulation (Osnato et al., 2012), which also indicates that GAs act both in LD and in SD.
In conclusion, TEM genes link the photoperiod- and GA-dependent flowering pathways, controlling the floral transition under inductive and non-inductive daylengths (Fig. 2).
OTHER RAV FAMILY MEMBERS MAY AFFECT FLOWERING
Results with RAV1 antisense lines suggest that RAV1 may be a flowering repressor in arabidopsis (Hu et al., 2004). However, it has not been shown whether the full-length antisense construct used to generate these antisense lines is specific for RAV1. Levels of other RAV family transcripts should be checked in these plants to discard the possibility that the early flowering is due to off-target effects on TEM1 and/or TEM2. It is also possible that RAV1 antisense plants flower a few days earlier than wild-type plants as a result of differences in the rate of leaf production (Hu et al., 2004).
When GmRAV, a soybean (Glycine max) TEM/RAV homologue, is overexpressed in tobacco (Nicotiana tabacum) it delays flowering. This suggests that GmRAV, similar to TEM1 and TEM2, can act as a flowering repressor. Although soybean flowering is promoted by SD, GmRAV shows higher expression under SD than under LD (Zhao et al., 2008). They proposed that the repression of flowering by GmRAV in tobacco may indirectly result from negative effects on photosynthesis and other aspects of plant physiology. Further research should determine whether GmRAV is a regulator of flowering.
RAV GENES ARE REGULATED BY DIFFERENT FLOWERING PATHWAYS
Age-dependent flowering pathway
Genes involved in several flowering pathways regulate TEM/RAV genes. Several AP2 family genes are targets of the miRNA miR172 and encode floral repressors that act in the photoperiod- and the age-dependent flowering pathways. In arabidopsis these repressors include AP2 itself, TARGET OF EAT1 (TOE1), TOE2, TOE3, SCHLAFMÜTZE (SMZ) and SCHNARCHZAPFEN (SNZ) (Zhu and Helliwell, 2011). AP2 and SMZ bind TEM1 chromatin in ChIP-chip experiments (Mathieu et al., 2009; Yant et al., 2010), suggesting that they may induce TEM1 expression. However, TEM1 mRNA levels are not altered in the leaves and the shoot meristem of an activation-tagged smz-D mutant, which flowers later than the wild type (Mathieu et al., 2009). By contrast, TEM2 is upregulated in smz-D, despite not being bound by SMZ in ChIP-chip experiments (Mathieu et al., 2009). These observations suggest that TEM1 and TEM2 may mediate at least part of the effects of AP2 and SMZ on flowering, although additional experiments are required to demonstrate this.
TOPLESS (TPL) and TPL-related (TPR) proteins constitute a family of five members that interact with diverse transcription factors and act as transcriptional co-repressors in arabidopsis (Long et al., 2006; Szemenyei et al., 2008). TOE1, TOE2 and AP2 are among these TPL/TPR-interacting transcription factors (Arabidopsis Interactome Mapping Consortium, 2011; Causier et al., 2012; Krogan et al., 2012). Overexpression of TOE1 delays flowering and TPL is required for this phenotype, suggesting that TPL, and perhaps also TPRs, acts as a co-repressor of flowering (Causier et al., 2012). Interestingly, all members of the RAV family, with the exception of RAV1L, also interact with TPL/TPR proteins. The RLFGV or MLFGV domains present in all RAV proteins (see above) are required for the interaction of at least RAV1 and RAV3L with TPL (Causier et al., 2012). Therefore, RAV proteins probably act in complexes with TPL/TPR to repress transcription of floral regulators. The action of TPL and its homologues in mammals and yeast involves histone deacetylation and chromatin condensation (Long et al., 2006; Krogan et al., 2012; Turki-Judeh and Courey, 2012; Wang et al., 2013). It will be interesting to determine whether the mechanism of transcriptional repression by RAV proteins also implies chromatin remodelling through the recruitment of TPL/TPR.
Ambient temperature pathway
Changes in ambient temperature affect flowering and low temperatures delay the floral transition in arabidopsis (Blazquez et al., 2003). EARLY FLOWERING 3 (ELF3) is a repressor of flowering involved in this response (Strasser et al., 2009). elf3 mutants flower earlier and are less sensitive to temperature than wild-type plants, such that the delay caused by low temperature is smaller in elf3 than in the wild type. TEM2 is downregulated in elf3 both at 16 and at 23 °C (Strasser et al., 2009), which correlates with the early flowering phenotype at both temperatures. In addition, the downregulation of TEM2 in elf3 is more dramatic at 16 than at 23 °C (Strasser et al., 2009), consistent with a bigger difference in flowering time between elf3 and the wild type at 16 than at 23 °C. This suggests that the repression of flowering by ELF3 may be mediated at least in part by an increase in TEM2 expression. RAV1 shows lower transcript levels in elf3 than in the wild type at 16 °C, but higher expression at 23 °C, indicating that RAV1 expression is also regulated by ELF3.
