Abstract
Gene silencing through sequence-specific targeting of mRNAs by RNAi has enabled genome-wide functional screens in cultured cells and in vivo in model organisms. These screens have resulted in the identification of new cellular pathways and potential drug targets. Considerable progress has been made to improve the quality of RNAi screen data through the development of new experimental and bioinformatics approaches. The recent availability of genome-editing strategies, such as the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 system, when combined with RNAi, could lead to further improvements in screen data quality and follow-up experiments, thus promoting our understanding of gene function and gene regulatory networks.
RNAi is an endogenous cellular process, first identified in Caenorhabditis elegans and conserved in most eukaryotic species, which involves targeted transcript cleavage and degradation following binding of a sequence-specific siRNA1. For more than 15 years, researchers have harnessed RNAi activity as a research tool by introducing into cells or whole organisms RNAi reagents (such as synthetic siRNAs, endoribonuclease-prepared siRNAs (esiRNAs)) or siRNA precursors (such as short hairpin RNAs (shRNAs) or long double-stranded RNAs (dsRNAs))2–6 (FIG. 1) that are designed to target endogenous mRNA transcripts. Importantly, RNAi has enabled high-throughput gene silencing (knockdown) in cells and organisms, as this had been a challenge with classical genetic approaches. At its best, RNAi screening combines the power of genetic screens with phenotypic assays — the use of which had previously been limited, at least in cultured cell lines, to small-molecule screens. RNAi screening has made it possible to identify new genes, or gene networks, that are involved in a wide variety of biological processes2,3, including assays relevant to signal transduction, cell viability, cell or organelle morphology, organelle or protein localization and/or function, drug resistance, and responses of host cells to pathogens (for reviews, see REFS 5,7–10).
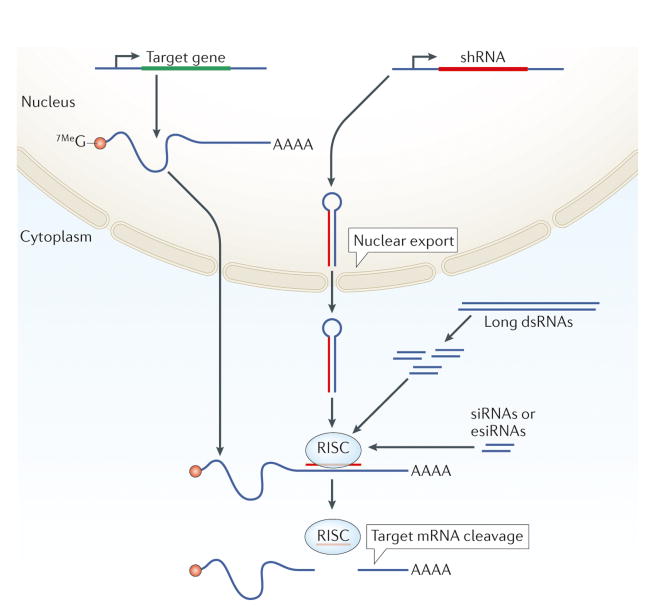
Figure 1. Gene silencing by RNAi.
RNAi reagents can be introduced into cells through different routes: siRNAs or endoribonuclease-prepared siRNAs (esiRNAs) can be transfected into mammalian (and other) cells; short hairpin RNAs (shRNAs) can be virally transduced into mammalian (and other) cells; double-stranded RNAs (dsRNAs) in solution can be applied to Drosophila melanogaster cells resulting in their uptake; dsRNAs (D. melanogaster or C. elegans) or shRNAs (D. melanogaster or mice) can be expressed from transgenic constructs; dsRNAs can be microinjected (C. elegans, D. melanogaster and some non-model insects); and Escherichia coli expressing dsRNAs can be fed to living animals (C. elegans or Planaria). Once in the cells, reagents such as dsRNAs are recognized by DICER (not shown), which processes them into siRNAs of 21–23 nucleotides in length. The synthetic or endogenously processed siRNA molecules are then incorporated into the RNA-induced silencing complex (RISC) and mediate gene silencing through target mRNA cleavage (if perfect sequence complementarity exists between the target mRNA and the siRNA) or translational interference (if the complementarity is partial; not shown).
To facilitate large-scale screens, a number of genome-wide RNAi libraries comprised of one or more types of RNAi reagents were developed by academic and commercial entities, with new libraries emerging as our understanding of the most effective strategies for the design and delivery of RNAi reagents improved (for information about available libraries and technological improvements to reagents, see REFS 4,6,7,11–14). Readers unfamiliar with RNAi screens are referred to past reviews on assay development and optimization2,3,7,15,16, high-throughput cell-based pooled format RNAi screens and arrayed format RNAi screens2,15,17, in vivo screening4,12,14,18,19, and screen data analysis2,7,20. So far, hundreds of large-scale, cell-based RNAi screens have been carried out in Drosophila melanogaster, mouse and human cells. RNAi has also been used for largescale in vivo screening in C. elegans and D. melanogaster (reviewed in REFS 2,12,14,19), as well as Planaria21–23, trypanosomes24 and mice. Furthermore, a number of databases are now available that support the browsing and analysis of results from large-scale RNAi screens (BOX 1).
Box 1. Databases for browsing and analysing RNAi screen dat.
Although no one database has been accepted as the established repository for RNAi data, several public databases have been developed as resources for sharing data from RNAi screens (see the table). RNAi data made public in this way can be used to help annotate gene function, be integrated with other large-scale data sets to investigate or provide support for new hypotheses, and provide helpful information to improve RNAi reagent design. To be most useful, RNAi data sets deposited in public repositories should include complete sequences for all RNAi reagents used, as well as detailed documentation of experimental and data analysis protocols and results.
| Database | URL | Organisms | RNAi reagents |
|---|---|---|---|
| FLIGHT | http://flight.icr.ac.uk/ | Drosophila melanogaster | Double-stranded RNA (dsRNA) |
| FlyRNAi.org | http://www.flyrnai.org/index.html | D. melanogaster | dsRNA |
| GenomeRNAi | http://genomernai.org/ | Human and D. melanogaster | dsRNA, siRNA and short hairpin RNA (shRNA) |
| PubChem BioAssay | http://www.ncbi.nlm.nih.gov/pcassay | Human, rat and D. melanogaster | siRNA, shRNA and dsRNA |
| RNAiDB | http://www.rnai.org/ | Caenorhabditis elegans | dsRNA |
| WormBase | http://www.wormbase.org | C. elegans and other nematodes | dsRNA |
The initial burst of excitement about RNAi was somewhat tempered by the finding that RNAi screens, like all screening approaches, are associated with false discovery (false-positive and false-negative results). For RNAi, the most prominent concern is false positives that are due to sequence-specific off-target effects (OTEs)20,25 (FIG. 2). The availability of RNAi data sets (BOX 1) has made a number of meta-analyses possible, including those that aim to compare on-target findings and/or OTEs between screens26–30. These studies have explored overlap among gene sets or pathways identified in related screens, which has helped to improve estimates of false discovery rates30; they have also revealed ‘frequent hitters’ — that is, genes that frequently score as positive hits across different assays, such as genes involved in ubiquitous processes that might exert relevant but relatively indirect effects, perhaps most notably genes encoding components of the ribosome or proteosome26; and they have provided new information regarding the specificity and relevance of primary screen hits27,28,31. Moreover, new experimental approaches and the use of novel genome-engineering systems to validate RNAi results are allowing better and faster identification of OTEs. Conversely, RNAi screening in mammalian cells has paved the way for innovation in related areas, including the use of microRNA mimics (miRNA mimics) and inhibitors (BOX 2), the use of RNAi or mutagenesis in three-dimensional (3D) mammalian culture systems (BOX 3) and the development of in vivo disease models in mice.
