Abstract
The American College of Rheumatology position statement on ANA testing stipulates the use of IIF as the gold standard method for ANA screening1. Although IIF is an excellent screening test in expert hands, the technical difficulties of processing and reading IIF slides – such as the labor intensive slide processing, manual reading, the need for experienced, trained technologists and the use of dark room – make the IIF method difficult to fit in the workflow of modern, automated laboratories.
The first and crucial step towards high quality ANA screening is careful slide processing. This procedure is labor intensive, and requires full understanding of the process, as well as attention to details and experience.
Slide reading is performed by fluorescent microscopy in dark rooms, and is done by trained technologists who are familiar with the various patterns, in the context of cell cycle and the morphology of interphase and dividing cells. Provided that IIF is the first line screening tool for SARD, understanding the steps to correctly perform this technique is critical.
Recently, digital imaging systems have been developed for the automated reading of IIF slides. These systems, such as the NOVA View Automated Fluorescent Microscope, are designed to streamline the routine IIF workflow. NOVA View acquires and stores high resolution digital images of the wells, thereby separating image acquisition from interpretation; images are viewed an interpreted on high resolution computer monitors. It stores images for future reference and supports the operator’s interpretation by providing fluorescent light intensity data on the images. It also preliminarily categorizes results as positive or negative, and provides pattern recognition for positive samples. In summary, it eliminates the need for darkroom, and automates and streamlines the IIF reading/interpretation workflow. Most importantly, it increases consistency between readers and readings. Moreover, with the use of barcoded slides, transcription errors are eliminated by providing sample traceability and positive patient identification. This results in increased patient data integrity and safety.
The overall goal of this video is to demonstrate the IIF procedure, including slide processing, identification of common IIF patterns, and the introduction of new advancements to simplify and harmonize this technique.
Keywords: Bioengineering, Issue 88, Antinuclear antibody (ANA), HEp-2, indirect immunofluorescence (IIF), systemic autoimmune rheumatic disease (SARD), dense fine speckled (DFS70)
Introduction
The term antinuclear antibody (ANA) describes a variety of autoantibodies that react with constituents of cell nuclei including DNA, proteins and ribonucleoproteins1,2. The HEp-2 cell, a native protein array with hundreds of antigens, provides an ideal substrate for the detection of ANA1. The detection of ANA in human serum is an important screening tool for connective tissue diseases, and IIF is the reference method for ANA testing1. Recently, IIF on HEp-2 cells has been replaced in some laboratories with antigen-specific immunoassays and multiplex methods. Due to concerns over false negative results and the lack of transparency to clinicians, the American College of Rheumatology formed a Task Force that concluded that IIF using HEp-2 cells should be the “gold standard” for ANA screening1.
Due to the subjective nature of ANA screening, the quality of HEp-2 cells is integral to accurate and confident reporting of results. The presence of a high number of mitotic cells, optimal cell morphology, sufficient confluency, and expression of relevant antigens are particularly important. IIF pattern recognition serves as an important tool to aid in patient diagnosis. Understanding the significance of various patterns enables the clinicians and laboratory personnel to perform the appropriate follow-up testing to confirm diagnosis. For example, homogeneous ANA pattern can occur in the presence of anti-dsDNA or chromatin antibodies, and may be associated with systemic lupus erythematosus (SLE)3. From the other side, it has recently been described that the so called dense fine speckled pattern (DFS) that is commonly seen in up to 12% of routine samples, has mostly been associated with anti-DFS70 antibodies. These autoantibodies, when found in isolation (without other disease-specific ANA) are not associated with systemic autoimmune rheumatic diseases4-9. Thereby confirmatory testing for anti-DFS70 antibodies can help reduce unnecessary reflex testing, offer considerable cost savings, and ease patient anxiety.
Given that IIF is the first line screening methodology to detect autoantibodies, it is paramount that the user understands how the selection of reagents and tissue substrates can affect results. Since the IIF technique is inherently subjective, it is important that the reagents used provide the highest quality results.
