Abstract
Background
The expression of the rodent phosphoinositide-specific phospholipase C δ-4 (PLCδ4) has been found to be elevated upon mitogenic stimulation and expression analysis have linked the upregulation of PLCδ4 expression with rapid proliferation in certain rat transformed cell lines. The human homologue of PLCδ4 has not been extensively characterized. Accordingly, we investigate the effects of overexpression of human PLCδ4 on cell signaling and proliferation in this study.
Results
The cDNA for human PLCδ4 has been isolated and expressed ectopically in breast cancer MCF-7 cells. Overexpression of PLCδ4 selectively activates protein kinase C-φ and upregulates the expression of epidermal growth factor receptors EGFR/erbB1 and HER2/erbB2, leading to constitutive activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) pathway in MCF-7 cells. MCF-7 cells stably expressing PLCδ4 demonstrates several phenotypes of transformation, such as rapid proliferation in low serum, formation of colonies in soft agar, and capacity to form densely packed spheroids in low-attachment plates. The growth signaling responses induced by PLCδ4 are not reversible by siRNA.
Conclusion
Overexpression or dysregulated expression of PLCδ4 may initiate oncogenesis in certain tissues through upregulation of ErbB expression and activation of ERK pathway. Since the growth responses induced by PLCδ4 are not reversible, PLCδ4 itself is not a suitable drug target, but enzymes in pathways activated by PLCδ4 are potential therapeutic targets for oncogenic intervention.
Background
Phosphoinositide-specific phospholipase C (PI-PLC) plays a role in the inositol phospholipid signaling by hydrolyzing phosphatidylinositol-4,5-bisphosphate (PIP2). This reaction produces two intracellular second messengers, inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG), which cause the increase of intracellular calcium concentration and the activation of protein kinase C (PKC), respectively [1-3]. In addition to hydrolyzing PIP2, PI-PLC can also utilize phosphatidylinositol (PI) or PI-4-phosphate as substrates.
The PLC family in murine or human species is comprised of 11 subtypes. On the basis of their structure, they have been divided into four classes, β (β1, 2, 3 and 4), γ (γ1 and 2), δ (δ1, 3 and 4), ε, and ζ types. Positive regulation mechanisms of PLC by association with membrane receptors are well characterized in β- and γ-type isozymes [2]. β-type isozymes are activated by the Gα or Gβγ subunit released from heterotrimeric Gαβγ proteins after ligand stimulation. γ-type isozymes are activated by the phosphorylation of specific tyrosine residues through the activation of receptor or nonreceptor tyrosine kinases. ε-type enzymes possess both PLC and ras dependent guanine nucleotide exchange (RasGEF) activity. As such, PLCε may mediate the effects of G protein-coupled receptors through two divergent pathways involving PI hydrolysis as well as direct activation of the Ras/MAP kinase pathway [4-6]. PLCζ, a sperm protein that shows similarity to a truncated PLCδ with the pleckstrin homology (PH) domain deleted, is involved in the triggering of Ca++ oscillations in eggs [7]; however, it remains to be documented whether PLCζ does have PLC enzymatic activity.
The regulation of δ-type PLC activity is less understood; however, certain isoforms of δ-type PLC such as δ1 can be regulated through interaction with a dual function G-protein (Gh) that also has transglutaminase activity [8,9]. Another δ-type PLC isozyme known as PLCδ4 has been implicated to have a key role in cell proliferation, as its mRNA is expressed in higher levels in regenerating liver than in normal resting liver and in tumor cells such as hepatoma and src-transformed cells than in non-transformed cells. Western blot analysis and immunohistochemical staining showed that the murine PLCδ4 is predominantly present in nuclei with its expression level markedly induced by serum in serum-starved murine cells, whereas the amounts of PLCβ1, PLCγ1, and PLCδ1 do not change significantly after serum stimulation [10]. The rat PLCδ4 level has also been found to be markedly elevated in a fast proliferating hepatoma H3924A cell line comparing to a slow growing hepatoma H7795 line; however, immunohistochemical staining and western analysis of subcellular fractions show rat PLCδ4 is mainly expressed in the cytoplasmic fraction [11]. These results suggest that PLCδ4 is expressed in response to mitogenic stimulation and plays important roles in cell growth and tumorigenesis. Splice variants of rat PLCδ4 with enzymatic PLC activities or with dominant-negative activity [12,13], and the promoter region of murine PLCδ4 [14] have also been described. Gene knockout by homologous recombination shows PLCδ4-/- sperms are not able to initiate the acrosome reaction required for egg fertilization [15].
Despite the extensive characterization of the murine or rat PLCδ4 enzyme, the effect of overexpression of the human form of PLCδ4 in cells has not been characterized. This paper reports the molecular cloning of human PLCδ4 and examines the signaling pathways that are affected by ectopic expression of PLCδ4 in human breast cancer MCF-7 cells.