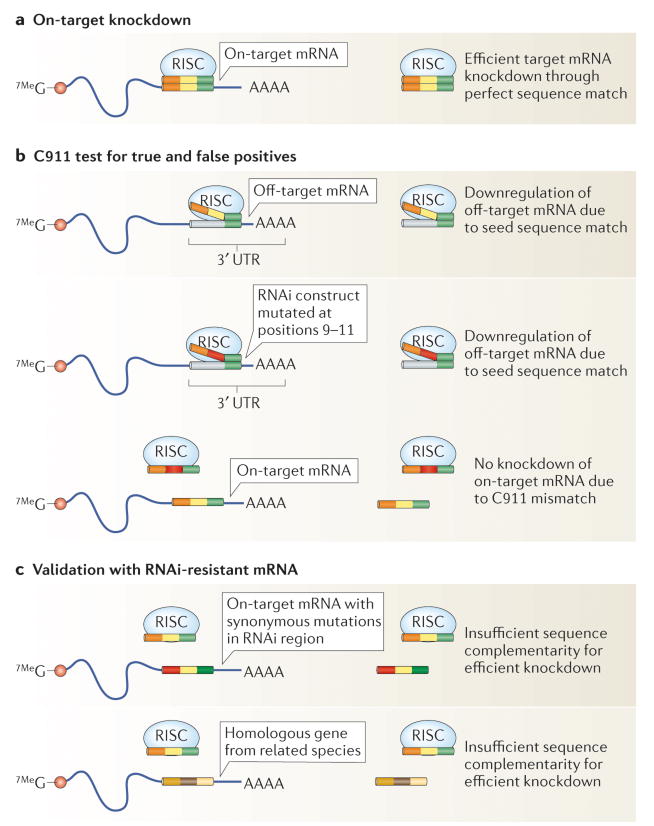
Figure 2. Strategies for validating RNAi screen results.
a | RNA-induced silencing complex (RISC)-incorporated siRNAs mediate target mRNA cleavage upon perfect sequence complementarity in either the coding region or the 3′ untranslated region (UTR) of the mRNA (depending on the siRNA design). For the siRNA shown, the 5′ seed region is in green, the middle region is in yellow and the 3′ end is in orange. b | Testing for potential off-target effects of a given siRNA can be carried out using the C911 method37. siRNA bases 9–11 are mutated while the seed region (bases 2–8) remains intact. This maintains off-target interactions mediated by seed region matches but perturbs on-target silencing. c | On-target specificity by phenotypic rescue can be demonstrated by the co-expression of RNAi-resistant versions of the target mRNA. Synonymous mutations in the siRNA-targeted region of the mRNA can be introduced to prevent RISC-mediated silencing while preserving function. Alternatively, a homologous gene from a related species that has sufficient sequence divergence in the siRNA targeting region to be RNAi resistant, but also sufficient similarity to elicit function, can be used to test on-target specificity of the RNAi construct.
Box 2. Targeting non-coding RNA.
In recent years, screening strategies that were originally developed for targeting mRNAs with short hairpin RNA (shRNA) and siRNA have been applied to non-coding RNAs, notably using libraries of reagents that inhibit or mimic microRNAs (miRNAs). Several such libraries have been developed and commercialized, enabling functional high-throughput, unbiased screens to be performed. This format has facilitated the identification of miRNAs that contribute to a variety of diseases and physiological responses, including viral infection95, breast cancer96,97 and drug treatment responses98,99. A study involving both miRNA mimics and siRNA screening using the same assay, identified miRNAs involved in cisplatin resistance, as well as the kinases targeted by these miRNAs100. Libraries of miRNA mimics are used more frequently than libraries of miRNA inhibitors, because inhibitors will only have an effect if the targeted miRNA is expressed during the assay.
As the miRNAs that are involved in a biological process are often unknown before the screen, it can be challenging to identify positive and negative screen controls. Fortunately, the use of screening strategies for miRNAs similar to those used for mRNAs has enabled investigators to use siRNA controls in miRNA screens, selecting siRNAs that are known to elicit the desired phenotype or phenotypes of the screen as positive controls until corresponding miRNA reagent controls are identified. When analysing the results from miRNA reagent screens, potential hits usually have weaker phenotypes than siRNA screen hits, probably the consequence of functional redundancy among miRNAs. Therefore, it can be helpful to determine how many mimics of miRNA belonging to the same miRNA family elicit similar phenotypes. This provides an indication of true target mRNAs, the determination of which is an essential follow-up step.
Recently, RNAi libraries targeting long non-coding RNAs (lncRNAs) have been generated101. It is still unclear how effective siRNA libraries will be in knocking down lncRNAs, particularly those localized to the nucleus, and whether this experimental strategy will further our understanding of how lncRNAs influence cellular processes102.
Box 3. RNAi screens in 3D cell culture.
Recent technological advances have enabled gene silencing in three-dimensional (3D) cell culture systems. Thus far, 3D RNAi screening has only been implemented on a small scale103, but its adaptation for use in large-scale screening is plausible. The appeal of screening in three dimensions is the ability to produce phenotypes that are more physiologically relevant than those obtained in two dimensional (2D) cell cultures, as some aspects of tissue and tumour growth are not reproduced in two dimensions. To ensure the access of RNAi reagents to all cells in a 3D culture, they are typically introduced by viral infection or transfection of homogeneous 2D cell cultures before inducing 3D structure formation. As with all RNAi screens, the most appropriate reagent for the assay depends on the question being asked. If the length of the experiment is short (for example, up to 5 days), siRNAs can be used, as was recently demonstrated in a study to identify genes influencing the ability of breast cancer cells to grow in an anchorage-independent manner104. The authors determined that the oestrogen receptor 1-positive MCF7 cells were inhibited by oestrogen receptor 1 knockdown, whereas the growth of HER2 (also known as ERBB2)-positive SK-BR-3 cells was suppressed by the knockdown of HER2. In addition, both cell lines exhibited reduced colony growth in soft agar in the presence of siRNAs targeting β-actin, a result not observed in standard 2D cultures. If the experiment requires a longer time course (greater than 5 days), stable expression of short hairpin RNA (shRNA) reagents is more appropriate in order to ensure target gene silencing throughout the course of the experiment. As with RNAi screens carried out in 2D cultures, various assay readouts are possible. High content image-based readouts are likely to preserve the most information because the different locations within a 3D structure can be analysed independently to determine how physical location and the 3D structure of the microenvironment (for example, hypoxic versus normoxic microenviroments) affect the observed phenotype.
