Abstract
Background
WOX (Wuschel-like homeobOX) genes form a family of plant-specific HOMEODOMAIN transcription factors, the members of which play important developmental roles in a diverse range of processes. WOX genes were first identified as determining cell fate during embryo development, as well as playing important roles in maintaining stem cell niches in the plant. In recent years, new roles have been identified in plant architecture and organ development, particularly at the flower level.
Scope
In this review, the role of WOX genes in flower development and flower architecture is highlighted, as evidenced from data obtained in the last few years. The roles played by WOX genes in different species and different flower organs are compared, and differential functional recruitment of WOX genes during flower evolution is considered.
Conclusions
This review compares available data concerning the role of WOX genes in flower and organ architecture among different species of angiosperms, including representatives of monocots and eudicots (rosids and asterids). These comparative data highlight the usefulness of the WOX gene family for evo–devo studies of floral development.
Keywords: WOX genes, HOMEOBOX, flower development, WUSCHEL, PRS/WOX3, MAW/WOX1, EVERGREEN/WOX9, Petunia × hybrida, Arabidopsis thaliana, dicots, monocots, plant evo–devo
WOX GENES ARE HOMEOBOX GENES
The >250 000 wild species of flowering plants display an incredible diversity of flower shapes (Krizek and Fletcher, 2005), whose architectural traits (such as fused versus free-standing petals and large versus narrow petals) can be very different from one species to another. Despite the fact that the genetic basis of organ identity in the flower is well understood nowadays, thanks to the development of the ABCE model of flower development, mainly based on the MADS BOX gene family (Bowman et al., 2012; Heijmans et al., 2012; Smaczniak et al., 2012), little is known about organ shape and the general morphology of the flower, for which a general model is still lacking. Interestingly, whereas MADS BOX genes are involved in organ identity at the flower level in plants, organ identity in animals is based on a completely different class of genes, the HOMEOTIC BOX (or HOMEOBOX) genes (Holland, 2013). First discovered in the fruit fly Drosophila melanogaster (Carroll, 1995; Castelli-Gair, 1998), HOMEOBOX genes derive their name from William Bateson's concept of homeosis, since mutations in these genes may lead to transformation of one part of the embryo into another during development (Robert, 2001). At the molecular level, HOMEOBOX proteins are characterized by the HOMEODOMAIN, composed of 60 amino acids on average and arranged in space with an N-terminal arm plus three α helixes able to bind DNA (Wolberger, 1996). At least 14 different classes of HOMEOBOX genes (where specific conserved domains, in addition to the shared HOMEODOMAIN, can be found) have been described in plants, from angiosperms to red algae, and many of them have been shown to play a role in plant development (Mukherjee et al., 2009).
Plant HOMEOBOX genes sharing sequence identity with the gene WUSCHEL (At-WUS) from arabidopsis are referred to as WOX ((Wuschel-related homeobOX) genes. At-WUS was identified as a central player in stem cell maintenance in the shoot apical meristem (SAM), although it is not required for SAM initiation (Laux et al., 1996; Liu et al., 2011). The name wuschel apparently derives from the bristled and bushy phenotype of the mutants, in which ectopic meristems are repetitively produced and prematurely terminated (Laux et al., 1996).
Since the discovery of At-WUS, several WOX genes have been characterized in different species. Other members of the WOX family are usually referred as ‘WOX’ followed by an Arabic numeral (with a few exceptions), and can be grouped in different subfamilies or clades (van der Graaff et al., 2009; Vandenbussche et al., 2009).
WOX GENES PLAY DIFFERENT ROLES IN PLANT DEVELOPMENT
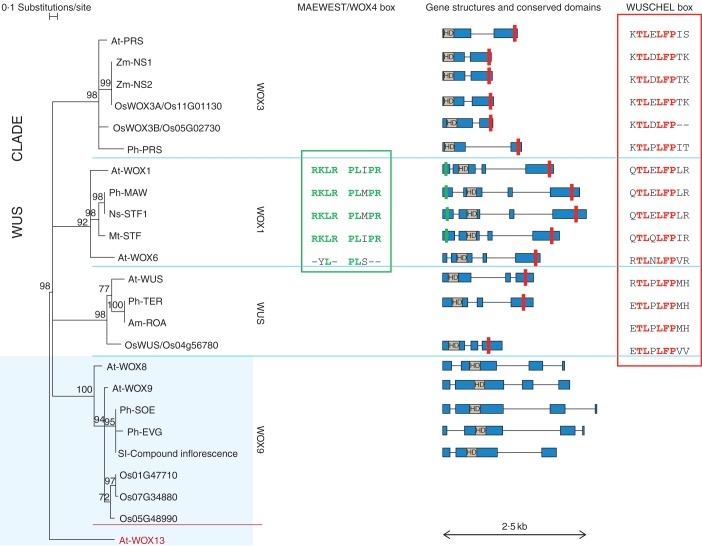
In arabidopsis, WOX genes have been shown to play a broad role in plant development, from stem cell maintenance at the meristem level (WUSCHEL in shoot meristem, WOX4 in cambium, WOX5 in root meristem) till embryo patterning (Laux et al., 1996; Haecker et al., 2004; Sarkar et al., 2007; Ji et al., 2010). We know from Picea abies that all the major WOX subfamilies, with the exception of the MAW/WOX1 subfamily, probably originated before the separation of angiosperms from gymnosperms (Hedman et al., 2013). A relationship between WOX gene number and body pattern complexity among different species, all from the ‘green lineage’ (a grouping of land plants and green algae), has also been proposed. In fact, WOX genes can be divided into three different lineages that are supposed to reflect their ancestry: an ancient lineage (comprising At-WOX10, At-WOX13, At-WOX14 and their homologues), an intermediate lineage (comprising At-WOX8, 9, 11 and 12 and their homologues) and a new or ‘WUS’ lineage (comprising AtWOX1–7, including WUSCHEL, and their homologues) (Haecker et al., 2004; Nardmann and Werr, 2012). Moreover, WOX genes from the WUS lineage are absent from green algae, bryophytes and ferns (with the exception of Leptosporangiatae). Further diversification, sub-functionalization and recruitment in different stem cell niches of these genes in angiosperms (but also gymnosperms), has been considered as contributing to the body plan diversity and evolutionary success of these groups (Nardmann and Werr, 2012). At the molecular level, the acquisition of repressive activity by proteins from the modern lineage, mainly due to an amino acid domain called the ‘WUSCHEL box’ (see red box on gene pictograms in Fig. 1), has been proposed to play a major role in this process (Lin et al., 2013). In this review we will further focus on the implication of WOX gene function during floral development.