This goal of this video protocol is to familiarize the user with the correct steps needed to perform the IIF method, the common patterns associated with ANA, and to introduce new advancements which can streamline the laboratory workflow and standardize results.
Protocol
1. Sample Preparation and Substrate Selection
Remove reagents from packaging and allow each item to come to room temperature. Prepare reagents and dilute patient sera according to the direction insert. Note: NOVA Lite slides are barcoded and can be easily integrated into automated systems in conjunction with the Laboratory Information System LIS. This procedure illustrates manual slide processing; high throughput laboratories, however, may opt for automated slide processing equipment.
2. Addition of Controls and Samples
Dispense 1 drop of positive control and 1 drop of negative control to the appropriate slide wells. Pipette 20-25 μl of diluted patient serum to the remaining wells. Process one slide at a time.
Place the slide(s) in a staining container with a damp paper towel on the bottom. Cover the container and incubate the slide(s) for 30 min. Note: The humid conditions will prevent the substrate from drying out. Drying of the wells could result in artifactual staining. During this incubation period, the anti-nuclear antibodies in the patient’s serum will bind to antigens of the cells that are fixed onto each well.
After the incubation period, rinse off the serum using a gentle stream of wash buffer. In order to avoid damaging the substrate and prevent cross-over of samples between wells, angle the slide slightly to avoid directing the stream directly onto the wells. Tap off excess wash buffer and place the slide into a Coplin jar containing wash buffer for approximately 5 min.
3. Addition of Fluorescent Conjugate
One by one, remove the slides from the wash buffer and gently tap to remove the excess wash buffer. Apply 1 drop of fluorescent conjugate (FITC labeled anti-human IgG) onto each well. Incubate the slides for 30 min in the humidified container, and be sure to replace the staining container cover. Note: For ANA testing, the use of an IgG Fc specific conjugate is recommended. The conjugate is light sensitive, and the cover will protect the slides from exposure to light in addition to maintaining the humidified environment. During this incubation period, the conjugate will bind to the patient’s anti-nuclear antibodies that have bound to the cell antigens. This conjugate binding results in the presence of fluorescence in the wells.
Rinse and wash the slide as described above.
Place a coverslip on a paper towel and apply mounting medium in a continuous line to the bottom edge of the coverslip.
Remove each slide from the wash buffer and tap the slide gently to remove the excess wash buffer. Touch the lower edge of the slide to the edge of the coverslip.
Gently lower the slide onto the coverslip in such a way that the mounting medium on the coverslip flows to the top edge of the slide without air bubble formation. Note: Coverslipping, including using the optimal amount of mounting medium, is a technique that takes practice to perfect.
4. Identification of Positive and Negative Results
Manual Interpretation
View slides with a fluorescent microscope, located in a dark room. Perform scanning of the entire well with a 20X or 25X objective to assess cell distribution and uniformity of fluorescence
Switch to a 40X objective to make the final interpretation regarding positivity and pattern.
Look at the positive and negative controls. Grade positivity using a reactivity grading scale from 1+ to 4+. Note: The negative control may not appear completely dark, but will often display low level nonspecific fluorescence. The positive control will display bright apple green fluorescence in the nucleus (Figure 1).
Automated Interpretation
Note: In addition to manual interpretation, load and scan slides by NOVA View Automated Fluorescence Microscope or other automated digital imaging instrument; no darkroom is necessary. After creating a project by selecting the appropriate slide type, the instrument acquires and stores high resolution digital images of cells in each well. Additionally, NOVA View measures fluorescent light intensity, and categorizes results as positive or negative, and provides pattern recognition for positive samples. Images are viewed by the operator on a high resolution computer monitor, allowing for final interpretation, revision and confirmation of NOVA View result. Reports can be generated on confirmed results. Digital microscopy should be used in conjunction with an experienced laboratory technician as it is intended to aid in interpretation of results.