Results
Human PLCδ4 expression in normal and tumor tissues
Based on sequence homology with the rat PLCδ4, we have assembled a full-length cDNA for human PLCδ4. More recent search of the Genbank database showed the coding region of our human PLCδ4 cDNA matches 100% with an annotated protein product from the Mammalian Gene Collection (GenBank# BC006355), suggesting no PCR errors were introduced during the assembly of our full length clone. Alignment of the PLCδ4 cDNA sequence with human genome http://genome.ucsc.edu/cgi-bin/hgGateway shows PLCδ4 gene to be located at chromosome 2q35 and contains 16 exons that span about 30 kbp. In addition to the full length PLCδ4 clone, a splice variant of PLCδ4, PLCδ4b, with exons 7, 8, and 9 deleted was isolated, resulting in the introduction of a premature stop codon after amino acid residue 274. Scanning of protein motifs [16] within the human PLCδ4 coding sequence identified a pleckstrin homology (PH) domain, EF-hand domains, PI-PLC-X and PI-PLC-Y catalytic domains, and a C2 domain that are commonly found in PLCδ proteins, whereas PLCδ4b contains only the coding regions for the PH and EF-hand domains (Figure 1). Northern blot analysis of normal tissues using human PLCδ4 cDNA as a probe shows PLCδ4 is most highly expressed in skeletal muscle and kidney tissues, and at moderate level in intestinal tissue (Figure 1, lower left panel). To assess the relative abundance of full length PLCδ4 and splice variant PLCδ4b, primers located upstream and downstream of the coding region for exons 7, 8, and 9 were used to detect PCR products generated from cDNA samples derived from various normal human tissues. For all tissues where full length PLCδ4 (800 bp product) can be detected, the splice variant PLCδ4b (300 bp product) can also be detected readily (Figure 1, lower right panel). To test whether PLCδ4 may be differentially expressed in certain normal vs tumor cells, we examined the expression of PLCδ4 using Cancer Profiling Arrays (BD Clontech, Palo Alto, CA) that contain hundreds of genetically matched cDNA pairs isolated from tumor and normal tissue samples (Figure 2). Consistent with data from Northern analysis, normal tissues from kidneys show the highest expression of PLCδ4, and the moderate expression of PLCδ4 in normal intestinal tissues can also be extended to stomach, colon, and rectal tissues from the rest of the alimentary tract. PLCδ4 expression is found to be downregulated in a high percentage of these tumor tissues as well as in prostate, vulva, thyroid, skin, and pancreatic tissues, whereas PLCδ4 expression is found to be upregulated in a high percentage (>25%) of breast and testicular tumor tissues and in a low percentage (<5%) of lung and uterine tumor tissues, when compared to the corresponding normal tissues.
Figure 1.
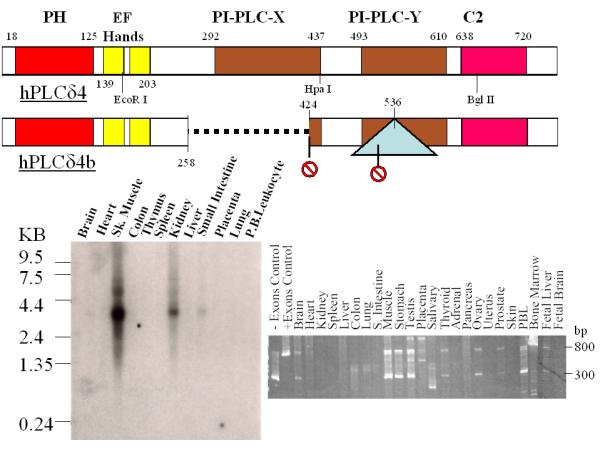
Linear map of hPLCδ4 isoforms and expression in normal tissues. The top panel depicts linear map of the full length and an alternatively spliced form of hPLCδ4 proteins. The dotted line refers to exons deleted, the triangle refers to intron inserted, and the stop signs refer to premature stop codons introduced as a result of alternative splicing. The lower panels show Northern blot (left panel) and PCR analysis (right panel) of PLCδ4 expression in various normal tissues.
Figure 2.
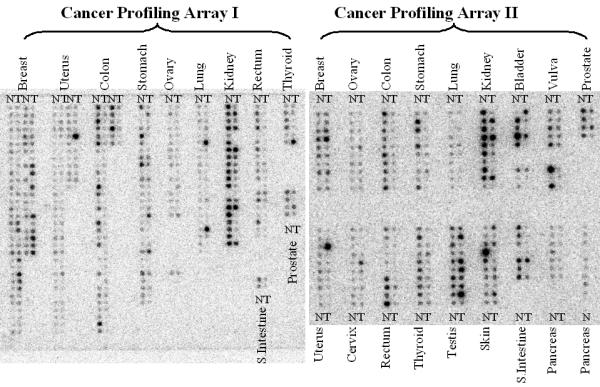
Cancer Profiling Array analysis of PLCδ4 expression using genetically matched cDNA pairs derived from tumor and normal tissue samples. Nylon membranes containing cDNA samples derived from various matched tumor and normal tissues on Cancer Profiling Array I and II (Clontech, Palo Alto, CA) were hybridized in ULTRAhyb™ (Ambion, Austin, TX) with a 2000 bp PLCδ4 coding fragment labeled with [α-32P]dCTP using a Strip-Ez labeling kit (Ambion, Austin, TX). Detailed description of each sample in Cancer Profiling Array I (left panel) and Cancer Profiling Array II (right panel) can be found at http://www.bdbiosciences.com/clontech/products/families/Disease_Profiling/index.shtml.
Generation of human PLCδ4 overexpressing MCF-7 cells
Since profiling of tissue arrays showed PLCδ4 to be overexpressed in breast tumor tissues (Figure 2), PLCδ4 was ectopically expressed in a breast cancer cell line of low tumorigenicity, MCF-7, to study whether PLCδ4 overexpression would affect any transformation phenotypes of the cells. When using PIP2 (Figure 3, panel A) as the substrate, PLCδ4 overexpressing clones in MCF-7 cell extracts show about 3 to 5 fold increase in PLC activity when compared to cells (~25 nmol/min/mg) transfected with a control vector expressing EGFP (Clonetech, Palo Alto, CA); whereas with PI as the substrate, PLCδ4a overexpressing clones (Figure 3, panel B) show about 20 to 50 fold increase in PLC activity when compared to untransfected MCF-7 cells (~6 nmol/min/mg) or cells transfected with a control vector. These results show the human PLCδ4 cDNA isolated does encode an active enzyme with PLC activity. The more substantial increase in activity when using PI as opposed to using PIP2 as the substrate may be due to the preference of human PLCδ4 for PI over PIP2 and/or the preference of the endogenous PLC isoforms present in MCF-7 for PIP2 over PI. Similar to other PLCs, PLCδ4 can therefore use both PI and PIP2 as substrates, although the preference for PIP2 to PI may vary from one PLC isoforms to another [17].