Fig. 1.
Phylogenetic tree of WOX sequences. WOX sequences from different species (Am, Antirrhinum majus; At, Arabidopsis thaliana; Mt, Medicago truncatula; Ns, Nicotiana sylvestris, Ph, Petunia × hybrida; Os, Oryza sativa; Sl, Solanum lycopersicum; Zm, Zea mays) are clustered into different subfamilies (WOX3, WOX1, WUS, WOX9) after phylogenetic analysis. The phylogenetic tree is based on the HOMEODOMAIN sequences. The WOX13 sequence (dark red) from arabidopsis is used as outgroup. To support WOX relationships, 1000 bootstrap samples were obtained using the software TREECON (Van de Peer and De Wachter, 1994). Bootstrap values <65 % are not shown and corresponding branches are displayed as unresolved. On the right of the tree, gene structure is displayed for most of the WOX gene sequences (solid blue bars are used for exons and thin black lines for introns), also depicting conserved amino acidic boxes (HD, HOMEODOMAIN; red rectangle, WUSCHEL box; green rectangle, MAEWEST/WOX4 box, as in Vandenbussche et al., 2009). In addition, conserved amino acid residues are displayed for the MAEWEST/WOX4 box and the WUSCHEL box. Note that WOX sequences also display characteristic C-terminal motifs with functional properties (not shown), as illustrated in Vandenbussche et al. (2009). Where not specified, accession numbers are the same as in Vandenbussche et al. (2009); Mt-STF (AEL30892.1), Nt-STF1 (AEL30893.1) and Sl-Compound Inflorescence (NP_001234072.1) are from GenBank.
AT-WUSCHEL IS REQUIRED FOR STEM-CELL MAINTENANCE IN THE FLOWER (WUS SUBFAMILY)
WUSCHEL (WUS) is the founding member of the WOX family and is also representative of a clade, the ‘WUS clade’ (Fig. 1). WUS was initially isolated in arabidopsis (Laux et al. 1996) and its function has been thoroughly investigated in this model species.
WUS promotes the identity and maintenance of stem cells, a pool of undifferentiated and continuously dividing cells located in the central zone of both the SAM and the flower meristem (FM) (Laux et al., 1996; Besnard et al., 2011). Thus, on a wus genetic background, the SAM, instead of producing new organs throughout the life of the plant, stops functioning prematurely in an aberrant flat morphology. However, wus plants are still able to initiate a secondary meristem, but it fails to self-maintain, resulting in plants with a highly disorganized, bushy architecture. Similarly, wus flowers display many fewer stamens (usually one or two) and no carpels, consistent with precocious FM termination (Laux et al., 1996). WUS is therefore necessary for meristem maintenance, but is not required for their initiation. Consistent with this function, WUS expression is restricted to a small domain, the organizing centre, located in the basal part of the central zone, beneath the L3 layer in the SAM and beneath the L2 layer in the FM (Mayer et al., 1998). Mechanistically, it is known now that WUS acts non cell-autonomously to both promote stem cell identity and directly activate CLAVATA3 (CLV3) expression within the central zone (Schoof et al., 2000; Yadav et al., 2011). In turn, the CLE peptide CLV3 diffuses outside of the central zone, binds to the CLV1 and CLV2/CORYNE receptor kinases and thus triggers the signalling pathway that eventually leads to the restriction of WUS expression within the organizing centre (Brand et al., 2000; Schoof et al., 2000; Lenhard and Laux 2003; Katsir et al., 2011; Nimchuk et al., 2011). Much evidence suggests that POLTERGEIST and POLTERGEIST LIKE1 are signalling intermediates between CLV3 perception and WUS regulation (Yu et al., 2000; Song et al., 2006). WUS is thus part of a negative genetic feedback loop that ensures the homeostasis of the meristem. Within this loop, it is interesting to note that WUS also directly represses CLV1 expression (Busch et al., 2010).
Identification of additional direct WUS targets, such as the A-type Arabidopsis Response Regulator7 (ARR7), shed light on the way WUS specifies stem cell identity (Leibfried et al., 2005). By repressing the expression of ARR7, WUS counteracts the inhibitory activity of ARR7 on cytokinin signalling in the centre of the SAM (To et al., 2004; Leibfried et al., 2005). WUS can therefore act both as an activator and a repressor of transcription (Ikeda et al., 2009; Busch et al., 2010), and the WUS box has been reported to be absolutely required for these two types of activity (Ikeda et al., 2009). The role of WUS as a transcriptional repressor was further underscored by its interaction with two co-repressors, WSIP1/TOPLESS and WSIP2 (Kieffer et al., 2006; Long et al., 2006).