In this Review, we discuss state of the art RNAi screening, with an emphasis on new experimental and bioinformatics approaches to data validation, screening reagents and systems. We also discuss the intersection between RNAi screening and complementary approaches such as CRISPR–Cas9-mediated genome editing (FIG. 3).
Figure 3. Genome-engineering approaches offer new opportunities for assay development, screening and validation.
A number of points of intersection exist between RNAi screening (or other types of large-scale screening, such as overexpression of open reading frame (ORF) clones, or microRNA (miRNA) mimics or inhibitors (BOX 2)) and genome-engineering technologies (step 1) such as TAL effector nucleases (TALENs) and the CRISPR (clustered regularly interspaced short palindromic repeats)– Cas9 systems (highlighted in blue). Genome engineering can be used to create robust, well-controlled assays (step 2) in cell lines and model organisms by introducing various mutations such as gene knockouts, diseaseassociated mutations and knock-in of selectable markers or in-frame fusions or reporter genes. CRISPR–Cas9-mediated knockouts (step 3) in mouse or human cells have been reported as an effective method for pooled-format screens. These can be performed in parallel to RNAi screens, followed by comparison of results from the two types of screens. Independently of the screening approach, genome engineering can be used to modify cells or organisms for follow-up studies of specific gene candidates (step 4). In this case, the CRISPR–Cas9 system or TALENs can be used to knock out genes identified in RNAi screens, with a concordance of the knockdown and knockout phenotypes providing a high degree of confidence in the results. They can also be used to create other types of modifications useful for follow-up studies, for example, transcriptional upregulation or downregulation, using modified forms of Cas9, or introducing fluorescence tags or reporters using a knock-in approach.
Strategies for improving RNAi result
Sequence-specific OTEs occur when RNAi reagents bind to RNAs other than their intended target owing to partial complementarity (FIG. 2). It is fairly straightforward, using sequence alignment, to identify the subsets of OTEs that occur due to extended regions of complementarity between RNAi reagents and genes other than the target, such as regions common to the target gene and its paralogues. As gene annotations change (for example, following the identification of new alternative splice forms or the extension of the 5′ or 3′ untranslated region (UTR)), the interpretation of what constitutes on-targets and off-targets can also change, as can other relevant predictions, such as whether a reagent might target all isoforms of a gene. Improved approaches are now available for the re-annotation of RNAi reagents (for example, see UP-TORR13 and GenomeRNAi26), thus facilitating the identification of reagents that no longer meet quality standards. However, eliminating reagents with extended complementarity from the library is not sufficient to fully address sequence-specific OTEs. To help address these concerns, new and improved approaches to identifying OTEs using bioinformatics, as well as experimental strategies for limiting OTEs, have recently been developed and successfully applied.
Addressing off-target effects with bioinformatics
miRNAs, which are encoded by endogenous genes, are short transcripts that bind mRNAs, particularly in their 3′ UTRs, and inhibit their translation32,33. This is mediated by incomplete complementarity between the miRNA and its target. Importantly, translational inhibition through incomplete complementarity is mechanistically independent from mRNA cleavage in the canonical RNAi pathway, which depends on extended sequence complementarity. Most sequencespecific OTEs are thought to occur when RNAi reagents function like miRNAs when incorporated into the RNA-induced silencing complex (RISC). In such cases, target recognition is not mediated by the binding of the full length of the siRNA to the 3′ UTRs and to other regions of the target mRNA (FIG. 2a), but is mediated by the binding of only a short seed region in the siRNA (nucleotide positions 2–8) (FIG. 2b). A recent report suggested that in three siRNA screens carried out in human cells, the majority of the primary hits could be attributed to seed region binding miRNA-like OTEs34. This highlights the importance of addressing OTEs and validating primary screen results, as discussed below. Most commercial siRNA libraries incorporate chemical modifications to the siRNA seed region to help reduce OTEs (as reviewed in REF. 35), but data analysis and experimental follow-up of screen results are essential to identify false-positive hits due to OTEs. Because seed regions are short, any particular seed sequence might be present in hundreds of transcripts, making it difficult to predict computationally potential off-targeted transcripts using seed sequence alignments, or to design RNAi reagents with seeds that might result in fewer off-targets. Software tools that can be used to identify seed-dependent OTEs in RNAi data sets include Genome-wide Enrichment of Seed Sequence matches (GESS)27 and Haystack28. GESS and Haystack use two different strategies to identify potential off-targete transcripts and both produce their best results when larger (genome-wide) data sets are used. Publicly accessible tools exist for the analysis of data sets by these methods: Online GESS36 and Haystack28.
Experimental approaches addressing off-target effects
The C911 RNAi reagent controls, which can be generated for any RNAi reagent by replacing bases 9–11 with their complement bases (hence the name)37, are experimental tools that enable specific concerns about seedbased OTEs to be addressed (FIG. 2b). A C911 control has the same siRNA seed region (bases 2–8) as the original RNAi reagent, but perfect complementarity with the intended target gene is destroyed. C911 versions of falsepositive siRNAs maintain their phenotype when assayed, whereas C911 versions of true-positive siRNAs do not37. The C911 control strategy should also be informative for shRNA experiments, as the endogenously processed shRNAs also have the potential to cause seed sequencemediated miRNA-like effects. Because C911 controls are easily designed for all RNAi reagents, for example, by using the online C911 calculator, it is feasible to test many RNAi hits using this strategy. A related strategy is to test seed region controls — RNAi reagents that have been designed with the seed sequences of the RNAi hits, but also with randomized nucleotide sequences outside of the seeds34.
The most common and straightforward experimental strategy to validate RNAi screen hits is to test multiple RNAi reagents for each gene, as different reagents will have different seed sequences. For siRNA screens, seven or more independent reagents per gene might be assayed; for pooled shRNA screens, some investigators screen libraries using more than 15 constructs per gene38,39. The greater the number of independent RNAi reagents per gene that reproduce the desired phenotype, the higher the confidence that the gene is a true hit in a screen. This is not always feasible, however, as it is not possible to design multiple independent RNAi reagents for some genes. Moreover, a potential caveat to this strategy is that potent reagents might be fairly rare, as suggested by the results of a large-scale study of shRNA effectiveness40. In the study, fluorescence protein-encoding sensors with shRNA binding sites were used to monitor knockdown effectiveness. In total, 20,000 shRNAs targeting nine transcripts were assayed using the sensors, and fewer than 2,000 remained following enrichment for target-specific shRNAs. Moreover, shRNAs conferring robust knockdown constituted less than 3% of the total tested40. If this proves true for most RNAi reagents, then even when large numbers of reagents are tested, only a small subset might score as hits.
A definitive experimental strategy for testing the specificity of an RNAi reagent is to show that the knockdown phenotype can be rescued by the expression of an RNAi-resistant version of the targeted gene (FIG. 2c). However, this has not been routinely carried out, probably because of the effort that was required until recently to design and produce such RNAi-resistant constructs, and because the interpretation of rescue experiments is complicated when rescue constructs are expressed at non-physiological levels 20. Several groups have used homologous genes from related species expressed in bacterial artificial chromosomes (BACs) or similar genomic fragments to rescue RNAi knockdown, for example, the use of a mouse homologue to rescue an siRNA phenotype in human cells41, or the use of a Drosophila pseudoobscura homologue to rescue a phenotype in D. melanogaster42,43. This approach has the advantage that the homologous proteins are more likely to be expressed at physiological levels as they remain in their genomic context. However, this strategy only works when the homologous gene can functionally replace the tested gene.