Two MADS-box transcription factors, FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP), form a complex that represses flowering during vegetative growth (Li et al., 2008). FLC and SVP have both overlapping and distinct functions (Balasubramanian et al., 2006; Lee et al., 2007b; Li et al., 2008). Both are involved in responses to ambient temperature. SVP is important for the repression of flowering at low ambient temperature, while FLC suppresses the induction of flowering by high temperatures (Balasubramanian et al., 2006; Lee et al., 2007b). FLC also plays an important role in vernalization, a response to long periods of cold that induces flowering after winter has passed (Song et al., 2012). In addition, FLC acts in the autonomous flowering pathway (Simpson, 2004). ChIP-seq experiments revealed that FLC binds to the promoter of TEM1, although TEM1 mRNA levels were not altered in an flc mutant (Deng et al., 2011). TEM1 and TEM2 chromatin is also bound by SVP, which up-regulates expression of these two genes (Tao et al., 2012). Therefore, the FLC–SVP complex may positively regulate at least TEM1 through direct binding to the TEM1 promoter. It would be interesting to test whether TEM1 and/or TEM2 affect the response of flowering to ambient temperature and/or vernalization. Although SVP and FLC had initially been described as transcriptional repressors (Hepworth et al., 2002; Gregis et al., 2006), they also seem capable of inducing transcription, including that of other flowering repressors in addition to TEM1 and TEM2 (Deng et al., 2011; Tao et al., 2012). The mechanism of this positive regulation remains unknown, but probably contributes to reinforce the repression of flowering under unfavourable conditions.
Another MADS-box protein with an important role in flowering-time control, SOC1, regulates TEM1 and TEM2 expression, but in the opposite way to the regulation by SVP. Regulatory regions of the TEM1 and RAV1 genes are bound by SOC1, indicating that the effect of SOC1 on at least TEM1 is probably direct (Tao et al., 2012). The repression of TEM1 and TEM2 by SOC1 is consistent with the induction of flowering by SOC1.
Brassinosteroids
Brassinosteroids (BRs) are a class of steroid hormones that regulate many developmental processes throughout plant life, such as vascular development, senescence and flowering. Mutants with altered content in endogenous BRs, such as deetiolated2 or dwarf4, flower late, indicating that components of the BR pathway also affect flowering time (reviewed by Li et al., 2010). Treatment with BR reduces RAV1 and GmRAV transcript levels in arabidopsis and in soybean, respectively (Hu et al., 2004; Zhao et al., 2008), indicating that BR down-regulates these genes. In arabidopsis, the effect of BR on RAV1 seems independent of the BR receptor BRI1 (Hu et al., 2004), suggesting that other BR receptors may be involved. The effect of BR on flowering might therefore be mediated by RAV family members. Given that BR affects many aspects of plant development and growth, additional research is required to determine in which aspect TEM/RAV genes may be involved.
Although the rice SVP group of genes seems not to be involved in flowering, they do affect BR responses (Duan et al., 2006; Lee et al., 2008). This, together with the regulation of TEM1 and TEM2 by SVP, the regulation of FLC expression by BR and the binding of FLC to TEM1 DNA (Domagalska et al., 2007; Deng et al., 2011; Tao et al., 2012), establishes another possible link between BR and RAV genes.
Light intensity and quality
In addition to photoperiod, light intensity and quality affect floral induction, as well as many other aspects of plant development and growth (Chen et al., 2004; Thomas, 2006). Several results indicate that RAV genes may be involved in light responses.
ELONGATED HYPOCOTYL 5 (HY5) is a transcription factor that promotes photomorphogenesis downstream of several photoreceptors (Oyama et al., 1997). In addition, HY5 represses flowering, as shown by the early flowering of hy5 mutants (Goto et al., 1991; Holm et al., 2002). TEM2 expression is positively regulated by HY5, which binds to TEM1, TEM2 and RAV1 chromatin, suggesting that the regulation of TEM2 is direct (Lee et al., 2007a). Therefore, TEM2 is a good candidate to link HY5 with the regulation of flowering in response to light signals.
EFFECT OF RAV FAMILY MEMBERS ON OTHER ASPECTS OF PLANT DEVELOPMENT
RAV genes regulate hypocotyl elongation
Transcriptomic analyses of arabidopsis seedlings grown in continuous white light and in the dark have shown that TEM2 is up-regulated in the hypocotyl and root in response to light, whereas RAV1 is down-regulated in cotyledons of light-grown seedlings (Ma et al., 2005). TEM1, RAV1 and RAV1L are rapidly repressed upon exposure of dark-grown seedlings to red light (Monte et al., 2004; Leivar et al., 2009; Shin et al., 2009). Moreover, TEM2 expression is induced by a short exposure to far-red light (Tepperman et al., 2004). These data indicate that RAV genes show specificity in their response to different light conditions in different organs.