Figure 3.
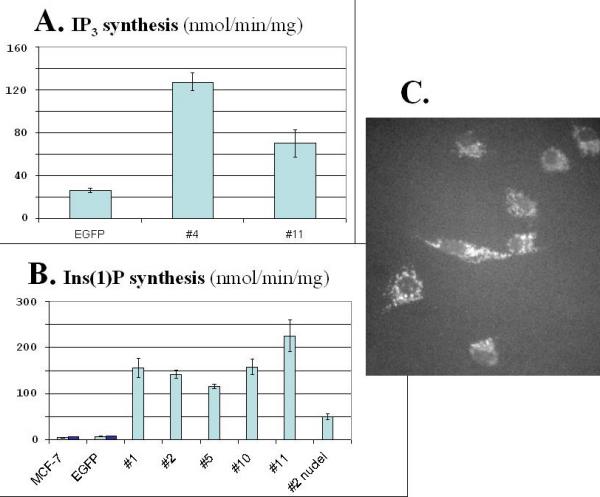
PLC assays in PLCδ4 overexpressing cells and expression of GFP-PH domain/PLCδ4 fusion protein in MCF-7 cells. Assay of enzymatic activities of PLCδ4 overexpressing clones in MCF-7 cell extracts were carried out using PIP2 (panel A) or PI (panel B) as the substrate. EGFP refers to cells transfected with a control vector expressing EGFP (Clonetech, Palo Alto, CA); MCF-7 refers to untransfected cells; #1, 2, 4, 5, 10, and 11 refer to activities derived from total cell extracts of independent isolates of PLCδ4 overexpressing clones. #2 nuclei refers to activities derived from nuclear extracts of PLCδ4 overexpressing clone #2. Assays were done in triplicates with error bars indicating ± standard deviation (SD). Fusion protein comprised of the PH domain of PLCδ4 and GFP was localized mainly in the cytoplasmic region (panel C).
Since the murine PLCδ4 has been found to be expressed predominantly in nuclei [10], nuclear fraction from a PLCδ4 overexpressing clone (Sample#2, Figure 3, panel B) was prepared to examine the distribution of PLC activity in nuclear vs whole cell extracts. The nuclear extract was found to contain about 1/3 of the total PLC activity when compared to whole cell extract, suggesting that, unlike murine PLCδ4 [10] but similar to rat PLCδ4 [11], human PLCδ4 is mainly found in cytoplasmic fractions. The structure of the PH domain is the major factor that determines the subcellular localization of PLC [18,19]. Consistent with cytoplasmic location based on cell fractionation data, fusion protein with green fluorescent protein (GFP) linked to the PH domain of PLCδ4 overexpressed in cells are associated with cytoplasmic membrane structures (Figure 3, panel C). GFP fusion constructs for PH domain of PLCβ1 have also been found in cytoplasmic membranes [19], whereas that of PLCδ1 have been found mainly in plasma membranes [18]. Including conservative amino acid changes, the matches between the PH domains of PLCδ1 and PLCδ4 exceed 80%, suggesting subtle changes in the PH domain can affect protein localization.
PLCδ4 selectively activates protein kinase C-φ in MCF-7 cells
Since DAG is a key lipid second messenger generated from hydrolysis of PI or PIP2 by PLC and protein kinase C (PKC) is the major family of proteins that are activated by DAG, we examined whether PLCδ4 overexpression would lead to activation of PKCs in MCF-7 cells. Analysis of PKC activation (Figure 4, left panel) using a phospho-PKC (pan) antibody that can detect PKCα, βI, βII, δ, ε, or η isoforms when a serine near the carboxy-terminus is autophosphorylated showed some or all of these PKC isoforms were already activated in cells transfected with control vector, pCivirPu, and no enhancement of overall phosphorylation was detected in cells overexpressing PLCδ4. Similarly, no enhancement of PKC phosphorylation was observed using an antibody specific for the detection of the subset of PKCα or PKCβII when thr638 or thr641 was phosphorylated respectively. On the contrary, PKCφ was activated in all four independent isolated lines of PLCδ4 overexpressing cells, whereas no phosphorylation was detected in vector control cells using an antibody specific for the detection of the PKCφ when thr538 was phosphorylated. No consistent change in phosphorylation trend was seen between PLCδ4 overexpressing cells and control cells when using an antibody specific for the detection of the subset of PKCζ or PKCλ when thr410 or thr403 was phosphorylated respectively, suggesting that the two outliers, one in the PLCδ4 overexpressing samples and one in the vector samples, may represent clonal variations. Hence PKCφ is the isoform of PKC that shows significant accentuation of activation in PLCδ4 overexpressing MCF-7 cells. Another method to assess PKC activation is to follow the phosphorylation of serine in PKC substrates using an antibody specific for the consensus motif of PKC substrates that detects phosphorylated serineresidues when surroundedby Arg or Lys at the -2 and +2 positions anda hydrophobic residue at the +1 position in proteins. Samples from MCF-7 transfected with vector controls (Figure 4, right panel) showed two major phosphorylated bands, whereas PLCδ4 overexpressing cells showed a few additional weak phosphorylated bands, consistent with the data that only a limited subset of PKCs are activated with PLCδ4 overexpression.
Figure 4.