Under inductive conditions, the vegetative SAM can switch to an inflorescence SAM (iSAM or IM), which, instead of producing leaves on its flanks, generates FM. All the data gathered on WUS function cannot be generalized to the FM, as exemplified for instance with TOPLESS RELATED1 and 2, which are repressed by WUS in the SAM but activated in the FM (Busch et al., 2010), further confirming the complex regulatory interaction reported earlier (Ikeda et al., 2009). However, the majority of data are common to both meristems. This is especially true for the stem cell maintenance process and the WUS/CLV negative feedback loop, with some minor differences, such as reduced sensitivity to changes in CLV signalling in the FM compared with the SAM (Laux et al., 1996; Clark et al., 1997; Mayer et al., 1998; Schoof et al., 2000; Müller et al., 2006; Yadav et al., 2011). In arabidopsis, the WUS/CLV loop is absent in incipient floral primordia but it is rapidly set up, with activation of WUS expression at stage 1, followed by that of CLV3 at stage 2–2½. However, and contrary to what happens in the SAM, stem cell maintenance is only transient in the FM (Prunet et al., 2009). Indeed, once all floral organs have been initiated, activity of the FM stops and the flower becomes determinate. The mechanism controlling FM termination has been described mainly in arabidopsis. It has been shown to rely on a second genetic feedback loop that implies WUS and AGAMOUS (AG) (Lenhard et al., 2001; Lohmann et al., 2001). AG encodes a C-class MADS box protein that also controls the identity of stamens and carpels, the male and female reproductive organs, respectively (Yanofsky et al., 1990; Bowman et al., 1991). The feedback loop starts with the activation of AG transcription, at stage 3, by WUS together with LEAFY (LFY), which acts in a partially redundant way in this process (Yanofsky et al., 1990; Lohmann et al., 2001), and ends with the repression of WUS in the centre of the FM, at stage 6, concomitantly with or immediately after carpel initiation. This second part of the loop absolutely requires AG, making AG the main developmental switch to FM termination. Thus, on an ag genetic background, flowers are indeterminate and keep producing floral organs in their centre, and this phenotype coincides with the maintenance of WUS expression within the FM organizing centre (Bowman et al., 1991; Lenhard et al., 2001; Lohmann et al., 2001). FM termination is therefore closely linked to initiation of carpel development. However, these two processes are not coincident and are uncoupled, although both are controlled by AG (Mizukami and Ma, 1995; Ji et al., 2011). Very interestingly, the fact that organizing centre cells retain a molecular identity distinguishable from that of surrounding cells even after the cessation of WUS expression further confirms the separation of the two processes but also demonstrates that organizing centre cells persist after FM termination and are not incorporated into carpels (Liu et al., 2011). Recently, two different mechanisms of repression of WUS by AG have been reported. They both explain why AG does not repress WUS expression from stage 3. In the first mechanism, AG represses WUS expression indirectly by activating KNUCKLES (KNU, a C2H2-type zinc finger transcription factor) expression, which in turn represses WUS expression directly or indirectly (Sun et al., 2009). In this model, KNU expression is blocked by repressive marks that are removed in an AG-dependent manner at stage 6. In the second mechanism, AG also directly represses WUS expression by recruiting polycomb group proteins to WUS (Liu et al., 2011). In this model, AG recruits polycomb group proteins to WUS earlier than in the first model, at stage 5. These two mechanisms are probably coordinated and act in parallel to each other in terminating floral stem cell maintenance.
From this detailed analysis in arabidopsis, it is clear now that to make a flower with a fixed number of floral organs it is of crucial importance that WUSCHEL is switched off at very precise moments during development of the floral bud. Not all flowering species have such a rigidly controlled floral organ number within their flowers, which seems to be a character acquired later in angiosperm evolution. It would be interesting to investigate whether changes in the WUS regulatory network have occurred during evolution that might have led to increased robustness of the system, resulting in fully determined flower architectures. Likewise, one can question whether floral meristem termination occurs at the same moment in species with different placentation topologies (Colombo et al., 2008). For example, in Petunia and rice, which belong to the central placentation types, the floral meristem remains active after carpel primordia have been produced, because the placenta (and later on the ovules) develops directly from the floral meristem centre between the carpels. By contrast, in parietal placentation types such as arabidopsis, the placenta and ovules differentiate from the medial regions of the carpels after the FM has terminated. Interestingly, in both rice and Petunia, it has been shown that both D- and C-clade AG-like MADS box proteins participate in floral meristem termination (Dreni et al., 2011; Heijmans et al., 2012), with the D-lineage proteins being strongly expressed during placenta development, while the D-clade gene STK in arabidopsis does not seem to be involved at all in determinacy control (Pinyopich et al., 2003). This regulatory difference might be a direct consequence of different placentation topologies.
Besides arabidopsis, loss of function mutants for WUS orthologues have been described so far only in Petunia (terminator) and snapdragon (rosulata) (Stuurman et al., 2002; Kieffer et al., 2006), confirming their role in maintenance of the SAM. Unfortunately, ter and roa mutants never develop flowering branches, and the roles of TER and ROA in floral meristem control have therefore not yet been analysed. Expression studies of WUS orthologues are available for a wider range of species. Perhaps the most remarkable findings have been presented by Nardmann and Werr (2006), who showed in grasses that none of the isolated WUS orthologues exhibited an organizing centre-type expression pattern in the vegetative SAM, as in arabidopsis. Instead, it has been shown that the WOX4 orthologue in rice, Os-WOX4, is involved in SAM maintenance, along with cytokinins (Ohmori et al., 2013). Moreover, in rice, mutant plants for the LONELY GUY gene, which codes for a cytokinin-activating enzyme, are also affected at the SAM and the inflorescence and floral meristems (Kurakawa et al., 2007). Taken together, these facts suggest major differences in WUS function in grass species compared with dicots.
THE PRS/WOX3 SUBFAMILY
The second WOX gene that was found to play a role in flower development is Arabidopsis PRESSED FLOWER (At-PRS, also called WOX3). Mutants for this gene have flowers with a flattened appearance (hence the name) because lateral sepal development is affected: they are usually smaller, sometimes with a filamentous appearance, or can be completely absent (Matsumoto and Okada, 2001). Although the size of the abaxial and adaxial sepals is normal, marginal regions show defects. At-PRS was shown to act independently of organ identity and meristem size. The expression of At-PRS was detected at the lateral regions of all lateral organs at very early stages, including leaves, flower primordia and floral organ primordia, despite the fact that phenotypic defects were much more restricted. Because of its expression pattern and mutant phenotype, At-PRS was proposed to regulate the lateral axis-dependent development of arabidopsis flowers (Matsumoto and Okada, 2001). Later, it was reported that arabidopsis prs mutants also lacked lateral stamens, and were additionally affected at the leaf level because of the absence of stipules at the leaf base (Nardmann et al., 2004).
Initially, floral mutant phenotypes had not been described for PRS/WOX3 homologues in species other than arabidopsis. Instead, it was shown that the NARROW SHEATH 1 and 2 genes in maize are PRS/WOX3 homologues (Nardmann et al., 2004) and that they perform a crucial role in leaf margin development, with the ns1 ns2 double mutant displaying a severely reduced leaf blade (Scanlon et al., 1996, 2000; Scanlon, 2000). A very similar leaf phenotype was found in nal2 nal3 double mutants in rice, with NAL2 and NAL3 (OsWOX3A) being homologous to the maize NS1 and NS2 genes (Cho et al., 2013; Ishiwata et al., 2013). Interestingly, the widths of the lemma and palea were also significantly reduced in nal2 nal3 mutants (Cho et al., 2013). Since the lemma and palea are considered to be equivalent to eudicot sepals, this indicates that the function of PRS/WOX3 proteins during floral development is conserved between monocots and dicots. Rice contains a third WOX3 copy, called OsWOX3B/DEP, but this functions in the regulation of trichome formation in leaves and glumes (Angeles-Shim et al., 2012). It therefore seems that the PRS/WOX3 subfamily in rice has further functionally diverged.