Representative Results
When evaluating cell staining results for HEp-2, five major nuclear patterns are most commonly reported: homogenous, speckled, centromere, nucleolar, and nuclear dots. These patterns are the result of autoantibody binding to specific constituents of the nucleus. Although ANA testing is specific to nuclear structures, cytoplasmic patterns associated with autoantibodies against cytoplasmic structures may also be observed. In some cases, several autoantibodies may be present in a sample, and the image may appear as a mixed pattern.
There are other, less frequently observed, and often cell cycle specific patterns that can be detected by the IIF method; most recently, a so called dense fine speckled pattern with special clinical significance has been described in some ANA positive sera.
Homogeneous Pattern
To identify a homogeneous pattern, scan the well and identify mitotic or dividing cells. The condensed chromatin of the mitotic cells (Figure 2, orange arrow) exhibits solid, uniform fluorescence which is often more pronounced than in the resting cell nuclei.In the homogeneous pattern, the resting cells exhibit uniform, diffuse fluorescence of the entire nucleus (Figure 2, white arrow). This characteristic pattern is often the result of anti-dsDNA antibodies10.
Speckled Pattern
In the coarse speckled pattern, the mitotic cells show no staining of the condensed chromosomal regions (Figure 3, orange arrow). The resting cells exhibit granular fluorescence throughout the entire nucleus. The nuclear speckling can be defined as coarse or fine. The Coarse speckling pattern is often the result of anti-Sm and anti-RNP11.
In the fine speckled pattern, the resting cells show fine or diffuse speckling throughout the nuclei, typically in uniform distribution (Figure 4, white arrow). The fine speckled pattern is often the result of anti-SSAand anti-SSB12.
Dense Fine Speckled Pattern
It is now recognized that the dense fine speckled pattern (DFS) is common in routine testing, and as much as 12% of samples are positive for anti-DFS70 autoantibodies. However, these autoantibodies, when found in isolation, are not associated with systemic autoimmune rheumatic diseases4-6. These antibodies are prevalent in healthy individuals and patients bearing diseases unrelated to SARD6. Confirmatory testing for anti-DFS70 antibodies can help reduce unnecessary reflex testing, offer considerable cost savings, and ease patient anxiety.
Since the DFS pattern is difficult to identify and ANA positivity can cause anxiety to both patients and physicians, it is highly recommended to perform a confirmatory test after identifying a pattern suggestive of anti-DFS70 antibodies to clarify the clinical significance of ANA positivty5,6.
In this pattern, mitotic cells (Figure 5, orange arrow) show positive speckled staining of the metaphase chromatin, while resting cell nuclei (Figure 5, white arrow) exhibit uniformly distributed fine speckles throughout the nucleus.
Centromere Pattern
To identify the centromere pattern, scan the wells and identify mitotic or dividing cells. In mitotic cells (Figure 6, orange arrow), these discrete speckles become closely associated in what is often described as a “metaphase bar”. In the centromere pattern, resting cells (Figure 6, white arrow) show approximately 40-60 discrete speckles distributed throughout the nuclei. The centromere pattern is the result of anti-CENP A,B,C antibodies13.
Nucleolar Pattern
Some nucleolar patterns are associated with diffuse cytoplasmic staining in mitotic cells (Figure 7, orange arrow) and negative chromosomal region, while other nucleolear patterns display positive staining of the chromosomal region. The nucleolar pattern is associated with homogenous or speckled staining of the nucleoli (Figure 7, white arrow), along with weak speckled or homogenous staining of the nucleoplasm. These patterns are associated with anti-RNA polymerase III, anti-fibrillarin, anti-Th/To and anti-PM/Scl antibodies14,15.
Nuclear Dot Pattern
The nuclear dot pattern is associated with negative metaphase mitotic cells (Figure 8, orange arrow)and with few discrete speckles in the resting cell nuclei (Figure 8, white arrow). This characteristic pattern is often the result of sp-100, PML, or p80 colin autoantibodies. These antibodies are associated with primary biliary cirrhosis and autoimmune hepatitis16.