PHYTOCHROME INTERACTING FACTORS (PIFs) play important roles in the regulation of light responses by the photoreceptors phytochrome A (PHYA) and PHYB (Leivar and Quail, 2011). The repression of RAV1 and RAV1L by red light requires the function of at least PIF3 (Monte et al., 2004), and other PIFs are involved in transcriptional regulation of TEM1 and TEM2 (Leivar et al., 2009). ChIP-seq experiments have identified TEM2 as a gene bound by PIF5 in plants subjected to low red/far-red light ratio, a condition that simulates shade (Hornitschek et al., 2012). Although the relevance of this binding for TEM2 expression is not yet clear, it suggests that PIF5 might be involved in the regulation of TEM2 by shade. A quadruple mutant lacking PIF1, PIF3, PIF4 and PIF5 (pifq) shows shorter hypocotyls and, under certain conditions, higher TEM1 and TEM2 transcript levels than wild-type plants (Leivar et al., 2008, 2009). Consistent with this, tem mutants and plants overexpressing TEM1 have longer and shorter hypocotyls than wild-type plants, respectively, under SD (Osnato et al., 2012). It remains to be shown whether PIFs affect TEM2 and/or TEM1 under this photoperiod, but the fact that PIFs promote hypocotyl growth under SD (Nozue et al., 2007) makes this hypothesis plausible. Therefore, TEM1 and TEM2 might play a role in light-regulated growth downstream of PIFs.
RAV genes might inhibit plant growth
Overexpression of TEM1 or TEM2 in arabidopsis causes dwarfism (Osnato et al., 2012). Tobacco plants overexpressing GmRAV (GmRAV-OX) also exhibit smaller leaves and roots and shorter internodes than wild-type plants. Soybean growth is reduced under SD compared with LD, inversely correlated with higher GmRAV levels under SD than LD (Zhao et al., 2008). Also, GmRAV causes a reduction in chlorophyll content and photosynthetic rate when overexpressed in tobacco (Zhao et al., 2008), which may explain the reduced growth of these plants. These findings suggest that TEM1, TEM2 and GmRAV might repress plant growth. This is consistent with the fact that BR treatment down-regulates GmRAV (Zhao et al., 2008). A detailed analysis of plant growth in loss-of-function tem mutants and GmRAV-silenced lines would be useful to demonstrate whether these genes play a role as growth regulators.
GmRAV might also be involved in root development, as tobacco GmRAV-OX plants develop fewer roots than wild-type plants (Zhao et al., 2008). Again, silencing of GmRAV in soybean would help to determine its biological function. Although overexpression of RAV1 causes a reduction in the number of lateral roots and probably in the rate of leaf production, suggesting that RAV1 may be a negative regulator of plant growth, down-regulation of RAV1 by an antisense construct does not have a significant effect on these processes (Hu et al., 2004).
RAV1 might regulate leaf senescence
Leaf senescence, a physiological mechanism affected by many internal and external factors (Lim et al., 2007), is strongly regulated by several genes to provide optimal plant fitness. This maximum plant fitness is obtained by remobilizing nutrients from senescent leaves (Woo et al., 2010). In silico technology has allowed identification of a subset of genes named as the SENESCENCE-ASSOCIATED GENES (SAGs). Among these SAGs, RAV1 was isolated due to the fact that not only RAV1 but also other RAV genes have been associated with leaf maturation and senescence. RAV1 expression is triggered at a mature stage, reaching maximum expression at an early senescence stage and decreasing at later stages. A similar expression pattern is found for TEM1, while for RAV1L the expression remains at high level until late senescence (Woo et al., 2010). These similar expression patterns during leaf development and senescence suggest a possible redundant role among this family in this aspect. However, neither single loss-of-function mutants of these genes nor the rav1 tem1 and rav1 rav1l double mutants show any significant alteration of the senescence process. By contrast, arabidopsis plants overexpressing RAV1 under a constitutive promoter show an early age-dependent leaf senescence phenotype as well as one induced by artificial dark (Woo et al., 2010). The main senescence-associated physiological markers, such as the degree of leaf yellowing, chlorophyll content and photochemical efficiency, are altered. Moreover, the expression of two senescence marker genes (SEN4 and SAG12) is upregulated in plants overexpressing RAV1, whereas RAV1 expression is induced by senescence-accelerating hormones such as jasmonic acid (JA) and ethylene. Similar results are found in transgenic plants that express RAV1 under an inducible promoter.
Consequently, these data suggest that at least RAV1 might play a role during leaf senescence initiation by the activation and/or repression of genes involved in the successful execution of the leaf senescence process (Woo et al., 2010). This control could be done by integrating the age-dependent aspects of leaf senescence with senescence-accelerating hormones and environmental influences. Moreover, tobacco GmRAV-OX plants show accelerated senescence in response to abscisic acid (ABA) and dark treatments (Zhao et al., 2008). Because the analyses of single and double mutants do not demonstrate a role of RAV genes in senescence, additional work is required to test whether other family members may control this process in a redundant manner with RAV1 or GmRAV.