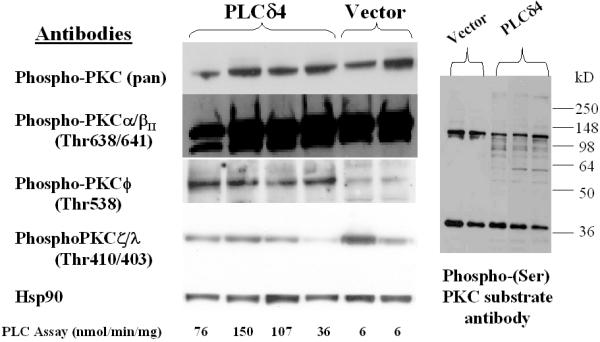
Overexpression of PLCδ4 selectively activates PKC-φ in MCF-7 cells. Western blot analysis of MCF-7 cell extracts derived from independent isolates overexpressing PLCδ4 and isolates transfected with control vectors using antibodies against various isoforms of phospho-PKC (left panel) and against the phosphorylated motif of PKC substrates (right panel). Antibodies were from Cell Signaling Technology (Beverly, MA). Blotting was done using the ECL Plus™ detection reagents from Amersham Biosciences (Piscataway, NJ).
Constitutive activation of extracellular signal-regulated kinase (ERK) pathway in PLCδ4 overexpressing cells
To determine whether PLCδ4 overexpression and/or PKCφ activation would affect signal transdutions pathways essential for cell proliferation in MCF-7 cells, phospho-specific antibodies against various key kinases were used to assess the activation of multiple pathways. Analysis of Erk1/2 or p44/42 MAPK activation (Figure 5) using an antibody specific for dually phosphorylated Erk1/2 at Thr202 and Tyr204 showed Erk1/2 was activated in all four independent isolated lines of PLCδ4 overexpressing cells, whereas little phosphorylation was detected in vector control cells. The increase in activated Erk1/2 in PLCδ4 overexpressing cells is due to modifications in protein phosphorylation rather than to change in Erk1/2 proteins level, as similar levels of expression were found using an antibody for the detection of total levels of endogenous Erk1/2 proteins in all samples. Similarly, kinases upstream of the Erk signaling pathway such as MAPK or Erk kinases (MEK1/2) as well as downstream of the Erk pathway such as such as the 90 kDa ribosomal S6 kinase (p90RSK) were also activated in PLCδ4 overexpressing cells based on western blot analysis with antibodies specific for MEK1/2 when phosphorylated at Ser217/221 and for p90RSK when phosphorylated at Thr359/Ser363. On the contrary, key proteins, such as stress activated protein kinase (SAPK) or Jun-N-terminal kinase (JNK), the 38 kDa mitogen activated kinase (p38 MAPK), and Akt or protein kinase B, involved in two alternative mitogen activated protein kinases (MAPK) pathways and the phosphoinositide-3-phosphate kinase (PI3K) pathway, respectively, showed no change in activation in PLCδ4 overexpressing cells based on western blot analysis with antibodies specific for p46 and 54 SAPK when dually phosphorylated at Thr183/Tyr185, for p38 MAPK when dually phosphorylated at Thr180/Tyr182, and for Akt when phosphorylated at Ser473. Hence PLCδ4 overexpression selectively activates the Erk or the p44/42 MAPK pathway constitutively with minimal effects on the SAPK/JNK, the p38 MAPK, and the PI3K pathways.
Figure 5.
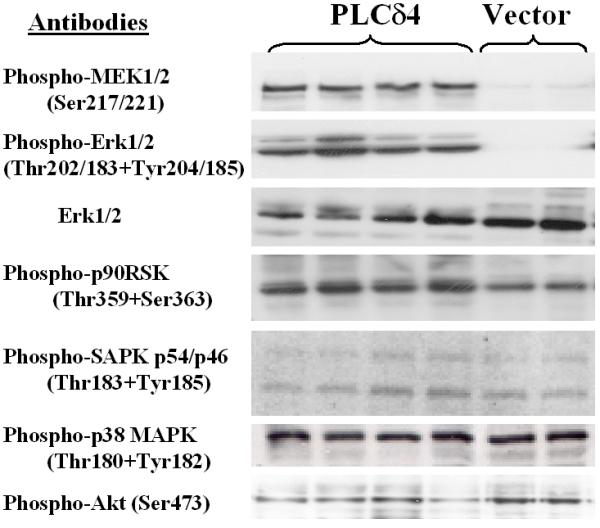
Overexpression of PLCδ4 activates Erk1/2 pathway in MCF-7 cells. Western blot analysis of MCF-7 cell extracts derived from 4 independent isolates overexpressing PLCδ4 (left 4 lanes) and 2 isolates transfected with control vectors (right 2 lanes) using antibodies against various phosphorylated forms of kinases and non-phosphorylated Erk1/2. Antibodies were from Cell Signaling Technology (Beverly, MA). Blotting was done using the ECL Plus™ detection reagents from Amersham Biosciences (Piscataway, NJ).
Upregulation of EGFR/ErbB1 and HER2/ErbB2 in PLCδ4 overexpressing cells
In breast cancer, the hyperactivation of the Erk pathway is often associated with the upregulation of EGFR expression and the downregulation of estrogen receptor (ERα) expression [20,21]. Accordingly, we examined the expression levels of EGFR and ERα in PLCδ4 overexpressing MCF-7 cells. Analysis of ERα expression and ERα activation (Figure 6) using an antibody specific for ERα protein and an antibody specific for ERα phosphorylated at Ser167 showed no significant differences between PLCδ4 overexpressing cells and vector control cells after adjusting for samples loading differences using the amount of heat shock protein 90 (Hsp90) detected as loading controls. On the contrary, analysis of EGFR/erbB1 total protein level or EGFR activation using antibodies specific for EGFR protein or for EGFR phosphorylated at Tyr992 and/or Tyr1068 showed both EGFR overall protein level and the phosphorylated forms of EGFR were substantially elevated in all four independent isolated lines of PLCδ4 overexpressing cells, whereas little EGFR and phosphorylated EGFR were detected in all five independent isolated lines of vector control cells and in untransfected MCF-7 cells. As similar extents of elevation were found for both EGFR and the phosphorylated forms of EGFR, it is likely that PLCδ4 overexpression upregulates EGFR at the genomic or at the transcriptional level rather than at the post-translational modification level.