Validating RNAi screen results with genome editing
The CRISPR–Cas9 system or other genome-editing approaches could be used to engineer RNAi-resistant versions of endogenous genes by introducing synonymous changes that abolish the RNAi target sequence, providing another means to assess potential OTEs. It is also possible to use genome-engineering approaches to create complete knockout (loss-of-function) alleles as follow-ups to functional screens (FIG. 3), providing a different type of validation of observed phenotypes. At least one recent study has reported the use of engineered knockouts to validate RNAi screen results44. In the study, FAT1 was identified as a negative regulator of apoptosis in a genome-wide siRNA screen of a human glioblastoma cell line. CRISPR–Cas9-mediated knockout of FAT1 conferred sensitivity to death receptor-induced apoptosis, which was consistent with the screen result44. It is conceivable that using the CRISPR–Cas9 system or other genome-engineering approaches to knock out nonessential genes will become a routine means of verifying RNAi screen results. The successful implementation of genome-editing technologies in several species45 suggests that this will be a relevant tool for follow-up studies in many types of cell lines and model systems.
Genome-engineering approaches such as the CRISPR– Cas9 system are not without caveats. For example, careful attention must be paid to the design of genomeengineering vectors in order to maximize the chance of gene disruption and to minimize the potential for introducing DNA breaks in regions other than the target gene. A recent study demonstrated that, although some short guide RNAs (sgRNAs) can target Cas9 to thousands of ectopic sites in the genome, target cleavage only occurs at sites with extended complementarity46. On the basis of these data, the authors proposed a two-step model for Cas9 binding and cleavage. Given the gaps in our overall understanding of how the CRISPR–Cas9 system functions in eukaryotic cells, it seems likely that we do not yet fully understand all the potential experimental problems of applying this technique. In addition, even when genome engineering can be used to induce effects that are strictly gene specific, RNAi knockdown, in which mRNA levels are typically reduced but not completely eliminated, might result in a weaker, incomplete or distinct phenotype compared with a gene knockout, which results in the full elimination of function. As a result, the two phenotypes might not appear identical in some cases even when both strategies are indeed exerting on-target effects47. Nevertheless, when the RNAi and knockout phenotypes are concordant, the results will be of high confidence.
Parallel screening in multiple species
Another effective approach to validate RNAi results is to carry out related screens in different model systems, such as screening for related phenotypes using RNAi in D. melanogaster and mammalian cells (for example, see studies focused on dengue virus–host cell interactions48, actin regulators49 or androgen receptor function50), or using RNAi in D. melanogaster cells and genetic screening in yeast (for example, see studies focused on the identification of genes required for nucleolar size regulation51). High-throughput comparative analysis of phenotype conservation can identify genes and protein complexes that have been evolutionarily repurposed or that are part of more complex, redundant networks. This strategy has been used successfully to study the conservation of genetic interactions across species52,53, as well as the conservation of mechanisms that control subcellular structures or features49,51. Taking a comparative approach can also help to overcome speciesspecific limitations, such as incomplete genome coverage of screening reagents or sequence-specific OTEs. The hits that correspond to cellular processes and complexes for which gene ontology terms are consistently enriched in the data set of both species have a higher probability of being true positive hits. In addition, single genes that score as positive hits in both species can also be considered to be high confidence hits, as they have been independently confirmed by different screen reagents, methodologies and organisms.
Screening multiple phenotypes and gene
In cell-based RNAi screens, specific phenotypes can be characterized by screening for multiple features or parameters, such as by screening the same library on multiple cell lines or under different treatment conditions, and/or using assay readouts in which the phenotype monitored is comprised of multiple parameters. High-content imaging, such as standard or confocal fluorescence imaging of multiple cellular features, provides an opportunity to include hundreds of parameters in defining the phenotypes of interest, allowing the detection and quantification of cellular and subcellular changes, as well as the classification of subphenotypes that might correspond to specific biological functions. Recent multi-parametric image-based screens have contributed to our understanding of several cell functions, including homologue pairing and cell morphology in D. melanogaster cells54,55, endocytosis in human cells56, epigenetic regulators of human colon cancer cells57 and the responses of human macrophage primary cells to the pathogen Mycobacterium tuberculosis58.
Single-cell analysis approaches are at the cutting edge of high-content, image-based analysis. Individual cells within a cell population might behave differently both as a consequence of intrinsic differences (for example, in cell cycle stage) and as a consequence of their unique microenvironment (for example, differences in local cell densities within a well of a micro-well plate) at the onset of or during a screen. Thus, the phenotypic responses of individual cells might differ. Moreover, in some cases, only a subset of cells might take up the RNAi reagent, such that cells with efficient knockdown will be interspersed with wild-type cells. The use of single-cell analyses to identify phenotypic differences among cells, as well as for filtering out wild-type-appearing cells within a population, can help to address these problems. A recent analysis of individual cell image data from several related cell-based RNAi screens provided direct evidence that the cell microenvironment affects RNAi reagent uptake and response59. This approach suggests that it is feasible to differentiate phenotypes that are directly attributable to gene silencing from phenotypes that are attributable to indirect effects originating from changes in the microenvironment, such as increased cell growth that leads to a higher cell density59. We anticipate that image-based screens will become less expensive and easier to develop and apply now that genome-engineering approaches can be used to create custom cell lines with fluorescent reporters or in-frame tags at endogenous loci (FIG. 3), circumventing the need for immunostaining, which can be more costly and result in higher screen result variability.
Even when effective and on-target knockdowns are achieved and the assay is robust, knock down of a single gene might not result in a discernable phenotype, for example, due to gene or pathway redundancy. Combinatorial RNAi screens, in which two genes are silenced simultaneously (double knockdowns), can be used to identify these phenotypes and uncover functional relationships between genes. The concept has been exemplified by the results of large-scale, combinatorial genetic studies in Saccharomyces cerevisiae60,61. Combinatorial RNAi screens can also facilitate the identification of suppressive effects, in which the knock down of a gene eliminates or reduces a phenotype that is associated with the knock down of another gene, or the identification of synthetic effects, in which the knock down of two genes has a synergistic effect. These include screens querying all possible double knockdown combinations, for example, a recent screen of cell numbers and nuclear features in D. melanogaster that involved the pairwise knock down of 70,000 combinations of 93 genes involved in signal transduction, resulting in the identification of more than 600 potential interactions62.
Another approach to combinatorial screening involves using RNAi, small molecules or genetic alterations to generate a sensitized cell background, which is then used in a large-scale RNAi screen. Differences in the genes conferring lethality in various isogenic cell lines — for example, with or without the expression of a specific oncogene — were reported in two studies related to oncogenic RAS signalling63,64. Similarly, some very large-scale RNAi screens have been carried out with the aim of uncovering cancer vulnerabilities through the identification of genes that are essential in various cancer cell lines. The results of these studies show promise for the identification of new drug targets for cancer therapy65–67. In the future, genome-engineering approaches could be used to generate sets of related cell lines, differing only in a specific compromised cellular pathway or process (FIG. 3). This will allow specifically controlled parallel RNAi screens that might uncover synergistic or new effects caused by the perturbation of more than one gene.