The three outer whorls of the flowers in arabidopsis, sepals, petals and stamens, are also organs that senesce and shed after pollination (Chen et al., 2011). The time of senescence and organ abscission is controlled by diverse hormones; one of the most important is ethylene, which accelerates this process (Roberts et al., 2002). It is known that FOREVER YOUNG FLOWER (FYF), a MADS transcription factor, acts as a repressor of the ethylene response controlling floral senescence and abscission (Chen et al., 2011). Recently, it was found that TEM1 and TEM2, which were previously characterized as downstream genes in the ethylene signalling pathway (Alonso et al., 2003), are significantly down-regulated in 35S::FYF plants. Interestingly, the FYF expression pattern is opposite to that of TEM1 and TEM2 during flower development. Therefore, these results suggest that FYF controls senescence and organ abscission by inactivating downstream genes in the ethylene response such as TEM1 and TEM2 (Chen et al., 2011).
RAV genes control bud outgrowth in trees
RAV homologous genes have also been identified in trees. A RAV gene from chestnut (Castanea sativa), CsRAV1, has recently been characterized. The closest relatives to CsRAV1 are two poplar (Populus thricocarpa) RAVs, PtRAV1 and PtRAV2, and all group with the arabidopsis TEM1 and TEM2 genes (Moreno-Cortés et al., 2012). Trees are known to have a long juvenile phase when they are still not able to flower. Trees usually form lateral buds that undergo dormancy in the winter period and these buds will grow out the following spring after the cold period. In poplar, sylleptic branching, i.e. outgrowth of branches in the same season in which the buds were formed, is produced and is mainly associated with juvenility (Ceulemans et al., 1990; Cooke et al., 2005). Tree breeders have long desired to shorten the juvenile phase to speed up breeding and to increase sylleptic branching to obtain a higher woody biomass (Novaes et al., 2009; Rae et al., 2009). The possibility that TEM genes might be involved in the age-dependent pathway in arabidopsis and that this could be conserved across species is of great interest for biomass production in trees. Moreover, the CO/FT module is conserved in Populus and, in addition to flowering, regulates bud-set and growth cessation (Böhlenius et al., 2006; Hsu et al., 2011). This suggests that poplar TEM orthologues could be involved in those processes. Although there is still no information on the function of poplar RAV genes, the chestnut CsRAV1 is induced during winter dormancy and in response to low temperatures, which might suggest a role in bud-set and growth cessation; however, more experiments are needed to confirm this. In addition, when CsRAV1 is overexpressed in hybrid poplar it induces extensive sylleptic branching that it is not observed in control trees (Moreno-Cortés et al., 2012). This extra branching greatly increases the biomass of these transgenic trees, which is consequently of agronomic and commercial interest.
RAV GENES AS INTERACTORS OF RESPONSES TO BIOTIC AND ABIOTIC STRESS
Plants, using a complex system, defend themselves against both biotic and abiotic stresses. Plants are able to adapt and survive under several types of biotic and abiotic stresses, such as drought, high salinity, high/low temperatures or pathogen attacks. Worldwide crop productivity and quality are threatened by this wide variety of stresses, and therefore a better understanding of the complex and interconnected systems of plant defence and adaptation to these stresses is crucial. It is known that plants respond to such stresses by inducing morphological, physiological and biochemical changes through crosstalk among different genetic pathways (Zhuang et al., 2011). The activation of plant defence responses is first initiated by the recognition/identification of primary pathogen-derived elicitors by plant cell receptors (Yang et al., 1997; Kim and Martin, 2004). This triggers signal transduction pathways regulated by the hormones ethylene, salicylic acid (SA) and JA (Glazebrook, 1999; Lee et al., 2005), which induce the expression of plant defensive genes that produce defensive compounds, such as pathogen-related (PR) proteins, chitinase and/or enzymes involved in the biosynthesis of protective secondary metabolites (Gu et al., 2002; Koo et al., 2007).
In recent years, it has been discovered that RAV family members from different plant species not only are induced by ethylene but also play essential roles in biotic and abiotic environmental stresses (Alonso et al., 2003; Feng et al., 2005; Kim et al., 2005; Sohn et al., 2006; Zhuang et al., 2011). For instance, RAV1 and TEM2 expression in arabidopsis is upregulated by touch-related stimuli such as touch, wind and water spray, suggesting that these genes may function for developmental adaptation in response to different environmental stimuli (Kagaya and Hattori, 2009). In fact, it was found that expression of both genes is induced in arabidopsis after treatment with biotic and abiotic stresses such as bacterial pathogens, SA, mannitol, high salinity and wounding (Feng et al., 2005; Sohn et al., 2006). In addition, RAV1 is rapidly induced by cold and this response is regulated by the circadian clock (Fowler & Thomashow, 2002; Fowler et al., 2005). Galegae orientalis is a nitrogen-fixing legume used for forage production and soil improvement in scandinavian agriculture (Varis, 1986). Similarly to other plant species, GoRAV expression is induced by cold, exogenous ABA, high salinity and drought (Chen et al., 2009). Moreover, BnaRAV-1-HY15, a RAV orthologue in Brassica napus, an important agricultural-oil crop, is also induced by cold, NaCl and polyethylene glycol treatments (Zhuang et al., 2011).