THE MAW/WOX1 SUBFAMILY
The evolutionary invention of petals, the usually brightly coloured organs of the flower, is generally believed to have played a major role in the evolution of pollination syndromes. In many taxa throughout the angiosperms, the petals fuse partly or completely to form a tubular structure, thereby creating a protective barrier enclosing the reproductive organs and nectaries in the centre of the flower. The maewest (maw) mutant in Petunia was isolated in a genetic screen for mutants with defects in petal fusion (Vandenbussche et al., 2009). Morphological analysis of maw flowers showed that petal fusion defects were mainly due to reduced lateral outgrowth of the initially separate petal primordia, which subsequently fail to fuse properly. Similar defects were found in carpels, resulting in partly unfused carpels, and sepals were narrower than wild-type. In addition, leaf blade outgrowth was considerably reduced along the lateral axis, as observed in floral organs, indicating that MAW plays a general role in the lateral outgrowth of organs. MAW was shown to encode a member of the WOX1 subfamily of WOX transcription factors (Vandenbussche et al., 2009). Similar phenotypes in leaf and flower development were found for mutants of MAW/WOX1 homologues in Medicago truncatula and Nicotiana sylvestris (McHale and Marcotrigiano, 1998; Lin et al., 2013; Tadege et al., 2011a). In addition, mutants for MAW/WOX1 homologues in two other species, narrow organs1 in Lotus japonicus and lathyroides in Pisum sativum (garden pea), have also been shown to be affected in lateral outgrowth of organs such as leaves and petals (Zhuang et al., 2012), further showing a broadly conserved role for MAW/WOX1 genes among different dicot species. In contrast, the dramatic maw/wox1 phenotypes found in Petunia, Medicago, Nicotiana, Lotus and pea are absent in arabidopsis wox1, wox6, and wox1 wox6 double mutants, showing that WOX1 function is redundant (Vandenbussche et al., 2009), and that other factors can compensate for the loss of WOX1/6 function in arabidopsis.
FUNCTIONAL OVERLAP BETWEEN MAW/WOX1 AND PRS/WOX3 SUBFAMILIES
Because mutants of members of both the PRS/WOX3 and MAW/WOX1 subfamilies in arabidopsis display a much less severe or no phenotypic difference compared with homologous mutants in other species (see the two previous paragraphs), and because PRS and WOX1 overlap in expression pattern, it was hypothesized that arabidopsis WOX1 and PRS genes might overlap in function. This was indeed confirmed by the phenotype of wox1 prs double mutants, consistent with their overlapping expression domains at the adaxial–abaxial boundary layer and at the organ margins (Vandenbussche et al., 2009; Nakata et al., 2012). In contrast to prs single-mutant flowers, all sepals (not only the lateral ones) in prs wox1 flowers displayed reduced blade outgrowth, as was the case also for the petals. This phenotype was also found in leaf development, with wox1 prs leaves displaying obvious defects in blade outgrowth, while prs mutants were only lacking stipules. These results clearly indicate that, despite the fact that the PRS/WOX3 and MAW/WOX1 subfamilies are structurally different (Fig. 1), their proteins share a common function in organ development along the lateral axis. However, note that the carpel fusion defects found in Petunia, Nicotiana and Medicago wox1 mutants (Vandenbussche et al., 2009; Tadege et al., 2011a) were not observed in wox1 prs mutants (Vandenbussche et al., 2009). So far, functional data for both the PRS/WOX3 and the MAW/WOX1 subfamily are only available in arabidopsis, and it will be interesting to investigate whether this functional overlap also exists in species in which maw/wox1 single mutants do display a strong phenotype on their own. Along the same lines, loss of wox3/prs function in monocots results in severe leaf blade reduction, but, remarkably, grasses (including wheat, maize, rice and Brachypodium) do not have WOX1 representatives (Nardmann and Werr, 2006; Nardmann et al., 2007; Vandenbussche et al., 2009), while all other WOX subfamilies are represented in their genomes. It would be very interesting to investigate whether the absence of the WOX1 subfamily in grasses has developmental implications related to differences in leaf development between monocots and dicots.
In arabidopsis, At-WOX1 and At-PRS have recently been proposed to define a so-called middle domain in leaf development, different from the classical adaxial and abaxial sides of the leaf, and able to drive blade outgrowth (Nakata et al., 2012). Furthermore, in this model At-WOX1 and At-WOX3 would be at the spatial and regulatory interface of adaxial (HD-ZIPIIIs, ASYMMETRIC LEAVES1 and 2)-, abaxial (KANADIs, ARFs)- or middle–abaxial (FILAMENTOUS FLOWER)-specifying genes (Nakata and Okada, 2012; Tsukaya, 2013). This may also imply the role of several hormones. For instance, ASYMMETRIC LEAVES1 and 2 regulate the expression of ARF3 (in both a direct and an indirect way) (Iwasaki et al., 2013), which probably controls the cytokinin biosynthetic pathway in its turn (Takahashi et al., 2013). At the same time, KANADI1 is linked to plant hormone pathways and leaf morphology, usually in a way antagonistic to HD-ZIPIII genes (Reinhart et al., 2013), such as the auxin pathway (Huang et al., 2014), but probably also the cytokinin pathway, by binding to the ASYMMETRIC LEAVES2 promoter (Merelo et al., 2013). On the other hand, a study of stenofolia (stf) mutants in Medicago and Nicotiana proposes a role in modulating phytohormone homeostasis and sugar metabolism, in this way playing a role in leaf development (Tadege et al., 2011a, b). Moreover, a role in cell proliferation along the adaxial–abaxial boundary has been shown for STF (Tadege et al., 2011), WOX1 and PRS (Nakata et al., 2012), and a recent paper describes the interaction between STF and ASYMMETRIC LEAVES2 with TOPLESS along the leaf margin (Zhang et al., 2014).