RNAi screening in vivo
Various approaches have been developed to harness the power of large-scale RNAi screening in mammalian cells in contexts that simulate in vivo environments. The development of one such approach — 3D culture systems for screening — is discussed in BOX 3. Even more physiologically relevant are screens carried out in living animals, where the effects of gene silencing can be assessed in defined populations of cells in their proper physiological context.
RNAi screening in vivo in mice
Systematic loss-offunction screening is becoming increasingly feasible in mammalian systems. For example, in vivo shRNA screens in mice have identified bromodomain-containing protein 4 (BRD4) as a therapeutic target in acute myeloid leukaemia68 and have identified novel regulators of oncogenic growth in a HrasG12V mouse model of skin tumorigenesis69. The approaches for carrying out in vivo screens vary, including ex vivo transduction of shRNA pools into mouse cells, which are then transplanted into specific tissues and organs70,71; direct viral infection into target cell populations in adult mice72; or infection of cells during embryogenesis for tissue-specific silencing during animal development69. The expression of shRNAs in vivo can be constitutive72 or inducible73.
Achieving robust results from in vivo screening is dependent on multiple factors69, including accurate shRNA quantification within each pool, a low per-cell transduction level of shRNAs (accomplished using low multiplicity of infection (MOI)), and testing of multiple shRNA reagents per gene. In vivo RNAi screening of multiple phenotypes requires less work than creating individual gene knockout lines. The results of in vivo RNAi experiments can then be translated to producing low-throughput knockout models for validation and follow-up studies. We note that the use of RNAi to make genetically engineered mouse models was recently reviewed elsewhere74.
RNAi screening in vivo in other organisms
A number of genome-wide RNAi libraries and related resources are available for investigating a variety of topics by in vivo screening in other model organisms, in particular in C. elegans and in D. melanogaster (reviewed in REFS 12,14,18,19). RNAi screens have also been carried out in some non-model organisms, such as in trypanosomes (for example, a study on quorum sensing signalling24) and Planaria (for example, studies on regeneration and stem cells21–23, as well as on the identification of a conserved factor required for Wnt secretion75), and have been proposed for others, such as parasitic nematodes4. Results from in vivo screens in flies and worms have frequently been translated to mammalian systems, and these data sets continue to be important for hypothesis generation and single gene mechanistic studies. For example, straightjacket, a D. melanogaster orthologue of the mammalian gene Cacna2d3, was identified in a genome-wide in vivo RNAi screen for mediators of heat nociception, and it was later shown that disruption of Cacna2d3 in mice is associated with heat pain sensitivity76. Other examples come from C. elegans, in which a number of RNAi screens have addressed cell biology processes related to neurodegenerative diseases; some genes identified in these screens have been studied in mammalian cells or in knockout mice (for a review, see REF. 14).
As is the case in cell-based screens, OTEs are relevant to interpreting genome-wide in vivo RNAi screens. The development of new resources, such as fly stock collections that harbour parts of the genomes of other species43, can facilitate validation by RNAi resistance. In addition, in some cases, RNAi reagents that are known to have a high degree of on-target specificity to ‘gene traps’ might be used to bypass unwanted OTEs in other genes. In D. melanogaster, for example, GFP-trap fly stocks are available in which a GFP-tagged gene replaces the endogenous gene, and so validated RNAi reagents that target GFP can be used to knock down the GFP-fused mRNAs: this was shown to elicit highly reproducible phenotypes77,78. Broad use of this method is currently limited to D. melanogaster, the only organism so far in which a number of homozygous viable GFP-trap stocks have been produced. Even in D. melanogaster, the number of GFP-trap stocks is small; however, a resource consisting of fly stocks designed for systematic generation of GFP (or other) tagged genes was recently established79, suggesting that the approach might prove useful for larger gene sets in the future. In principle, fusion targeting might be used in any system in which endogenous genes can be tagged with GFP or any other sequence that can be effectively targeted by RNAi.
One main advantage of using RNAi in a model organism such as D. melanogaster is that gene silencing can be restricted to specific tissues and developmental stages; for example, the use of the Gal4–UAS (Gal4–upstream activating sequence) system, in which the transcription factor Gal4 specifically binds the UAS enhancer and thus drives the expression of the cloned RNAi construct in a tissue-specific and spatiotemporalspecific manner12, which circumvents the problems associated with studying genes and pathways that function in multiple tissues and developmental stages. This is in contrast to C. elegans, in which RNAi is usually systemic and tissue-specific gene silencing can be accomplished only through complex genetic manipulation80,81. Recently, the expression of CRISPR–Cas9 in a tissue-restricted manner in D. melanogaster was shown to efficiently disrupt both alleles of the targeted fly gene in somatic tissues82. Although this approach provides, in principle, an alternative to analysing by RNAi phenotypes in somatic tissues in a targeted manner, it has limitations. For example, the approach is not 100% efficient and some sgRNA targeting will destroy the target site but will not cause a frameshift or other disruptive mutations (which will result in the production of a protein with wild-type function). As a consequence, gene disruptions will be present in only a subset of the cells in which the sgRNA is expressed (that is, present in only a subset of Gal4-expressing cells). Because gene disruption is not labelled, it will be difficult to identify the subset of cells in which gene activity has been disrupted unless an antibody against the gene product exists. This is in contrast to tissue-specific Gal4–UASmediated RNAi, in which the expression of the RNAi reagent is induced in all cells that express Gal4 and thus knock down is uniform throughout the Gal4-expressing tissue. Regardless, tissue-specific CRISPR–Cas9 activity can be used for large-scale screens and may complement RNAi approaches in D. melanogaster. Using CRISPR– Cas9 to enable mosaic analyses may be more beneficial in organisms such as C. elegans, where RNAi is systemic.
Data analysis and integratio
In addition to carrying out statistical analyses and experimental follow-up studies of RNAi screen data, the application of various bioinformatics approaches can greatly aid in distinguishing between high-confidence and low-confidence screen hits. One common approach is to analyse the data in aggregate using a gene-set enrichment or related algorithms, such as using the DAVID database (the database for annotation, visualization and integrated discovery) from the US National Institutes of Health83, COMPLEAT84 and other software to detect gene ontology terms, protein complexes or signalling pathways that are enriched in the screen hits compared to controls. These results can be used to confirm lowconfidence hits, such as hits with borderline statistical scores or hits for which not all RNAi reagents targeting the gene were positive and, conversely, to rule out hits that are the sole representatives of a category, which might suggest that they are false positives. Pathway enrichment can also help to address false-negative hits, as genes that were not represented among the screen hits but that were members of the selected gene ontology category, protein complex or pathway could be added to the list of genes to be included in follow-up studies.