A RAV orthologue (CaRAV1) from chili pepper (Capsicum annuum) is strongly induced during pathogen infection with Xanthomonas campestris, environmental stresses and abiotic elicitors (Sohn et al., 2006). Overexpression of CaRAV1 in arabidopsis induces several PR genes and enhances resistance not only against other pathogens such as Pseudomonas syringae, but also against osmotic stresses by high dehydration and salinity (Sohn et al., 2006). Solanum lycopersicum, tomato, is the second most consumed vegetable in the world. Ralstonia solanacearum causes the bacterial wilt disease, probably the most important bacterial vascular disease in tomato (Hai et al., 2008). Ectopic expression of SlRAV2 increases bacterial wilt tolerance in tomato plants by inducing the expression of PR genes such as SlERF5 and PR5 (Li et al., 2011).
Endogenous small RNA pathways and RNA silencing are major components of the plant response to different biotic and abiotic stresses. RNA silencing is a sequence-specific RNA degradation mechanism activated during viral infection that serves to protect plants against viruses (Ding and Voinnet, 2007). On the other side, plant viruses try to block the plant RNA silencing defence using different proteins (Diaz-Pendon and Ding, 2008). TEM2 is essential for suppression of RNA silencing by at least two unrelated plant viral proteins, potyviral HC-Pro and carmoviral P38, two potent viral suppressors of silencing that block primary and transitive RNA silencing (Endres et al., 2010). In tobacco, both viral repressors require NtRAV2 to block exclusively the activity of primary small interfering RNAs. NtRAV2 interacts physically with HC-Pro proteins and is required for HC-Pro suppression of virus-induced gene silencing (VIGS). Moreover, TEM2 induces the expression of FRY1 and CML38, two genes that act as endogenous suppressors of silencing in arabidopsis (Anandalakshmi et al., 2000). Consequently, TEM2 seems to be an essential control point in viral suppression of silencing. However, neither of the related arabidopsis genes RAV1 or TEM1 seems to have a redundant role in this specific aspect as they are not able to compensate for the loss of TEM2 to divert host defences toward responses that interfere with antiviral silencing (Endres et al., 2010). TEM2 may repress directly or indirectly the transcription of genes that encode proteins of the plant silencing machinery (Endres et al., 2010). Therefore, RAV orthologues from different plant species could function as key modulators of biotic and abiotic stress responses by integrating the regulation of diverse plant defence signalling pathways.
CONCLUSIONS
Despite RAV genes not being completely characterized, promising results obtained in recent years suggest strongly that RAV family members play important roles in many different physiological and developmental pathways in several plant species (Fig. 3). RAV genes act as repressors in the regulation of gene expression in various plant biological processes that may be of crucial importance for plant survival and crop production. Among these processes, floral transition is the best studied and TEM1 and TEM2 control at least the photoperiod- and GA-dependent flowering pathways. Moreover, RAV genes in different species may play important roles in other developmental processes and may also modulate some of the complex systems of response to diverse abiotic and biotic stresses. In conclusion, the RAV family, a unique family of transcription factors in plants, seems to integrate and control different physiological mechanisms that are affected by many internal and external factors. These essential controls should contribute to improve plant fitness, with the final outcome being optimal plant development and adaptation to environmental threats.
Fig. 3.
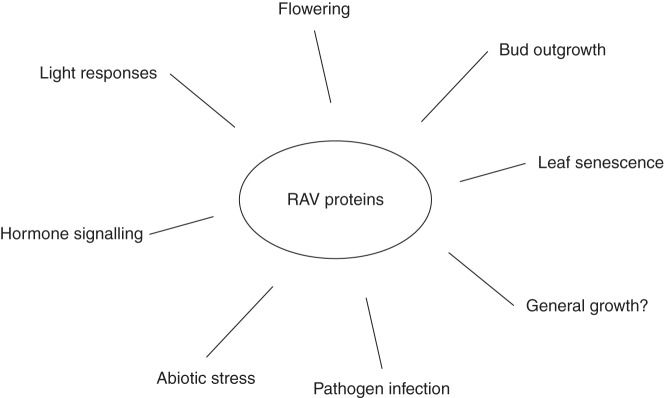
Summary of the processes regulated by RAV proteins in different plant species.