EVERGREEN IN PETUNIA IS INVOLVED IN INFLORESCENCE ARCHITECTURE (WOX9 SUBFAMILY)
The WOX9 subfamily is represented by two genes in both arabidopsis and Petunia (Fig. 2). The arabidopsis representatives are STIMPY (STIP, WOX9) and STIMPY-LIKE (STPL, WOX8) (Haecker et al., 2004; Wu et al., 2005), and the Petunia representatives are EVERGREEN (EVG) and SISTER OF EVERGREEN (SOE) (Rebocho et al., 2008). EVG in Petunia and COMPOUND INFLORESCENCE in tomato are essential for inflorescence development and architecture (Lippman et al., 2008; Rebocho et al., 2008). On an evg background, floral identity is not specified and apical floral meristems develop as inflorescence shoots instead (Fig. 3). Moreover, evg mutations display defects in the physical separation of the apical and lateral meristem, resulting in the formation of a fasciated meristem. Petunia displays a cymose inflorescence in which the apical meristem terminates by forming an FM and growth continues from the lateral or ‘sympodial’ meristem, which will generate a subsequent sympodial meristem before terminating in a flower. In Petunia, FM identity is mainly specified by ABERRANT LEAF AND FLOWER (ALF) and DOUBLE TOP (DOT), which are the homologues of LEAFY and UNUSUAL FLORAL ORGANS, respectively (Souer et al. 1998, 2008). Unexpectedly, EVG is not expressed in the apical floral meristem but in the sympodial incipient meristem (Rebocho et al., 2008). Mechanistically, the model assumes that EVG counteracts the effect of an unknown mobile factor that inhibits DOT expression in the FM, possibly indirectly by promoting proliferation of the lateral IM and separation from the apical FM.
Fig. 2.
Wox mutant flower phenotypes in petunia, Nicotiana, Medicago and arabidopsis (Vandenbussche et al., 2009; Tadege et al., 2011b; Lin et al., 2013). (A, C, D, E) Wild-type (WT) flower phenotypes for Petunia × hybrida, Nicotiana sylvestris, Medicago truncatula and Arabidopsis thaliana, respectively. (B) A Petunia wild-type pistil. (F, H, I, L) Mutant flowers: maw in Petunia (F), lam1 in Nicotiana (H), stf in Medicago (I) and wox1 prs in arabidopsis (L). (G) A strongly affected maw pistil (carpels unfused). (C, D, H, I) Courtesy of M. Tadege.
Fig. 3.

evergreen in Petunia (Rebocho et al., 2008). Petunia wild-type (A) and evg (B) inflorescences. Whereas the wild-type follows a typical zig-zag pattern, making a flower at each node and resulting in a cymose inflorescence (C), the evg mutant has a fasciated and bushy inflorescence (D), flowering only occasionally (terminal flowers).
In contrast, WOX8/STPL and WOX9/STIP in arabidopsis are required for embryo patterning and vegetative SAM maintenance but not for inflorescence development and architecture. WOX8/STPL and WOX9/STIP are expressed during the early stages of embryo development with overlapping and specific expression domains (Haecker et al., 2004). Briefly, only WOX8/STPL is expressed in the egg cell and zygote, whereas WOX8/STPL and WOX9/STIP are both expressed after the division of the zygote. However, their expression is restricted to the basal daughter cell, which will form the suspensor and the hypophysis (Haecker et al., 2004). Consistent with this expression pattern, weak wox9/stip alleles display fewer cells in the basal part of the embryo while embryo development of strong alleles stops at the globular stage, both phenotypes being due to a reduction or a complete arrest of the cell cycle (Wu et al., 2005, 2007). Interestingly, the wox9/stip phenotype can be rescued by the addition of exogenous sucrose, although developing carpels are not fully rescued, further confirming the role of WOX9/STIP in stimulating the cell cycle (Wu et al., 2005). WOX8/STPL was shown to functionally overlap with WOX9/STIP in promoting embryonic cell division (Wu et al., 2007; Breuninger et al., 2008). Later during development, WOX9/STIP promotes the growth of the vegetative SAM and is required for the maintenance of WUS expression at the shoot apex. In this regulatory network, WOX9/STIP acts downstream of the cytokinin signalling pathways (Skylar et al., 2010). More recently, WOX8/STPL has been shown to promote, along with the expression of WOX2, CUC2 and CUC3, the establishment of the cotyledon boundary (Lie et al., 2012).
It therefore seems that EVG, WOX8/STPL and WOX9/STIP have nothing in common. However, it is interesting to note that in both arabidopis and Petunia the constitutive expression of WOX9/STIP and EVG causes similar defects, suggesting that the proteins are functionally very similar and that diversification of EVG and WOX9/STIP might rely on alterations in their expression patterns.
SOE, the second member of the WOX9 clade in Petunia, displays an expression pattern very similar to those of WOX8/STPL and WOX9/STIP in arabidopsis (Rebocho et al., 2008). Furthermore, the constitutive expression of SOE in Petunia phenocopies those of EVG and WOX9/STIP, further indicating that these proteins are functionally similar. It has therefore been proposed that SOE and WOX8/STPL–WOX9/STIP represent an ancestral gene and that EVG is a duplicated gene that acquired a new function in inflorescence development and a key role in the evolution of cymes (Rebocho et al., 2008). This example illustrates how genes can be recruited upon duplication to undergo a neo- or sub-functionalization process.
EVOLUTION OF WOX GENE FUNCTION: PRIMARILY THROUGH CHANGES IN EXPRESSION PATTERNS?
Despite the fact that most of the different WOX subfamilies are structurally quite different from each other (differences in exon numbers, conserved peptide motifs specific for each subfamily, see Vandenbussche et al., 2009), proteins in a number of these subfamilies do seem to share some ancestral common function. A first example can be found in members of the WOX1 and PRS subfamilies in arabidopsis (Vandenbussche et al., 2009; Nakata et al., 2012). In this case, WOX1 and PRS expression overlaps and the phenotype of wox1 prs double mutants clearly shows that they also functionally overlap. In a series of other examples, it turns out that the protein sequences of different family members have retained similar capacities, even though their expression patterns have completely diverged and do not overlap any more. For example, WUS is able to complement prs and wox5 mutant phenotypes when expressed under their respective promoters (Sarkar et al., 2007; Shimizu et al., 2009). More recently, Lin and colleagues (2013) showed that arabidopsis WUS, WOX1, WOX2, WOX3, WOX4, WOX5 and WOX6 were all able to complement leaf blade and floral developmental defects in the Nicotiana lam1 mutant (lam1 is the Nicotiana wox1 homologue mutant) when expressed under the control of the Mt-STF promoter [promoter of the Medicago STENOFOLIA gene (WOX1 homologue)].