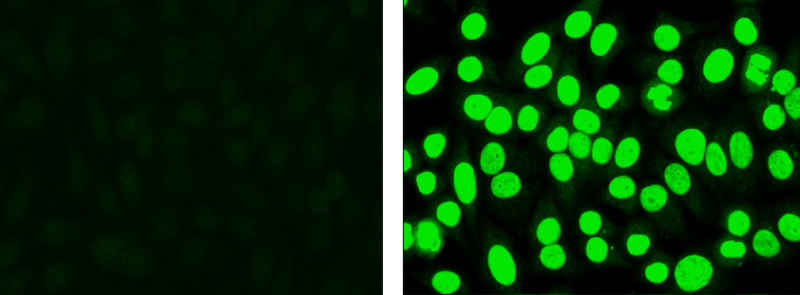 Figure 1. Negative control (left): Cells display low level non-specific fluorescence, but no specific nuclear staining. Positive control (right): Cells display apple green nuclear fluorescence.
Figure 1. Negative control (left): Cells display low level non-specific fluorescence, but no specific nuclear staining. Positive control (right): Cells display apple green nuclear fluorescence.
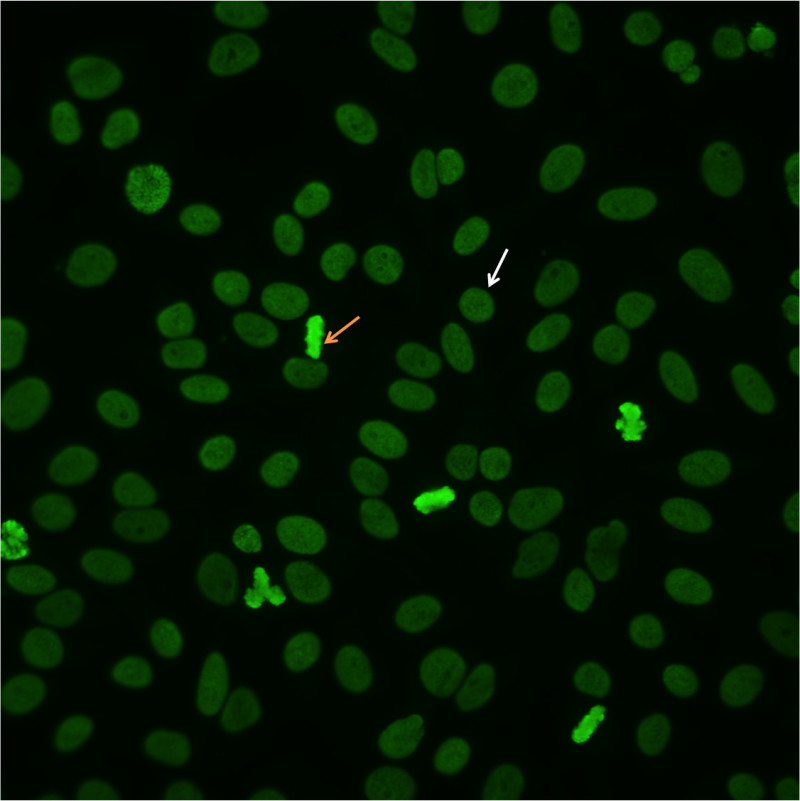 Figure 2.Homogenous pattern. Mitotic cells (orangearrow) show solid fluorescence. Resting cells show even, diffuse staining (whitearrow).
Figure 2.Homogenous pattern. Mitotic cells (orangearrow) show solid fluorescence. Resting cells show even, diffuse staining (whitearrow).