ACKNOWLEDGEMENTS
We thank Pablo Leivar for comments on the manuscript. Our work on floral development is supported by a grant from the Spanish MINECO (BFU2012–33746) and S.P.'s research group has been recognized as a Consolidated Research Group by the Catalonia Government (2009 SGR 697). A.E.A-J. is a predoctoral fellow of the investigator formation programme (FI) from the Catalonia Government.
LITERATURE CITED
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi R, Marathe R, Ge X, et al. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science. 2000;290:142–144. doi: 10.1126/science.290.5489.142. [DOI] [PubMed] [Google Scholar]
- Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genetics. 2006;2:980–989. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D. Integration of floral inductive signals in Arabidopsis. Nature. 2000;404:889–892. doi: 10.1038/35009125. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nature Genetics. 2003;33:168–71. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Current Biology. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. The TOPLESS Interactome: a framework for gene repression in Arabidopsis. Plant Physiology. 2012;158:423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans R, Stettler RF, Hinckley TM, Isebrands JG, Heilman PE. Crown architecture of Populus clones as determined by branch orientation and branch characteristics. Tree Physiology. 1990;7:157–167. doi: 10.1093/treephys/7.1-2-3-4.157. [DOI] [PubMed] [Google Scholar]
- Cooke JE, Martin TA, Davis JM. Short-term physiological and developmental responses to nitrogen availability in hybrid poplar. New Phytologist. 2005;167:41–52. doi: 10.1111/j.1469-8137.2005.01435.x. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annual Review of Genetics. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Chen MK, Hsu WH, Lee PF, Thiruvengadam M, Chen HI, Yang CH. The MADS box gene, FOREVER YOUNG FLOWER, acts as a repressor controlling floral organ senescence and abscission in Arabidopsis. Plant Journal. 2011;68:168–185. doi: 10.1111/j.1365-313X.2011.04677.x. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang Z, Wang X, Dong J, Ren J, Gao H. Isolation and characterization of GoRAV, a novel gene encoding a RAV-type protein in Galegae orientalis. Genes and Genetic Systems. 2009;84:101–109. doi: 10.1266/ggs.84.101. [DOI] [PubMed] [Google Scholar]
- Deng W, Ying H, Helliwell CA, Taylor JM, Peacock WJ, Dennis ES. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6680–6685. doi: 10.1073/pnas.1103175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Pendon JA, Ding SW. Direct and indirect roles of viral suppressors of RNA silencing in pathogenesis. Annual Review of Phytopathology. 2008;46:303–326. doi: 10.1146/annurev.phyto.46.081407.104746. [DOI] [PubMed] [Google Scholar]
- Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska MA, Schomburg FM, Amasino RM, Vierstra RD, Nagy F, Davis SJ. Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development. 2007;134:2841–2850. doi: 10.1242/dev.02866. [DOI] [PubMed] [Google Scholar]
- Duan K, Li L, Hu P, Xu SP, Xu ZH, Xue HW. A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant Journal. 2006;47:519–31. doi: 10.1111/j.1365-313X.2006.02804.x. [DOI] [PubMed] [Google Scholar]
- Endres MW, Gregory BD, Gao Z, et al. Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathogens. 2010;6:e1000729. doi: 10.1371/journal.ppat.1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Bohlenius H, Moritz T, Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. The Plant Cell. 2006;18:2172–2181. doi: 10.1105/tpc.106.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J-X, Liu D, Pan Y, et al. An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Molecular Biology. 2005;59:853–868. doi: 10.1007/s11103-005-1511-0. [DOI] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G. SnapShot: control of flowering in Arabidopsis. Cell. 2010;141 doi: 10.1016/j.cell.2010.04.024. 550, 550 e1–2. [DOI] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. The Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Lee K, Onouchi H, et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. The EMBO Journal. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiology. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. The Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. Genes controlling expression of defense responses in Arabidopsis. Current Opinion in Plant Biology. 1999;2:280–286. doi: 10.1016/S1369-5266(99)80050-8. [DOI] [PubMed] [Google Scholar]
- Goto N, Kumagai T, Koornneef M. Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long-day plant. Physiologia Plantarum. 1991;83:209–215. [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. The Plant Cell. 2006;18:1373–1382. doi: 10.1105/tpc.106.041798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, et al. Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. The Plant Cell. 2002;14:817–831. doi: 10.1105/tpc.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai TTH, Esch E, Wang JF. Resistance to Taiwanese race 1 strains of Ralstonia solanacearum in wild tomato germplasm. European Journal of Plant Pathology. 2008;122:471–479. [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. The EMBO Journal. 2002;21:4327–4337. doi: 10.1093/emboj/cdf432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma L-G, Qu L-J, Deng X-W. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes & Development. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. The Plant Journal. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Adams JP, Kim H, et al. FLOWERING LOCUS T duplication coordinates reproductive and vegetative growth in perennial poplar. Proceedings of the National Academy of Sciences USA; 2011. pp. 10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YX, Wang YH, Liu XF, Li JY. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Research. 