Together, this demonstrates that proteins of the WUS, MAW/WOX1, WOX2, PRS/WOX3, WOX4 and WOX5 subfamilies (together forming the WUS clade) still have some functional properties in common, despite their ancient origin. This further suggests that changes in cis-regulatory elements have constituted a major source of functional diversification within the WUS clade, obviously without excluding the possibility that changes in the protein sequence might also have contributed.
In contrast, WOX7, WOX9, WOX11 and WOX13 were not able to complement the lam1 mutant phenotype. Interestingly, all WOX proteins that were able to complement possess the WUS box (WUS clade), whereas all others lack this motif (Vandenbussche et al., 2009; Lin et al., 2013), highlighting the importance of repressive activity linked to the WUSCHEL box for leaf blade expansion. This was further confirmed by the observation that chimeric WOX7, WOX9 and WOX13 proteins fused with either the WUS box or an SRDX repressor domain could complement the lam1 phenotype (Lin et al., 2013). This shows that the acquisition of one or more transcriptional repressor domains in the members of WUS clade compared with the more ancient WOX9 and WOX13 clades has been instrumental in gaining their central function in organizing cell proliferation for meristem maintenance and lateral organ development.
WOX GENES AND FLOWER DEVELOPMENT: CONCLUDING REMARKS
Floral phenotypes thus far described for wox mutants include premature floral termination (wus), reduced lateral development of floral organs (MAW/WOX1 and PRS/WOX3 subfamilies), resulting in narrow organs with petal and carpel fusion defects, or the complete absence of flowering due to a defect in inflorescence meristem identity (evergreen, WOX9 subfamily). Except for the latter, the developmental defects found in the flower are part of a more general phenotype, which also includes defects in leaf blade expansion. Goethe (Goethe, 1790; Coen, 2001) proposed a long time ago that floral organs are in fact modified leaves, so it is perhaps not very surprising to find that WOX mutants are affected in both vegetative and floral development. Yet it is clear that nature has exploited WOX gene function during evolution for shaping floral architecture. Therefore, while the homeotic function of animal HOX genes is fulfilled in plant floral development by MADS box transcription factors, WOX genes contribute to general aspects of floral architecture and morphology. Classically, plant developmental biology has focused mainly on Arabidopsis thaliana as a model organism. Nevertheless, much of the progress made in our understanding of the function of different WOX genes comes from studies in different species. We consider this to be a strong argument in favour of the idea that plant developmental biology in general would benefit from reorientation towards a more multi-model approach.
ACKNOWLEDGEMENTS
M.V. is funded by an ATIP-AVENIR grant (Centre National de la Recherche Scientifique). We are grateful to Million Tadege (Oklahoma State University) and his team for providing pictures of lam1 and stf mutants in Nicotiana and Medicago. We would also like to thank the two anonymous reviewers, as well as the editor, for very helpful suggestions to improve the manuscript.
LITERATURE CITED
- Angeles-Shim RB, Asano K, Takashi T, et al. A WUSCHEL-related homeobox 3B gene, depilous (dep), confers glabrousness of rice leaves and glumes. Rice. 2012;5:28. doi: 10.1186/1939-8433-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard F, Vernoux T, Hamant O. Organogenesis from stem cells in planta: multiple feedback loops integrating molecular and mechanical signals. Cellular and Molecular Life Sciences. 2011;68:2885–2906. doi: 10.1007/s00018-011-0732-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Drews GN, Meyerowitz EM. Expression of the arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell. 1991;3:749–758. doi: 10.1105/tpc.3.8.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. The ABC model of flower development: then and now. Development. 2012;139:4095–4098. doi: 10.1242/dev.083972. [DOI] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. Differential expression of WOX genes mediates apical-basal axis formation in the arabidopsis embryo. Developmental Cell. 2008;14:867–876. doi: 10.1016/j.devcel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, et al. Transcriptional control of a plant stem cell niche. Developmental Cell. 2010;18:849–861. doi: 10.1016/j.devcel.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- Castelli-Gair J. Implications of the spatial and temporal regulation of Hox genes on development and evolution. International Journal of Developmental Biology. 1998;42:437–444. [PubMed] [Google Scholar]
- Cho S-H, Yoo S-C, Zhang H, et al. The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytologist. 2013;198:1071–1084. doi: 10.1111/nph.12231. [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/s0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- Coen E. Goethe and the ABC model of flower development. Comptes Rendus de l'Académie des Sciences. Série III, Sciences de la Vie. 2001;324:523–530. doi: 10.1016/s0764-4469(01)01321-x. [DOI] [PubMed] [Google Scholar]
- Colombo L, Battaglia R, Kater MM. Arabidopsis ovule development and its evolutionary conservation. Trends in Plant Science. 2008;13:444–450. doi: 10.1016/j.tplants.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, et al. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. The Plant cell. 2011;23:2850–2863. doi: 10.1105/tpc.111.087007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goethe JW, von . Versuch die Metamorphose der Pflanzen zu erklären. Gotha: C.W. Ettinger; 1790. [Google Scholar]
- Van der Graaff E, Laux T, Rensing SA. The WUS homeobox-containing (WOX) protein family. Genome Biology. 2009;10:248. doi: 10.1186/gb-2009-10-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haecker A, Gross-Hardt R, Geiges B, et al. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- Hedman H, Zhu T, von Arnold S, Sohlberg JJ. Analysis of the WUSCHEL-RELATED HOMEOBOX gene family in the conifer Picea abies reveals extensive conservation as well as dynamic patterns. BMC Plant Biology. 2013;13:89. doi: 10.1186/1471-2229-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans K, Morel P, Vandenbussche M. MADS-box genes and floral development: the dark side. Journal of Experimental Botany. 2012;63:5397–5404. doi: 10.1093/jxb/ers233. [DOI] [PubMed] [Google Scholar]