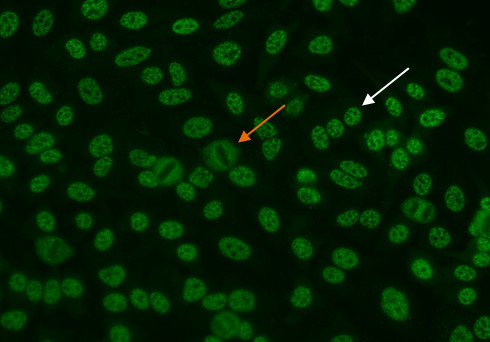 Figure 3.Coarse speckled pattern. Mitotic cells (orange arrow) show negative staining. Resting cells show a distinct speckled stain (white arrow).
Figure 3.Coarse speckled pattern. Mitotic cells (orange arrow) show negative staining. Resting cells show a distinct speckled stain (white arrow).
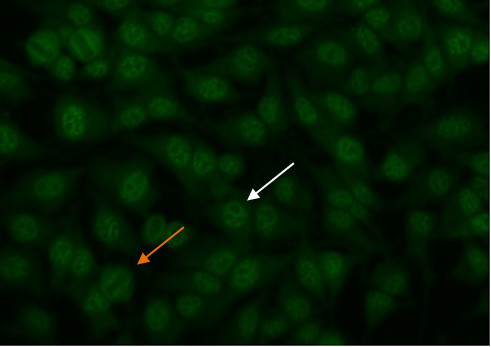 Figure 4.Fine speckled pattern. Mitotic cells (orange arrow) show negative staining. Resting cells show a distinct fine speckled staining (white arrow).
Figure 4.Fine speckled pattern. Mitotic cells (orange arrow) show negative staining. Resting cells show a distinct fine speckled staining (white arrow).
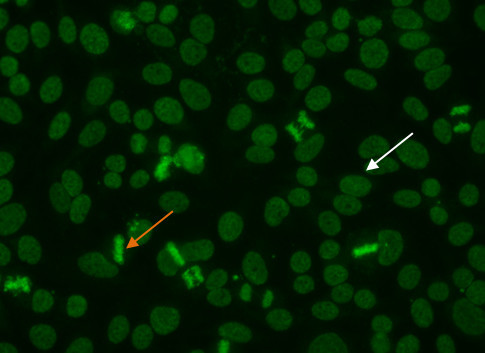 Figure 5.Dense fine speckled pattern. Mitotic cells (orange arrow) show a fine granular solid staining. Resting cells show a very fine, diffuse speckled stain (whitearrow).
Figure 5.Dense fine speckled pattern. Mitotic cells (orange arrow) show a fine granular solid staining. Resting cells show a very fine, diffuse speckled stain (whitearrow).
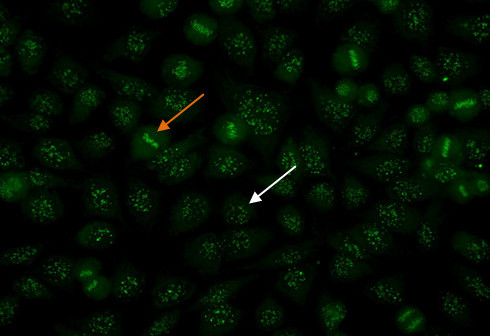 Figure 6. Centromere pattern. Mitotic cells (orangearrow) show uniform, discrete speckles. Resting cells (whitearrow) shows 40-60 discrete speckles per cell nucleus.
Figure 6. Centromere pattern. Mitotic cells (orangearrow) show uniform, discrete speckles. Resting cells (whitearrow) shows 40-60 discrete speckles per cell nucleus.
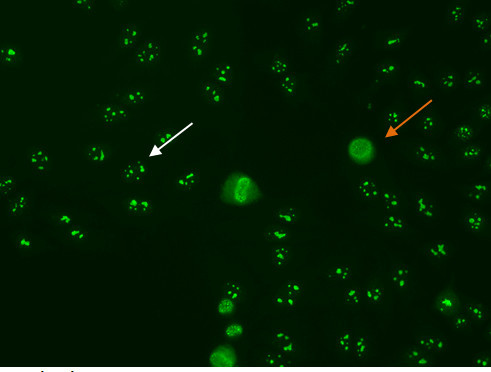 Figure 7. Nucleolar pattern. Mitotic cells (orangearrow) appear as large clusters of granular staining. Resting cell nucleishows fluorescence in the nucleoli(whitearrow).