2004;14:8–15. doi: 10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- Huijser P, Schmid M. The control of developmental phase transitions in plants. Development. 2011;138:4117–4129. doi: 10.1242/dev.063511. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Ohme-Takagi M. A novel group of transcriptional repressors in Arabidopsis. Plant and Cell Physiology. 2009;50:970–975. doi: 10.1093/pcp/pcp048. [DOI] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. FT protein acts as a long-range signal in Arabidopsis. Current Biology. 2007;17:1050–1054. doi: 10.1016/j.cub.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KCS, et al. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. The EMBO Journal. 2008;27:1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo JA, Piñeiro M. Timing is everything in plant development. The central role of floral repressors. Plant Science. 2011;181:364–378. doi: 10.1016/j.plantsci.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Hattori T. Arabidopsis transcription factors, RAV1 and RAV2, are regulated by touch-related stimuli in a dose-dependent and biphasic manner. Genes & Genetic Systems. 2009;84:95–99. doi: 10.1266/ggs.84.95. [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Research. 1999;27:470–478. doi: 10.1093/nar/27.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, et al. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kim S-Y, Kim Y-C, Lee J-H, et al. Identification of a CaRAV1 possessing an AP2/ERF and B3 DNA-binding domain from pepper leaves infected with Xanthomonas axonopodis pv. glycines 8ra by differential display. Biochimica et Biophysica Acta (BBA) – Gene Structure and Expression. 2005;1729:141–146. doi: 10.1016/j.bbaexp.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Martin GB. Molecular mechanisms involved in bacterial speck disease resistance of tomato. Plant Pathology Journal. 2004;20:7–12. [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Koo YJ, Kim MA, Kim EH, et al. Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Molecular Biology. 2007;64:1–15. doi: 10.1007/s11103-006-9123-x. [DOI] [PubMed] [Google Scholar]
- Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development. 2012;139:4180–4190. doi: 10.1242/dev.085407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim SH, Jung YH, et al. Molecular cloning and functional analysis of rice (Oryza sativa L.) OsNDR1 on defense signaling pathway. Plant Pathology Journal. 2005;21:149–157. [Google Scholar]
- Lee JH, He K, Stolc V, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell. 2007a;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes and Development. 2007b;21:397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Choi SC, An G. Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. The Plant Journal. 2008;54:93–105. doi: 10.1111/j.1365-313X.2008.03406.x. [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends in Plant Science. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Current Biology. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. The Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CW, Su RC, Cheng CP, et al. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiology. 2011;156:213–227. doi: 10.1104/pp.111.174268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L, et al. A repressor complex governs the integration of flowering signals in Arabidopsis. Developmental Cell. 2008;15:110–120. doi: 10.1016/j.devcel.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Li J, Li Y, Chen S, An L. Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. Journal of Experimental Botany. 2010;61:4221–4230. doi: 10.1093/jxb/erq241. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Gil Nam H. Leaf senescence. Annual Review of Plant Biology. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Lin M-K, Belanger H, Lee YJ, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. The Plant Cell. 2007;19:1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, et al. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. The Plant Cell. 2008;20:292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- Ma L, Sun N, Liu X, Jiao Y, Zhao H, Deng XW. Organ-specific expression of Arabidopsis genome during development. Plant Physiology. 2005;138:80–91. doi: 10.1104/pp.104.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E, Sjolander K, Hake S. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 2004;16:2265–2277. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Current Biology. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. Repression of flowering by the miR172 target SMZ. PLoS Biology. 2009;7:e1000148. doi: 10.1371/journal.pbio.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, et al. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. The Plant Journal. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Wright L, Fujiwara S, et al. Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. The Plant Cell. 2005;17:2255–2270. doi: 10.1105/tpc.105.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, et al. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proceedings of the National Academy of Sciences of the United States of America; 2004. pp. 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Suh S-S, Lee H, Choi K-R, Hong CB, Paek N-C, Kim S-G, Lee I. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. The Plant Journal. 2003;35:613–623. doi: 10.1046/j.1365-313x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Moreno-Cortés A, Hernández-Verdeja T, Sánchez-Jiménez P, González-Melendi P, Aragoncillo C, Allona I. CsRAV1 induces sylleptic branching in hybrid poplar. New Phytologist. 2012;194:98–90. doi: 10.1111/j.1469-8137.2011.04023.x. [DOI] [PubMed] [Google Scholar]
- Mutasa-Göttgens E, Hedden P. Gibberellin as a factor in floral regulatory networks. Journal of Experimental Botany. 2009;60:1979–1989. doi: 10.1093/jxb/erp040. [DOI] [PubMed] [Google Scholar]