Another powerful bioinformatics approach is to integrate results from RNAi screens with the results of ‘omics’ studies based on other methods, such as proteome or transcriptome analyses, which have different strengths and caveats and thus can complement RNAi. Genes or proteins identified on the basis of multiple lines of evidence can be assigned to higher confidence categories. For example, a combined proteomic and RNAi approach was used to functionally annotate putative protein complexes related to Hippo signalling85. The study identified at least one new component of the Hippo pathway, an α-arrestin protein family member, Leash, which is involved in the degradation of Yorkie, the fly orthologue of YAP1 (REF. 85). Importantly, whereas approaches such as gene ontology or pathway analysis are likely to bias the results towards what is already known from the literature, the integration of results from additional omics data sets or other functional screening approaches might help to uncover truly novel findings. It is therefore critically important to make complete and annotated screen data available to others in order to facilitate improved reagent and assay design, re-analysis of results, and integration of screen results with other studies86. When presenting RNAi screen hit lists, it is the responsibility of each investigator to make their level of confidence in their screen hits explicit by referencing the specific experiments and the statistical approaches that led to their conclusions.
New methods of mammalian cell screenin
An alternative strategy to RNAi screening in mammalian cultured cells has been to carry out transposonbased genetic screens in haploid mammalian cell lines. For example, a derivative of the KBM7 chronic myeloid leukaemia (CML) near-haploid mammalian cell line, together with gene-trap retroviruses that contain a strong splice acceptor site and a marker gene, could be used to identify genes that are involved in specific biological processes, such as survival in response to TRAIL (tumour necrosis factor-related apoptosisinducing ligand; also known as TNFSF10) and exposure to pathogens87,88. These reports showed that systematic loss-of-function screens in cultured cells, which until recently were thought to be feasible only in yeast61, can be applied to mammalian cells. Similarly, a gene-trap retrovirus approach is being used to generate large-scale knockout collections of human cells, with more than 3,396 genes tagged to date89.
With the demonstration that CRISPR–Cas9-based methods allow the efficient recovery of biallelic mutants in diploid cells90, large-scale knockout screens are no longer limited to haploid cell lines. Indeed, genomeengineering approaches45 not only offer new routes to assay development and validation of RNAi results (FIG. 3), but can also be used for high-throughput screening47. Recently, two publications reported using genomewide CRISPR–Cas9-based knockout libraries to carry out pooled-format screens in human cells. In one study, a screen of more than 70,000 unique sgRNAs (targeting ∼7,000 genes, with ten sgRNAs per gene) was performed in the presence of the nucleotide analogue 6-thioguanine, and the four mismatch repair pathway components expected to score as positive in the assay were indeed identified, with four or more sgRNAs per gene scoring as positive hits91. In another study, researchers identified genes essential for the survival of cancer cells and pluripotent stem cells, as well as for resistance to the BRAF inhibitor vemurafenib92. A mouse largescale pooled CRISPR–Cas9 knockout library was used in screens to determine the sensitivity of mouse cells to Clostridium septicum α-toxin or 6-thioguanine, resulting in the identification of four previously unknown gene candidates for sensitivity to these treatments93.
In each of the three CRISPR–Cas9 screen studies discussed here, several unique sgRNAs targeting the same gene produced comparable phenotypes, suggesting that the CRISPR–Cas9 system has an efficient recovery of on-target hits91–93. It is therefore plausible that CRISPR–Cas9-based screens will become an important complement to RNAi screens in the future, perhaps replacing a subset of pooled shRNA screens. As mentioned above, however, we do not yet fully understand how to design effective and on-target CRISPR–Cas9 reagents, and there can be biologically meaningful differences between gene knockdown and gene knockout. Presumably, some gene functions revealed by incomplete, RNAi-based gene disruption phenotypes would be missed in gene knockout screens47. As is the case for RNAi, there is a need to carefully follow-up on results from CRISPR-Cas9-based screens, including carrying out rescue assays to confirm that the phenotypes observed result only from on-target genomic knockouts.
Concluding remark
The availability of genome-wide RNAi screening platforms in several model organisms allows for systematic interrogation of gene function. Although many caveats apply to the design, analysis and interpretation of high-throughput RNAi studies, RNAi screens are clearly having an impact, perhaps most notably in the fields of cancer research, host–pathogen interactions and signal transduction. Recent developments in experimental controls and data analysis strategies to detect OTEs and to confirm on-target effects have provided increased confidence in the results obtained from large-scale RNAi screens. Multi-pronged approaches, such as performing related omics experiments in parallel with RNAi screening, or performing complementary screens in other systems, can also be used to generate high-confidence results and to promote network-level analysis.
Genome-engineering technologies intersect with RNAi in a number of ways, including in assay development, screening procedures and hit validation. However, these approaches also come with their own set of caveats, including the potential to introduce offtarget DNA breaks or chromosomal rearrangements that might be difficult to detect. The ability to use various TAL effector nucleases (TALENs) or Cas9 modifications as transcriptional repressors rather than as inducers of DNA breaks45,94, should help to reduce such unwanted effects. Importantly, there are no reports to date of using CRISPR–Cas9 knockouts in arrayed screening formats, so at least in the near future RNAi is likely to remain the method of choice for studies that require high-content image-based (and other) arrayed-format screening assays.
It is generally accepted that high-throughput RNAi screen data sets are insufficient for high-confidence annotation of gene function (for example, see REF. 30). However, with careful attention to reagent and assay design, data analysis, data integration and follow-up experimental validation, large-scale RNAi screens can be successful at uncovering new genes, signalling pathways and gene networks involved in various biological processes, and will continue to be a valuable experimental tool in many research areas for years to come.
Genetic screen.
Identification of the organisms or cells that display a mutant phenotype of interest following large-scale disruption of genes; for example, by using mutagens.
Pooled format RNAi screen.
Screens in which pools of reagents are introduced together into the cell population; for mammalian cell screening this is typically carried out with the goal of integrating just one of the short hairpin RNAs into each cell. Positive reagents are identified by comparing the abundance of any given reagent in the cell population before and after selection.
Arrayed format RNAi screen.
Screens in which each reagent (or a small pool of reagents directed against the same gene) is introduced separately into cells.
miRNA mimic
Short, double-stranded RNAs that, when introduced into cells, mimic endogenous microRNAs by activating post-transcriptional repression of target genes.
Seed regio
Base pairs 2–8 of the fully processed siRNA. It can mediate microRNA-like effects Even in the absence of perfect complementarity between the remainder of its sequence and that of the transcript.
Multiplicity of infectio.
(MOI). During a viral infection, the ratio of the number of infectious virions to the number of targeted cells.
Gene-trap retrovirus approac.
Screening using mutagenic retroviral integration into genes. The integrated retrovirus can be used as a trap for identifying the genes disrupted in each resulting phenotype.
Acknowledgments
The Drosophila RNAi Screening Center is supported by US National Institutes of Health (NIH) NIGMS R01 GM067761 (N.P.). S.E.M. receives additional support from the Dana Farber/Harvard Cancer Center, which is supported in part by NCI Cancer Center Support Grant NIH 5 P30 CA06516. C.E.S. and J.A.S. are supported by funds from Harvard Medical School. R.A.N. is supported by a Human Frontier Science Program long-term fellowship. N.P is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests statement: The authors declare no competing interests.