Figure 6.
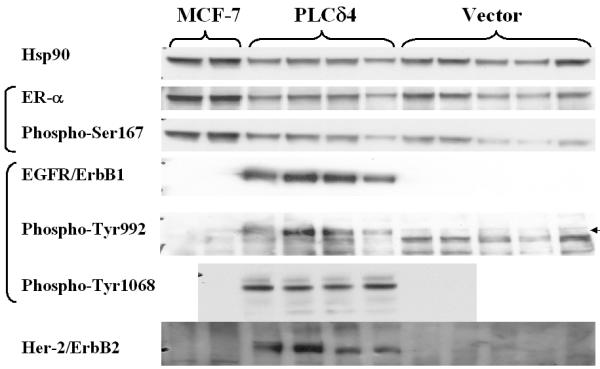
Overexpression of PLCδ4 upregulates ErbB1/2 but does not affect ER-α in MCF-7 cells. Western blot analysis of MCF-7 cell extracts derived from 2 untransfected samples (left 2 lanes), 4 independent isolates overexpressing PLCδ4 and 5 isolates transfected with control vectors (right 5 lanes) using antibodies against Hsp90 (Biosource International, Camarillo, CA), various non-phosphorylated and phosphorylated forms of ER-α and EGFR (Cell Signaling Technology, Beverly, MA), and non-phosphorylated ErbB2 (Upstate Cell Signaling Solutions, Charlottsville, VA). Blotting was done using the ECL Plus™ detection reagents from Amersham Biosciences (Piscataway, NJ).
EGFR can also form heterodimers with other members of erbB receptors such as the ligand-less erbB2 to promote tumor progression [22]. ErbB2/HER2 is overexpressed in 20%–30% of breast and ovarian cancer patients and is associated with aggressive tumor characteristics and poor prognosis. Analysis of erbB2 total protein level with anti-erbB2 showed erbB2 protein levels were also elevated in all four independent isolated lines of PLCδ4 overexpressing cells, whereas little ErbB2 was detected in all five independent isolated lines of vector control cells and in untransfected MCF-7 cells.
PLCδ4 overexpression enhances MCF-7 cells proliferation in low serum, colonies formation in soft agar, and spheroid density on hydrogel
To assess whether PLCδ4 overexpression in cells would lead to certain phenotypic changes that are commonly observed in transformed cells, the growth characteristics in low serum (0.5%) of MCF-7 cell lines overexpressing PLCδ4 and of lines transfected with vector control were compared. All three independent lines of MCF-7 cells carrying the vector overexpressing PLCδ4 have a higher proliferation rate than that of the parental MCF-7 cell line and those of the three vector control lines (Figure 7), despite any apparent growth disadvantage of cells carrying the control vector when compared to untransfected cells.
Figure 7.
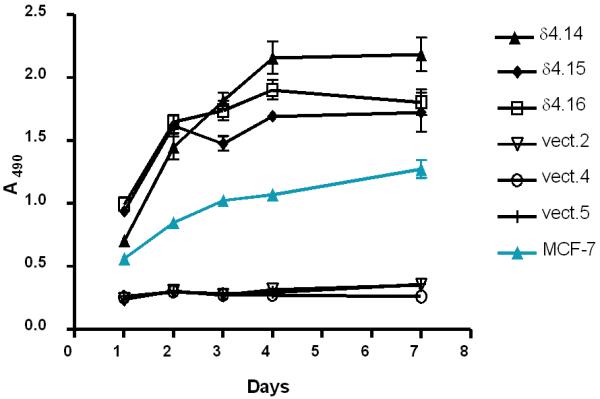
PLCδ4 overexpressing cells enhance cell proliferation in low (0.5%) serum. 3 independent isolates overexpressing PLCδ4 were designated as δ4.14, δ4.15, and δ4.16; 3 isolates transfected with control vectors as vect.2, vect.4, and vect.5; and untransfected cells as MCF-7. Cell proliferation assays were performed using the CellTiter 96® AQueous One Solution (Promega, Madison, WI) in triplicates with error bars indicating ± SD.
Another common division abnormality of cancer cells is their capacity to grow with less anchorage dependency and to divide when held in suspension without attachment or with minimal attachment to a rigid surface. This can either be assayed by the propensity of a given cell line to form colonies in soft agar or to form dangling spheroids on top of Costar® Ultra-Low Attachment Clusters plate surface made up of a neutrally charged hydrogel layer, as the presence of soft agar or hydrogel at the bottom of the culture dish eliminates or minimizes cell contact with the tissue culture plate surface conducive for cell attachment. MCF-7 cells overexpressing PLCδ4 can form colonies in soft agar (21 ± 3 colonies at an initial plating density of 105 cells/ml), whereas cells transfected with the control vector do not form colonies when seeded at similar densities (Figure 8). Although untransfected MCF-7 cells, vector control cells, and PLCδ4 overexpressing cells can all form dangling spheroids on hydrogel layers, the spheroids from PLCδ4 overexpressing cells appear to be more densely packed as evidenced by their more opaque structure under the light microscope (Figure 9). These data suggest that PLCδ4 overexpression may contribute to a transformation phenotype by promoting both anchorage dependent and independent cell growth.