Figure 7. Nucleolar pattern. Mitotic cells (orangearrow) appear as large clusters of granular staining. Resting cell nucleishows fluorescence in the nucleoli(whitearrow).
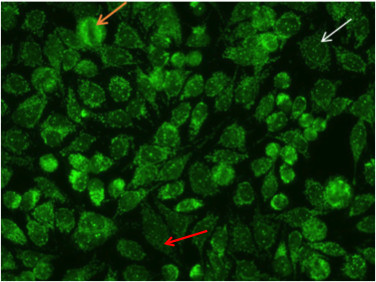 Figure 8. Nuclear dot pattern. Mitotic cells (orangearrow) appear negative. Resting cells show a few discrete speckles in the nucleus (whitearrow).Additionally, the nucelar dot pattern can coexist with cytoplasmic staining associated with autoantibodies to mitochondrial antigens (red arrow).
Figure 8. Nuclear dot pattern. Mitotic cells (orangearrow) appear negative. Resting cells show a few discrete speckles in the nucleus (whitearrow).Additionally, the nucelar dot pattern can coexist with cytoplasmic staining associated with autoantibodies to mitochondrial antigens (red arrow).
Discussion
Screening by HEp-2 cells is a critical first step in the diagnosis of patients with SARD. However, IIF methods lack harmonization. Sources of variability include preparation of slides, conjugate specificity, and efficiency of the fluorescent microscope and experience of the reader. Despite these concerns, IIF remains the “gold standard” for ANA testing. HEp-2 cells contain a large variety of autoantigens, and provide the ideal substrate for the detection of these autoantibodies. In some laboratories, IIF has been replaced with solid phase screening methods such as multiplex assay or ELISA. The shortcoming of such tests is that they do not display the full range of antigens to be sufficiently sensitive. As a consequence, true positive patients may be missed which can have deleterious consequences In addition to the issues of treatment delay and wrong diagnosis, additional healthcare costs may occur due to the repetition of confirmatory tests or unnecessary diagnostic investigations. Given the inherent challenges to IIF, it is paramount to perform this technique properly to avoid subjectivity in results.
To ensure accurate interpretation and reporting of results for ANA screening, it is vital to use the highest quality substrate. When selecting a HEp-2 substrate, it is critical that the cells be optimized to express clinically relevant epitopes in their native protein state. Transfected or otherwise modified HEp-2 cell lines may not allow proper antibody-antigen binding. In addition, when several different autoantibodies are present simultaneously, recombinant cells may mask one or more patterns. High number of mitotic cells should also be present in the substrate. Adequate number of mitotic cells allows quick and accurate identification of patterns.
In addition to the attributes of the substrate, the objective of this video protocol is to describe the IIF technique by showing critical steps such as the addition of sample, slide washing, addition of conjugate, cover slipping, and determination of positive and negative results. Results can be compromised if proper technique is not used for each of these steps. Proper washing is important to remove all unbound antibody. Some patients display very high amounts of autoantibodies and in these cases it is important to wash the serum from the wells such that it does not contaminate other wells.
Although IIF has traditionally been a very labor intensive and subjective laboratory method, new technologies such as slide processors, barcoded slides and automated digital microscopy can greatly simplify the workflow, increase the consistency and reduce the sources of variability of interpretation.
Disclosures
The authors Gabriella Lakos, Cassandra Bryant, Carol Buchner, and Anna Eslami are employees of INOVA Diagnostics.
Acknowledgments
We thank Cassandra Bryant for performing the IIF experiment and Carol Buchner for her expert technical review.