References
- 1.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nature Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 2.Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nature Rev Genetics. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 3.Grimm S. The art and design of genetic screens: mammalian culture cells. Nature reviews Genetics. 2004;5:179–189. doi: 10.1038/nrg1291. [DOI] [PubMed] [Google Scholar]
- 4.Zhuang JJ, Hunter CP. RNA interference in Caenorhabditis elegans: uptake, mechanism, and regulation. Parasitology. 2012;139:560–573. doi: 10.1017/S0031182011001788. [DOI] [PubMed] [Google Scholar]
- 5.Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nature Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- 6.Surendranath V, Theis M, Habermann BH, Buchholz F. Designing efficient and specific endoribonuclease-prepared siRNAs. Methods Mol Biol. 2013;942:193–204. doi: 10.1007/978-1-62703-119-6_11. [DOI] [PubMed] [Google Scholar]
- 7.Mohr S, Bakal C, Perrimon N. Genomic screening with RNAi: results and challenges. Annu Rev Biochem. 2010;79:37–64. doi: 10.1146/annurev-biochem-060408-092949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panda D, Cherry S. Cell-based genomic screening: elucidating virus-host interactions. Curr Opin Virol. 2012;2:784–792. doi: 10.1016/j.coviro.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherry S. What have RNAi screens taught us about viral-host interactions? Curr Opin Microbiol. 2009;12:446–452. doi: 10.1016/j.mib.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherry S. Genomic RNAi screening in Drosophila S2 cells: what have we learned about host-pathogen interactions? Curr Opin Microbiol. 2008;11:262–270. doi: 10.1016/j.mib.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nature Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrimon N, Ni JQ, Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harb Perspect Biol. 2010;2:a003640. doi: 10.1101/cshperspect.a003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, et al. UP-TORR: online tool for accurate and Up-to-Date annotation of RNAi Reagents. Genetics. 2013;195:37–45. doi: 10.1534/genetics.113.151340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sin O, Michels H, Nollen EA. Genetic screens in Caenorhabditis elegans models for neurodegenerative diseases. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbadis.2014.01.015. http://dx.doi.org/10.1016/j.bbadis.2014.01.015. [DOI] [PubMed]
- 15.Birmingham A, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nature Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 16.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nature Methods. 2009;6:569–575. doi: 10.1038/nmeth.1351. A review on approaches to assay optimization and data analysis relevant to mammalian siRNA and other arrayed RNAi screens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nature Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 18.Mohr SE, Perrimon N. RNAi screening: new approaches, understandings, and organisms. Wiley Interdiscip Rev RNA. 2012;3:145–158. doi: 10.1002/wrna.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanos ME, Bennett CF, Kaeberlein M. Genomewide RNAi longevity screens in Caenorhabditis elegans. Curr Genom. 2012;13:508–518. doi: 10.2174/138920212803251391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigoillot FD, King RW. Vigilance and validation: keys to success in RNAi screening. ACS Chem Biol. 2011;6:47–60. doi: 10.1021/cb100358f. This review article discusses many key issues related to RNAi screening in mammalian cells, with a special emphasis on off-target effects (including miRNA-like effects) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner DE, Ho JJ, Reddien PW. Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell. 2012;10:299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forsthoefel DJ, et al. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev Cell. 2012;23:691–704. doi: 10.1016/j.devcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rink JC, Vu HT, Sanchez Alvarado A. The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development. 2011;138:3769–3780. doi: 10.1242/dev.066852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mony BM, et al. Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei. Nature. 2014;505:681–685. doi: 10.1038/nature12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Echeverri CJ, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nature Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt EE, et al. GenomeRNAi: a database for cellbased and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res. 2013;41:D1021–D1026. doi: 10.1093/nar/gks1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigoillot FD, et al. A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nature Methods. 2012;9:363–366. doi: 10.1038/nmeth.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buehler E, et al. siRNA off-target effects in genomewide screens identify signaling pathway members. Scientif Rep. 2012;2:428. doi: 10.1038/srep00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhinder B, Djaballah H. Systematic analysis of RNAi reports identifies dismal commonality at genelevel & reveals an unprecedented enrichment in pooled shRNA screens. Comb Chem High Throughput Screen. 2013;16:665–681. doi: 10.2174/13862073113169990045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bushman FD, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaelin WG., Jr Molecular biology. Use and abuse of RNAi to study mammalian gene function. Science. 2012;337:421–422. doi: 10.1126/science.1225787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 34.Franceschini A, et al. Specific inhibition of diverse pathogens in human cells by synthetic microRNA-like oligonucleotides inferred from RNAi screens. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1402353111. http:dx.doi.org/10.1073/pnas.1402353111. [DOI] [PMC free article] [PubMed]
- 35.Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 36.Yilmazel B, et al. Online GESS: prediction of miRNAlike off-target effects in large-scale RNAi screen data by seed region analysis. BMC Bioinformat. 2014;15:192. doi: 10.1186/1471-2105-15-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buehler E, Chen YC, Martin S. C911: A bench-level control for sequence specific siRNA off-target effects. PloS ONE. 2012;7:e51942. doi: 10.1371/journal.pone.0051942. This paper describes the C911 mismatch control method to distinguish between true- and false-positive hits of a specific siRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassik MC, et al. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nature Methods. 2009;6:443–445. doi: 10.1038/nmeth.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman GR, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci USA. 2014;111:3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fellmann C, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Mol Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. This paper describes the development of a sensor assay to identify highly potent shRNAs that are predominantly missed by existing shRNA design algorithms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kittler R, et al. RNA interference rescue by bacterial artificial chromosome transgenesis in mammalian tissue culture cells. Proc Natl Acad Sci USA. 2005;102:2396–2401. doi: 10.1073/pnas.0409861102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo S, Booker M, Perrimon N. Cross-species RNAi rescue platform in Drosophila melanogaster. Genetics. 2009;183:1165–1173. doi: 10.1534/genetics.109.106567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langer CC, Ejsmont RK, Schonbauer C, Schnorrer F, Tomancak P. In vivo RNAi rescue in Drosophila melanogaster with genomic transgenes from Drosophila pseudoobscura. PloS ONE. 2010;5:e8928. doi: 10.1371/journal.pone.0008928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kranz D, Boutros M. A synthetic lethal screen identifies FAT1 as an antagonist of caspase-8 in extrinsic apoptosis. EMBO J. 2014;33:181–197. doi: 10.1002/embj.201385686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu X, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nature Biotech. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heintze J, Luft C, Ketteler R. A CRISPR CASe for high-throughput silencing. Frontiers Genet. 2013;4:193. doi: 10.3389/fgene.2013.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sessions OM, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohn JL, et al. Comparative RNAi screening identifies a conserved core metazoan actinome by phenotype. J Cell Biol. 2011;194:789–805. doi: 10.1083/jcb.201103168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imberg-Kazdan K, et al. A genome-wide RNA interference screen identifies new regulators of androgen receptor function in prostate cancer cells. Genome Res. 2013;23:581–591. doi: 10.1101/gr.144774.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neumuller RA, et al. Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci Signal. 2013;6:ra70. doi: 10.1126/scisignal.2004145. This report presents results obtained using related image-based nucleolar size assays, performed using genome-wide RNAi screening in D. melanogaster cells and genetic screening in yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dixon SJ, Andrews BJ, Boone C. Exploring the conservation of synthetic lethal genetic interaction networks. Commun Integr Biol. 2009;2:78–81. doi: 10.4161/cib.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frost A, et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell. 2012;149:1339–1352. doi: 10.1016/j.cell.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joyce EF, Williams BR, Xie T, Wu CT. Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet. 2012;8:e1002667. doi: 10.1371/journal.pgen.1002667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin Z, et al. A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nature Cell Biol. 2013;15:860–871. doi: 10.1038/ncb2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collinet C, et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- 57.Laufer C, Fischer B, Billmann M, Huber W, Boutros M. Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nature Methods. 2013;10:427–431. doi: 10.1038/nmeth.2436. [DOI] [PubMed] [Google Scholar]
- 58.Sundaramurthy V, et al. Integration of chemical and RNAi multiparametric profiles identifies triggers of intracellular mycobacterial killing. Cell Host Microbe. 2013;13:129–142. doi: 10.1016/j.chom.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 59.Snijder B, et al. Single-cell analysis of population context advances RNAi screening at multiple levels. Mol Systems Biol. 2012;8:579. doi: 10.1038/msb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Avery L, Wasserman S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baryshnikova A, et al. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol. 2010;470:145–179. doi: 10.1016/S0076-6879(10)70007-0. [DOI] [PubMed] [Google Scholar]
- 62.Horn T, et al. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nature Methods. 2011;8:341–346. doi: 10.1038/nmeth.1581. This paper describes a large-scale combinatorial RNAi screen in D. melanogaster cells by analysing synthetic genetic interactions. [DOI] [PubMed] [Google Scholar]
- 63.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steckel M, et al. Determination of synthetic lethal interactions in KRAS oncogene-dependent cancer cells reveals novel therapeutic targeting strategies. Cell Res. 2012;22:1227–1245. doi: 10.1038/cr.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci USA. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koh JL, et al. COLT-Cancer: functional genetic screening resource for essential genes in human cancer cell lines. Nucleic Acids Res. 2012;40:D957–D963. doi: 10.1093/nar/gkr959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. This paper describes an in vivo RNAi screen of known chromatin regulators in mice to find new therapeutic targets in acute myeloid leukaemia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beronja S, et al. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501:185–190. doi: 10.1038/nature12464. This paper describes an elegant, genome-wide in vivo RNAi screen for oncogenic growth in mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller PG, et al. In Vivo RNAi screening identifies a leukemia-specific dependence on integrin β 3 signaling. Cancer Cell. 2013;24:45–58. doi: 10.1016/j.ccr.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wuestefeld T, et al. A Direct in vivo RNAi screen identifies MKK4 as a key regulator of liver regeneration. Cell. 2013;153:389–401. doi: 10.1016/j.cell.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 73.Zuber J, et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated selfrenewal program as essential for leukemia maintenance. Genes Dev. 2011;25:1628–1640. doi: 10.1101/gad.17269211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Livshits G, Lowe SW. Accelerating cancer modeling with RNAi and nongermline genetically engineered mouse models. Cold Spring Harbor Protoc. 2013 doi: 10.1101/pdb.top069856. http://dx.doi.org/10.1101/pdb.top069856. [DOI] [PMC free article] [PubMed]
- 75.Bartscherer K, Pelte N, Ingelfinger D, Boutros M. Secretion of Wnt ligands requires Evi, a conserved transmembrane protein. Cell. 2006;125:523–533. doi: 10.1016/j.cell.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 76.Neely GG, et al. A genome-wide Drosophila screen for heat nociception identifies α2δ3 as an evolutionarily conserved pain gene. Cell. 2010;143:628–638. doi: 10.1016/j.cell.2010.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neumuller RA, et al. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics. 2012;190:931–940. doi: 10.1534/genetics.111.136465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pastor-Pareja JC, Xu T. Shaping cells and organs in Drosophila by opposing roles of fat body-secreted Collagen IV and perlecan. Dev Cell. 2011;21:245–256. doi: 10.1016/j.devcel.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Venken KJ, et al. MiMIC: a highly versatile transposon insertion resource for engineerin Drosophila melanogaster genes. Nature Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Firnhaber C, Hammarlund M. Neuron-specific feeding RNAi in C. elegans and its use in a screen for essential genes required for GABA neuron function. PLoS Genet. 2013;9:e1003921. doi: 10.1371/journal.pgen.1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qadota H, et al. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 2007;400:166–173. doi: 10.1016/j.gene.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1405500111. http://dx.doi.org/10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed]
- 83.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 84.Vinayagam A, et al. Protein complex-based analysis framework for high-throughput data sets. Sci Signal. 2013;6:rs5. doi: 10.1126/scisignal.2003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kwon Y, et al. The Hippo signaling pathway interactome. Science. 2013;342:737–740. doi: 10.1126/science.1243971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shamu CE, Wiemann S, Boutros M. On target: a public repository for large-scale RNAi experiments. Nature Cell Biol. 2012;14:115. doi: 10.1038/ncb2435. [DOI] [PubMed] [Google Scholar]
- 87.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 88.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burckstummer T, et al. A reversible gene trap collection empowers haploid genetics in human cells. Nature Methods. 2013;10:965–971. doi: 10.1038/nmeth.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. This report provides examples of large-scale CRISPR–Cas9-induced gene knockout screening in mammalian cells and includes a comparison of knockout versus shRNA results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPRguide RNA library. Nature Biotech. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 94.Terns RM, Terns MP. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet. 2014;30:111–118. doi: 10.1016/j.tig.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Santhakumar D, et al. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc Natl Acad Sci USA. 2010;107:13830–13835. doi: 10.1073/pnas.1008861107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Keklikoglou I, et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene. 2012;31:4150–4163. doi: 10.1038/onc.2011.571. [DOI] [PubMed] [Google Scholar]
- 97.Leivonen SK, et al. High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Oncol. 2014;8:93–104. doi: 10.1016/j.molonc.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Du L, et al. A high-throughput screen identifies miRNA inhibitors regulating lung cancer cell survival and response to paclitaxel. RNA Biol. 2013;10:1700–1713. doi: 10.4161/rna.26541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neijenhuis S, Bajrami I, Miller R, Lord CJ, Ashworth A. Identification of miRNA modulators to PARP inhibitor response. DNA Repair. 2013;12:394–402. doi: 10.1016/j.dnarep.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 100.Otto EA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nature Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mirkin EV, Mirkin SM. To switch or not to switch: at the origin of repeat expansion disease. Mol Cell. 2014;53:1–3. doi: 10.1016/j.molcel.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thoma CR, et al. A high-throughput-compatible 3D microtissue co-culture system for phenotypic RNAi screening applications. J Biomolecular Screen. 2013;18:1330–1337. doi: 10.1177/1087057113499071. [DOI] [PubMed] [Google Scholar]
- 104.Marhefka JN, Abbud-Antaki RA. Validation of the Cancer BioChip System as a 3D siRNA screening tool for breast cancer targets. PloS ONE. 2012;7:e46086. doi: 10.1371/journal.pone.0046086. [DOI] [PMC free article] [PubMed] [Google Scholar]