Figure 8.
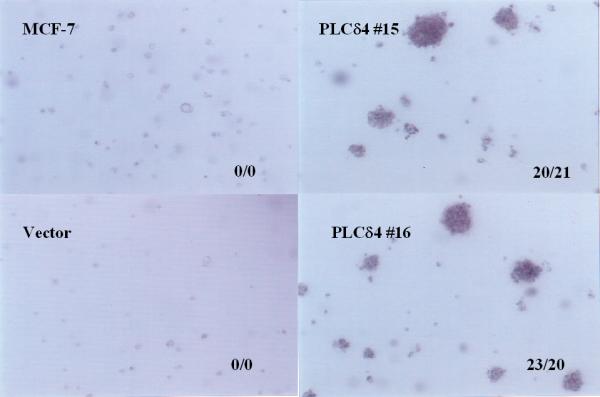
PLCδ4 overexpressing cells form colonies in soft agar. The cells were plated at 1 × 105 cells/ml in 6-well plates containing RPMI 1640 with 0.35% agar, 5% FBS, 10% tryptose phosphate broth, 100 nM 17-β-estradiol. The picture was taken on day 10 with 40× magnification. Samples were plated on duplicate wells. The two numbers separated by a slash mark shown at the lower right-hand corner of each panel represent the total number of colonies scored in each of the two wells.
Figure 9.
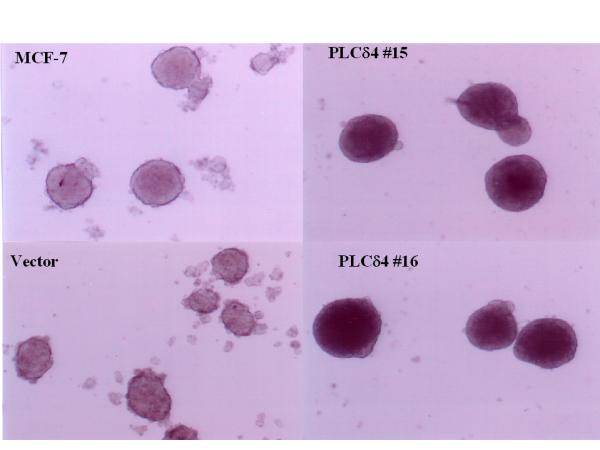
PLCδ4 overexpressing cells form more densely packed spheroids. Cells were grown in Costar® Ultra-Low Attachment Clusters Plates (Corning, Acton, MA) in RPMI supplemented with 10% FBS. The pictures were taken on day 5 with 40× magnification.
The induction of growth signaling response is not reversible by siRNA specific for PLCδ4
To test whether PLCδ4 is a druggable cancer target of which its effects on growth signaling can be reversed by a specific inhibitor for PLCδ4, we introduced siRNA into a cell line (PLCδ4-16) overexpressing PLCδ4 to see if there is any attenuation of signal responses. Western blottings show the expression levels of PLCδ4 in the lysates of overexpressing cells were reduced almost down to those of vector control cells using any three of the siRNAs specific for PLCδ4 (Figure 10, δ4.1, 2 or 3), whereas scrambled siRNA or siRNA specific for lamin A/C had no effect on PLCδ4 level. As expected, siRNAs for PLCδ4 had little effect on total Erk1/2 and Hsp90 level and siRNA for Lamin A/C knocked down only the level of Lamin C. Unexpectedly, β-actin and Lamin C were elevated in PLCδ4 overexpressing cells compared to vector control cells. Although frequently regarded as a housekeeping gene or an innocuous gene for siRNA control, respectively, β-actin and Lamin C can be upregulated in certain rapidly proliferating cells [23,24]. As determined previously, both ErbB1 expression level and Erk1/2 phosphorylation were elevated in PLCδ4 overexpressing cells compared to vector control cells; however, no reduction in these two growth signaling responses could be detected in the presence of PLCδ4 siRNAs despite >80% knockdown of PLCδ4 expression. Hence it is not possible to reverse the transformation phenotype induced by PLCδ4 using siRNA.
Figure 10.
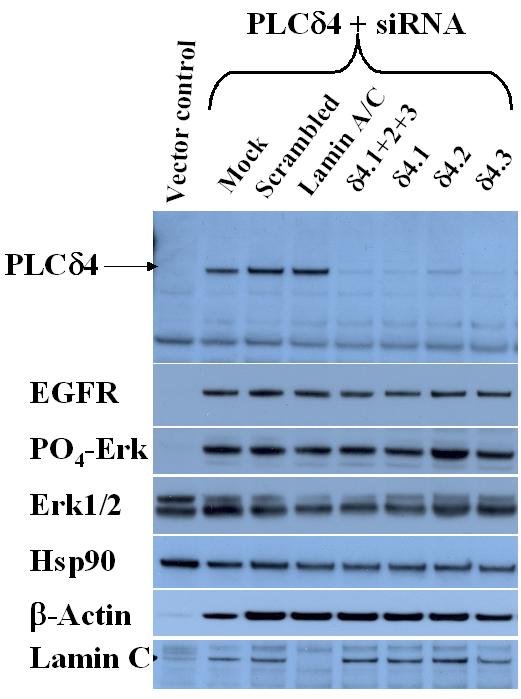
PLCδ4 siRNAs knock down PLCδ4 expression but do not affect the upregulation of EGFR and the activation of Erk1/2. SiRNAs against varaious targets were obtained from Dharmacon Research (Lafayette, CO). See Methods section for sequence specifics and siRNA transfection procedure. Anti-PLCδ4 polyclonal antibodies raised in rabbits immunized with a synthetic peptide WEQQQTMARHLTEI corresponding to amino acids 390–402 of human PLCδ4 were obtained from Covance Research Products (Berkeley, CA). Antibodies against phosphorylated and non-phosphorylated Erk1/2, EGFR, and Lamin A/C were from Cell Signaling Technology (Beverly, MA). Hsp90 antibodies were from Biosource International (Camarillo, CA). β-actin antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Western blot analyses were performed using the ECL Plus™ detection reagents from Amersham Biosciences (Piscataway, NJ).