- Novaes E, Osorio L, Drost DR, et al. Quantitative genetic analysis of biomass and wood chemistry of Populus under different nitrogen levels. New Phytologist. 2009;182:878–90. doi: 10.1111/j.1469-8137.2009.02785.x. [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proceeding of the National Academy of Sciences USA. 1997;94:7076–81. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato M, Castillejo C, Matías-Hernández L, Pelaz S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nature Communications. 2012;3:808. doi: 10.1038/ncomms1810. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Development. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Somers DE, Kim YS, et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- Rae AM, Street NR, Robinson KM, Harris N, Taylor G. Five QTL hotspots for yield in short rotation coppice bioenergy poplar: the Poplar Biomass Loci. BMC Plant Biology. 2009;9:23. doi: 10.1186/1471-2229-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biological Chemistry. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Hussain A, Taylor IB, Black CR. Use of mutants to study long-distance signalling in response to compacted soil. Journal of Experimental Botany. 2002;53:45–50. [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemistry Biophysics Research Communications. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Sawa M, Kay SA. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America; 2011. pp. 11698–11703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, et al. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proceedings of the National Academy of Sciences of the United States of America; 2009. pp. 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. The autonomous pathway: epigenetic and post-transcriptional gene regulation in the control of Arabidopsis flowering time. Current Opinion in Plant Biology. 2004;7:570–574. doi: 10.1016/j.pbi.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Sohn K, Lee S, Jung H, Hong J, Hwang B. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Molecular Biology. 2006;61:897–915. doi: 10.1007/s11103-006-0057-0. [DOI] [PubMed] [Google Scholar]
- Song J, Angel A, Howard M, Dean C. Vernalization – a cold-induced epigenetic switch. Journal of Cell Science. 2012;125:3723–3731. doi: 10.1242/jcs.084764. [DOI] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends in Plant Science. 2013;18:575–583. doi: 10.1016/j.tplants.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser B, Alvarez MJ, Califano A, Cerdán PD. A complementary role for ELF3 and TFL1 in the regulation of flowering time by ambient temperature. The Plant Journal. 2009;58:629–640. doi: 10.1111/j.1365-313X.2009.03811.x. [DOI] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. Nature. Vol. 410. 1116–1120; 2001. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. The Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. The Plant Cell. 2003;15:2856–2865. doi: 10.1105/tpc.016345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. The Plant Journal. 2012;70:549–561. doi: 10.1111/j.1365-313X.2012.04919.x. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hudson ME, Khanna R, et al. Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. The Plant Journal. 2004;38:725–739. doi: 10.1111/j.1365-313X.2004.02084.x. [DOI] [PubMed] [Google Scholar]
- Thomas B. Light signals and flowering. Journal of Experimental Botany. 2006;57:3387–3393. doi: 10.1093/jxb/erl071. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annual Review of Plant Biology. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Turki-Judeh W, Courey AJ. Groucho: a corepressor with instructive roles in development. In: Plaza S, Payre F, editors. Current topics in developmental biology. New York: Academic Press; 2012. pp. 65–96. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Varis E. Goat's rue (Galega orientalis L.) a potential pasture legume for temperate conditions. Journal of Agricultural Science Finland. 1986;58:83–101. [Google Scholar]
- Waltner JK, Peterson FC, Lytle BL, Volkman BF. Structure of the B3 domain from Arabidopsis thaliana protein At1g16640. Protein Science. 2005;14:2478–2483. doi: 10.1110/ps.051606305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proceedings of the National Academy of Sciences USA; 2013. pp. 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL. Gene networks controlling the initiation of flower development. Trends in Genetics. 2010;26:519–527. doi: 10.1016/j.tig.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, et al. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. The Plant Cell. 2006;18:2971–84. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HR, Kim JH, Kim J, et al. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. Journal of Experimental Botany. 2010;61:3947–3957. doi: 10.1093/jxb/erq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, et al. Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1. The Plant Cell. 2004;16:3448–3459. doi: 10.1105/tpc.104.026112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Shah J, Klessig DF. Signal perception and transduction in plant defense responses. Genes & Development. 1997;11:1621–1639. doi: 10.1101/gad.11.13.1621. [DOI] [PubMed] [Google Scholar]
- Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, et al. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. The Plant Cell. 2010;22:2156–2170. doi: 10.1105/tpc.110.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Luo Q, Yang C, Han Y, Li W. A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta. 2008;227:1389–1399. doi: 10.1007/s00425-008-0711-7. [DOI] [PubMed] [Google Scholar]
- Zhu Q-H, Helliwell CA. Regulation of flowering time and floral patterning by miR172. Journal of Experimental Botany. 2011;62:487–495. doi: 10.1093/jxb/erq295. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Sun CC, Zhou XR, Xiong AS, Zhang J. Isolation and characterization of an AP2/ERF-RAV transcription factor BnaRAV-1-HY15 in Brassica napus L. HuYou15. Molecular Biology Reports. 2011;38:3921–3928. doi: 10.1007/s11033-010-0508-1. [DOI] [PubMed] [Google Scholar]