References
- Meroni PL, Schur PH. ANA screening: an old test with new recommendations. Ann Rheum Dis. 2010;69(8):1420–1422. doi: 10.1136/ard.2009.127100. [DOI] [PubMed] [Google Scholar]
- Tan EM. Autoantibodies to nuclear antigens (ANA): Their immunobiology and medicine. Advances in immunology. 1982;33:167–240. doi: 10.1016/s0065-2776(08)60836-6. [DOI] [PubMed] [Google Scholar]
- Sack U, et al. Autoantibody detection using indirect immunofluorescence on HEp-2 cells. Ann NY Acad Sci. 2009;1173:166–173. doi: 10.1111/j.1749-6632.2009.04735.x. [DOI] [PubMed] [Google Scholar]
- Watanabe A, et al. Anti-DFS70 antibodies in 597 healthy hospital workers. Arthritis Rheum. 2004;50(3):892–900. doi: 10.1002/art.20096. [DOI] [PubMed] [Google Scholar]
- Mahler M, et al. Importance of the dense fine speckled pattern on HEp-2 cells and anti-DFS70 antibodies for the diagnosis of systemic autoimmune diseases. Autoimmun Revi. 2012;11(9):642–645. doi: 10.1016/j.autrev.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Mahler M, Fritzler MJ. The clinical significance of the dense fine speckled immunofluorescence pattern on HEp-2 cells for the diagnosis of systemic autoimmune diseases. Clin Dev Immunol. 2012. [DOI] [PMC free article] [PubMed]
- Miyara M, et al. Clinical phenotypes of patients with anti-DFS70/LEDGF antibodies in a routine ANA referral cohort. Clin Dev Immunol. 2013. [DOI] [PMC free article] [PubMed]
- Mariz H, et al. Pattern on the antinuclear antibody-HEp-2 test is a critical parameter for discriminating antinuclear antibody positive healthy individuals and patients with autoimmune rheumatic diseases. Arthritis Rheum. 2011;63(1):191–200. doi: 10.1002/art.30084. [DOI] [PubMed] [Google Scholar]
- Muro Y, et al. High concomitance of disease marker autoantibodies in anti-DFS70/LEDGF autoantibody-positive patients with autoimmune rheumatic disease. Lupus. 2008;17(3):171–176. doi: 10.1177/0961203307086311. [DOI] [PubMed] [Google Scholar]
- Agmon-Levin N, et al. International recommendations for the assessment of autoantibodies to cellular antigens referred to as anti-nuclear antibodies. Ann Rheum Dis. 2014;73(1):17–23. doi: 10.1136/annrheumdis-2013-203863. [DOI] [PubMed] [Google Scholar]
- Craft J, et al. The U2 small nuclear ribonucleoprotein particle as an autoantigen. J Clin Invest. 1998;81(6):1716–1724. doi: 10.1172/JCI113511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonsson S, et al. A serologic marker for fetal risk of congenital heart block. Arthritis Rheum. 2002;46(5):1233–1241. doi: 10.1002/art.10232. [DOI] [PubMed] [Google Scholar]
- Miyawaki S, et al. Clinical and serological heterogeneity in patients with anticentromere antibodies. J Rheumatology. 2005;32(8):1488–1494. [PubMed] [Google Scholar]
- Raijamkers R, et al. PM-Scl-75 is the main autoantigen in patients with the polymyositis/scleroderma overlap syndrome. Arthritis Rheum. 2004;50(2):565–569. doi: 10.1002/art.20056. [DOI] [PubMed] [Google Scholar]
- Yang JM, et al. Human scleroderma sera contain autoantibodies to protein components specific to the UC small nucleolar RNP complex. Arthritis Rheum. 2003;48(1):210–217. doi: 10.1002/art.10729. [DOI] [PubMed] [Google Scholar]
- Granito A, et al. Antinuclear antibodies as ancillary markers in primary biliary cirrhosis. Expert Rev Mol Diagn. 2012;12(1):65–74. doi: 10.1586/erm.11.82. [DOI] [PubMed] [Google Scholar]