Discussion
The expression of the rodent phosphoinositide-specific phospholipase C δ-4 (PLCδ4) has been found to be elevated upon serum stimulation in Swiss 3T3 cells and Northern blot analysis have linked the upregulation of PLCδ4 expression with rapid proliferation in hepatocytes and with oncogenic transformation in certain rodent cell lines [10], suggesting that PLCδ4 may be an enzyme target for oncogenic intervention. Accordingly, we have isolated the cDNA for human PLCδ4 to probe the expression of PLCδ4 using northern blot and cDNA expression panel derived from normal tissues (Figure 1) and Cancer Profiling Arrays (BD Clontech, Palo Alto, CA) derived from genetically matched cDNA pairs isolated from various tumor and normal tissue samples (Figure 2). The truncated splice variant of PLCδ4, PLCδ4b, was found to expressed in the same tissues where full length PLCδ4 could be detected (Figure 1, lower right panel), consistent with the recent discovery of widespread use of a mechanism that coupled alternative splicing with a premature stop codon for the downregulation of a transcript through nonsense-mediated mRNA decay [25]. A somewhat analogous splice variant with premature stop codon has been isolated for rat PLCδ4 with dominant-negative activity [13]; however, coexpression of human PLCδ4 along with PLCδ4b has not led to significant reduction of PLC activity in MCF-7 cells in our hands (data not shown) possibly due to subtle differences between the human and the rat sequences. Though PLCδ4 expression was found to be downregulated in the majority of tumor tissues, it was found to be upregulated in a high percentage (>25%) of breast and testicular tumor tissues. The overexpression of PLCδ4 in testicular tumor is of interest, as the only observed phenotype in PLCδ4-/- mice is having defective sperms [15], suggesting PLCδ4 plays an important role in testis and that deregulation of PLCδ4 expression in testis can lead to oncogenesis. With the exceptions of a few universal tumor antigens such as telomerase, survivin, mdm2, cytochrome P450 1B1 [26] and sphingosine kinase 1 [27] that are overexpressed in most tumor tissues, the differential overexpression of PLCδ4 in selected tumor tissues is consistent with the expression pattern of most individual oncogenes, where overexpression is only found in restricted tissues as opposed to all tumor tissues, despite the fact that various tumor tissues do share a common cluster of overexpressed genes related to cell cycling and DNA replication [28]. Hence experiments in addition to expression profiling are required to determine whether PLCδ4 is a gene with oncogenic potential.
We have studied the effects of overexpression of human PLCδ4 on cell signaling and proliferation in this study. MCF-7, a breast cancer cell line of low tumorigenicity, was chosen for ectopic expression of PLCδ4, since profiling of tissue arrays showed PLCδ4 can be overexpressed in breast tumor tissues (Figure 2). Constitutive overexpression of PLCδ4 selectively activates protein kinase C-φ (Figure 4, left panel), whereas the activation states of all other isoforms of PKC studied are not affected by PLCδ4 overexpression based on using phospho-specific PKC isoform-specific antibodies. The data is consistent with results from another phospho-specific antibody against the consensus motif of PKC substrates, showing slight enhancement of phosphorylation of PKC substrates with PLCδ4 overexpression (Figure 4, right panel).
Using antibodies that target signaling pathways for cell proliferation, we have found that PLCδ4 overexpression leads to constitutive activation of extracellular signal-regulated kinases 1 and 2 (ERK1/2) pathway in MCF-7 cells (Figure 5) and of kinases both upstream and downstream of the Erk signaling pathway such as Erk kinases (MEK1/2) and the 90 kDa ribosomal S6 kinase (p90RSK), whereas key proteins involved in two alternative MAPK pathways and the PI3K pathway, such as SAPK/JNK, p38 MAPK, and Akt, respectively, showed no change in activation in PLCδ4 overexpressing cells. PLCδ4 overexpression also upregulates the expression of epidermal growth factor receptors EGFR/erbB1 and HER2/erbB2 in MCF-7 cells (Figure 6). Of the four ErbB isoforms, cells co-expressing ErbB1 and ErbB2 have been found to be the most aggressive in forming tumors in mice [29]. Though upregulation of EGFR expression is often associated with the downregulation of estrogen receptor (ERα) expression in breast cancer cells [20,21], no significant change was found in ERα expression in PLCδ4 overexpressing MCF-7 cells (Figure 6). Since both EGFR transcription [21,30] and ErbB2 expression [31] have been found to be upregulated by phorbol ester, it is possible that the DAG generated from PLCδ4 overexpression may activate selective PKCs such as PKC-φ, leading to enhancement of EGFR and ErbB2 gene transcription and activation of the Erk signaling pathway.
We have examined several cell growth parameters to determine whether enhancement of ErbB1/2 expression and activation of Erk pathway in PLCδ4 overexpressing MCF-7 cells can be translated into a more proliferative state of cell growth. MCF-7 cells stably expressing PLCδ4 demonstrate several phenotypes of transformation; namely, enhanced rate of proliferation and saturating cell density in low serum (Figure 7), improved efficiency in forming colonies in soft agar (Figure 8), and capacity to form more densely packed spheroids in low-attachment plates (Figure 9). These data suggest that PLCδ4 overexpression may contribute to a transformation phenotype by promoting both anchorage dependent and independent cell growth.