- Holland PWH. Evolution of homeobox genes. Wiley Interdisciplinary Reviews: Developmental Biology. 2013;2:31–45. doi: 10.1002/wdev.78. [DOI] [PubMed] [Google Scholar]
- Huang T, Harrar Y, Lin C, et al. Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-element and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell. 2014;26:246–262. doi: 10.1105/tpc.113.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell. 2009;21:3493–3505. doi: 10.1105/tpc.109.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata A, Ozawa M, Nagasaki H, et al. Two WUSCHEL-related homeobox genes, narrow leaf2 and narrow leaf3, control leaf width in rice. Plant & Cell Physiology. 2013;54:779–792. doi: 10.1093/pcp/pct032. [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Takahashi H, Iwakawa H, et al. Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in arabidopsis. Development. 2013;140:1958–1969. doi: 10.1242/dev.085365. [DOI] [PubMed] [Google Scholar]
- Ji J, Shimizu R, Sinha N, Scanlon MJ. Analyses of WOX4 transgenics provide further evidence for the evolution of the WOX gene family during the regulation of diverse stem cell functions. Plant Signaling & Behavior. 2010;5:916–920. doi: 10.4161/psb.5.7.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L, Liu X, Yan J, et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in arabidopsis. PLoS Genetics. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Davies KA, Bergmann DC, Laux T. Peptide signaling in plant development. Current Biology. 2011;21:R356–R364. doi: 10.1016/j.cub.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Stern Y, Cook H, et al. Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell. 2006;18:560–573. doi: 10.1105/tpc.105.039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. Molecular mechanisms of flower development: an armchair guide. Nature Reviews. Genetics. 2005;6:688–698. doi: 10.1038/nrg1675. [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Laux T, Mayer KF, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JPC, Busch W, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Laux T. Stem cell homeostasis in the arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development. 2003;130:3163–3173. doi: 10.1242/dev.00525. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T. Termination of stem cell maintenance in arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell. 2001;105:805–814. doi: 10.1016/s0092-8674(01)00390-7. [DOI] [PubMed] [Google Scholar]
- Lie C, Kelsom C, Wu X. WOX2 and STIMPY-LIKE/WOX8 promote cotyledon boundary formation in arabidopsis. Plant Journal. 2012;72:674–682. doi: 10.1111/j.1365-313X.2012.05113.x. [DOI] [PubMed] [Google Scholar]
- Lin H, Niu L, McHale NA, Ohme-Takagi M, Mysore KS, Tadege M. Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proceedings of the National Academy of Sciences of the USA. 2013;110:366–371. doi: 10.1073/pnas.1215376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman ZB, Cohen O, Alvarez JP, et al. The making of a compound inflorescence in tomato and related nightshades. PLoS Biology. 2008;6:e288. doi: 10.1371/journal.pbio.0060288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim YJ, Müller R, et al. AGAMOUS terminates floral stem cell maintenance in arabidopsis by directly repressing WUSCHEL through recruitment of polycomb group proteins. Plant Cell. 2011;23:3654–3670. doi: 10.1105/tpc.111.091538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, et al. A molecular link between stem cell regulation and floral patterning in arabidopsis. Cell. 2001;105:793–803. doi: 10.1016/s0092-8674(01)00384-1. [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Okada K. A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of arabidopsis flowers. Genes & Development. 2001;15:3355–3364. doi: 10.1101/gad.931001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- McHale NA, Marcotrigiano M. LAM1 is required for dorsoventrality and lateral growth of the leaf blade in Nicotiana. Development. 1998;125:4235–4243. doi: 10.1242/dev.125.21.4235. [DOI] [PubMed] [Google Scholar]
- Merelo P, Xie Y, Brand L, et al. Genome-wide identification of KANADI1 target genes. PloS One. 2013;8:e77341. doi: 10.1371/journal.pone.0077341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Ma H. Separation of AG function in floral meristem determinacy from that in reproductive organ identity by expressing antisense AG RNA. Plant Molecular Biology. 1995;28:767–784. doi: 10.1007/BF00042064. [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Brocchieri L, Bürglin TR. A comprehensive classification and evolutionary analysis of plant homeobox genes. Molecular Biology and Evolution. 2009;26:2775–2794. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. Dynamic and compensatory responses of arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell. 2006;18:1188–1198. doi: 10.1105/tpc.105.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Okada K. The three-domain model: a new model for the early development of leaves in Arabidopsis thaliana. Plant Signaling & Behavior. 2012;7:1423–1427. doi: 10.4161/psb.21959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Matsumoto N, Tsugeki R, Rikirsch E, Laux T, Okada K. Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in arabidopsis. Plant Cell. 2012;24:519–535. doi: 10.1105/tpc.111.092858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J, Werr W. The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Molecular Biology and Evolution. 2006;23:2492–2504. doi: 10.1093/molbev/msl125. [DOI] [PubMed] [Google Scholar]
- Nardmann J, Werr W. The invention of WUS-like stem cell-promoting functions in plants predates leptosporangiate ferns. Plant Molecular Biology. 2012;78:123–134. doi: 10.1007/s11103-011-9851-4. [DOI] [PubMed] [Google Scholar]
- Nardmann J, Ji J, Werr W, Scanlon MJ. The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development. 2004;131:2827–2839. doi: 10.1242/dev.01164. [DOI] [PubMed] [Google Scholar]
- Nardmann J, Zimmermann R, Durantini D, Kranz E, Werr W. WOX gene phylogeny in Poaceae: a comparative approach addressing leaf and embryo development. Molecular Biology and Evolution. 2007;24:2474–2484. doi: 10.1093/molbev/msm182. [DOI] [PubMed] [Google Scholar]
- Nimchuk ZL, Tarr PT, Meyerowitz EM. An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell. 2011;23:851–854. doi: 10.1105/tpc.110.075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori Y, Tanaka W, Kojima M, Sakakibara H, Hirano H-Y. WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell. 2013;25:229–241. doi: 10.1105/tpc.112.103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Bioinformatics. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- Prunet N, Morel P, Negrutiu I, Trehin C. Time to stop: flower meristem termination. Plant Physiology. 2009;150:1764–1772. doi: 10.1104/pp.109.141812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebocho AB, Bliek M, Kusters E, et al. Role of EVERGREEN in the development of the cymose petunia inflorescence. Developmental Cell. 2008;15:437–447. doi: 10.1016/j.devcel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Liu T, Newell NR, et al. Establishing a framework for the Ad/abaxial regulatory network of arabidopsis: ascertaining targets of class III homeodomain leucine zipper and KANADI regulation. Plant Cell. 2013;25:3228–3249. doi: 10.1105/tpc.113.111518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert JS. Interpreting the homeobox: metaphors of gene action and activation in development and evolution. Evolution & Development. 2001;3:287–295. doi: 10.1046/j.1525-142x.2001.003004287.x. [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Scanlon MJ. NARROW SHEATH1 functions from two meristematic foci during founder-cell recruitment in maize leaf development. Development. 2000;127:4573–4585. doi: 10.1242/dev.127.21.4573. [DOI] [PubMed] [Google Scholar]
- Scanlon MJ, Schneeberger RG, Freeling M. The maize mutant narrow sheath fails to establish leaf margin identity in a meristematic domain. Development. 1996;122:1683–1691. doi: 10.1242/dev.122.6.1683. [DOI] [PubMed] [Google Scholar]
- Scanlon MJ, Chen KD, McKnight CC., IV The narrow sheath duplicate genes: sectors of dual aneuploidy reveal ancestrally conserved gene functions during maize leaf development. Genetics. 2000;155:1379–1389. doi: 10.1093/genetics/155.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. The stem cell population of arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- Shimizu R, Ji J, Kelsey E, Ohtsu K, Schnable PS, Scanlon MJ. Tissue specificity and evolution of meristematic WOX3 function. Plant Physiology. 2009;149:841–850. doi: 10.1104/pp.108.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skylar A, Hong F, Chory J, Weigel D, Wu X. STIMPY mediates cytokinin signaling during shoot meristem establishment in arabidopsis seedlings. Development. 2010;137:541–549. doi: 10.1242/dev.041426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muiño JM, et al. Characterization of MADS-domain transcription factor complexes in arabidopsis flower development. Proceedings of the National Academy of Sciences of the USA. 2012;109:1560–1565. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S-K, Lee MM, Clark SE. POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for arabidopsis shoot and floral stem cells. Development. 2006;133:4691–4698. doi: 10.1242/dev.02652. [DOI] [PubMed] [Google Scholar]
- Souer E, van der Krol A, Kloos D, et al. Genetic control of branching pattern and floral identity during Petunia inflorescence development. Development. 1998;125:733–742. doi: 10.1242/dev.125.4.733. [DOI] [PubMed] [Google Scholar]
- Souer E, Rebocho AB, Bliek M, Kusters E, de Bruin RAM, Koes R. Patterning of inflorescences and flowers by the F-Box protein DOUBLE TOP and the LEAFY homolog ABERRANT LEAF AND FLOWER of petunia. Plant Cell. 2008;20:2033–2048. doi: 10.1105/tpc.108.060871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman J, Jäggi F, Kuhlemeier C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes & Development. 2002;16:2213–2218. doi: 10.1101/gad.230702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Xu Y, Ng K-H, Ito T. A timing mechanism for stem cell maintenance and differentiation in the arabidopsis floral meristem. Genes & Development. 2009;23:1791–1804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Lin H, Bedair M, et al. STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris. Plant Cell. 2011a;23:2125–2142. doi: 10.1105/tpc.111.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Lin H, Niu L, Mysore KS. Control of dicot leaf blade expansion by a WOX gene, STF. Plant Signaling & Behavior. 2011b;6:1861–1864. doi: 10.4161/psb.6.11.17761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Iwakawa H, Ishibashi N, et al. Meta-analyses of microarrays of arabidopsis asymmetric leaves1 (as1), as2 and their modifying mutants reveal a critical role for the ETT pathway in stabilization of adaxial-abaxial patterning and cell division during leaf development. Plant & Cell Physiology. 2013;54:418–431. doi: 10.1093/pcp/pct027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JPC, Haberer G, Ferreira FJ, et al. Type-A arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. Leaf development. The arabidopsis book. 2013:e0163. doi: 10.1199/tab.0163. doi:10.1199/tab.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M, Horstman A, Zethof J, Koes R, Rijpkema AS, Gerats T. Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and arabidopsis. Plant Cell. 2009;21:2269–2283. doi: 10.1105/tpc.109.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolberger C. Homeodomain interactions. Current Opinion in Structural Biology. 1996;6:62–68. doi: 10.1016/s0959-440x(96)80096-0. [DOI] [PubMed] [Google Scholar]
- Wu X, Dabi T, Weigel D. Requirement of homeobox gene STIMPY/WOX9 for arabidopsis meristem growth and maintenance. Current Biology. 2005;15:436–440. doi: 10.1016/j.cub.2004.12.079. [DOI] [PubMed] [Google Scholar]
- Wu X, Chory J, Weigel D. Combinations of WOX activities regulate tissue proliferation during arabidopsis embryonic development. Developmental Biology. 2007;309:306–316. doi: 10.1016/j.ydbio.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. WUSCHEL protein movement mediates stem cell homeostasis in the arabidopsis shoot apex. Genes & Development. 2011;25:2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Yu LP, Simon EJ, Trotochaud AE, Clark SE. POLTERGEIST functions to regulate meristem development downstream of the CLAVATA loci. Development. 2000;127:1661–1670. doi: 10.1242/dev.127.8.1661. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang Y, Li G, Tang Y, Kramer EM, Tadege M. STENOFOLIA recruits TOPLESS to repress ASYMMETRIC LEAVES2 at the leaf margin and promote leaf blade outgrowth in Medicago truncatula. Plant Cell. 2014;26:650–654. doi: 10.1105/tpc.113.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L-L, Ambrose M, Rameau C, et al. LATHYROIDES, encoding a WUSCHEL-related Homeobox1 transcription factor, controls organ lateral growth, and regulates tendril and dorsal petal identities in garden pea (Pisum sativum L.) Molecular Plant. 2012;5:1333–1345. doi: 10.1093/mp/sss067. [DOI] [PubMed] [Google Scholar]