A criterion for validation and selection of new drug targets is to determine whether there is evidence of the reversal of the transformation phenotype using genetic means or drug leads. We have been able to knock down the expression of PLCδ4 in PLCδ4 overexpressing cells by transfecting siRNAs specific for PLCδ4 sequences (Figure 10); however, no reduction in ErbB1 expression level and Erk1/2 phosphorylation in PLCδ4 overexpressing cells in the presence of siRNA could be detected. The growth signaling responses induced by PLCδ4 are hence not reversible by siRNA. The inability for siRNAs for PLCδ4 to knock down growth signaling responses suggests PLCδ4 may play a role in the initiation of carcinogenesis by inducing other genetic changes, as a neoplastic state can sometimes be irreversible by becoming independent of the initiating oncogenic event through induction of genome destabilization. For instance, even transient induction of MYC activity was able to sustain tumorigenesis in certain cell types [32]. Overexpression of ErbB2, a receptor induced by PLCδ4 (Figure 6), has also been associated with genetic instability in cells [33]. These results suggest that PLCδ4 per se may be a difficult target to be manipulated with drugs, and the sites for intervention may lie instead on other enzyme targets in pathways activated by PLCδ4, such as the ErbB receptors and the Ras/Raf/MAP kinase cascade. The validation of some these targets has been confirmed with two approved drugs, Herceptin® specific for ErbB2 and Iressa® for EGFR tyrosine kinase, with numerous inhibitors for the ErbB receptor family in clinical trials [34], and with the studies of Raf inhibitor BAY 37-9751 and MEK inhibitor CI-1040 along the ERK pathway in clinical trials [35].
As activation of ErbB can activate another isoform of PLC, PLCγ [2], it is also possible that activation PLCγ may compensate for the knockdown of PLCδ4 by siRNA and that compounds capable of inhibiting multiple isoforms of PLC may be more suitable for oncogenic intervention. A black-box approach can also be used to screen for compounds by high throughput screening that would inhibit growth signaling responses induced by PLCδ4 with differential cytotoxcity in PLCδ4 overexpressing MCF-7 cells versus matched vector control cells. The use of human engineered tumor cells has been successful in identifying new as well as classical genotype-selective anti-tumor compounds [36].
Conclusions
Overexpression or dysregulated expression of PLCδ4 may initiate oncogenesis in certain tissues through upregulation of ErbB expression and activation of ERK pathway. PLCδ4 can therefore be a useful tumor marker for breast or testicular cancer tissues. Since the growth signaling responses induced by PLCδ4 are not reversible by siRNA, a surrogate for a PLCδ4-specific inhibitor, PLCδ4 itself is probably not a suitable drug target for development, but enzymes in pathways activated by PLCδ4 are potential therapeutic targets for oncogenic intervention. Alternatively, a black-box approach can be used to screen compounds with differential cytotoxcity in PLCδ4 overexpressing MCF-7 cells.
Methods
Isolation of human PLCδ4 cDNA
Search of the Genbank database [37] of expressed sequence tag (dbEST) using the rat PLCδ4 protein sequences [12] as probe identified an I.M.A.G.E. Consortium [LLNL] cDNA clone (GenBank# AI366170), pBSK-D4, isolated from a human oligodendroglioma library containing an insert of ~2500 bp with three separate stretches of sequences containing extensive homology to the rat PLCδ4 coding sequence. These three coding regions do not share a common reading frame and is most likely derived from a spurious splice variant of human PLCδ4 with exons deletion and intron insertion in two separate areas. This cDNA clone thus codes for a truncated PLCδ4 containing the first 258 amino acids of full-length PLCδ4 with another 16 amino acids and a termination codon added at the C-terminal end as a result of exons deletion.
To isolate a human PLCδ4 cDNA encoding an active PLC enzyme, PCR primers, oD4-2f, 5'-AGACACGTCC CAGTCTGGAA CC-3', and oD4-2r, 5'-CTGCTTCCTC TTCCTCATAT TC-3' (Qiagen, Alameda, CA) with sequences, respectively, corresponding to the coding strand upstream of a Eco RI site and to the complementary strand downstream of a unique Hpa I site were used to screen cDNAs derived from various human cell lines to identify PCR products that are longer (due to insertion of additional exons) than the product (expected to be ~300 bp) when using pBSK-D4 as the template. Templates from cDNAs derived from CA-HPV-10, Capan-1, Hs.7661, MiaPaCa, A549, MDA-MB-231, Panc-1, NCI-H460, LnCaP, MCF-7, and HNMEC cells all gave PCR products of ~800 bp upon amplification. The human normal mammary epithelial cells (HNMEC) was fron Clonetics (Walkersville, MD), whereas the rest of the cell lines were from American Type Culture Collection (Manassas, VA). The ~800 bp PCR product was then cleaved with Eco RI and Hpa I to generate a ~730 bp fragment for ligating into the vector pBSK-D4 that had been cleaved with Eco RI and Hpa I for the generation of the plasmid pBSK-D43X. DNA sequence analysis shows the cDNA insert in pBSK-D43X contains an additional ~500 bp region with extensive homology to the rat PLCδ4 sequence not found within the insert in pBSK-D4. pBSK-D43X lacked the 5'-end region of PLCδ4 cDNA upstream of Ecor RI, as the ~840 bp Eco RI fragment that spans the Eco RI site found in the 5'-cDNA adaptor sequence and the internal Eco RI site found in the PLCδ4 coding region was removed during preparation of the Eco RI/Hpa I pBSK-D4 vector.
Separately, PCR primers with sequences corresponding to the coding strand upstream of Hpa I of pBSK-D4, oD4-3f, 5'-CTGGTGAAGG GGAAGAAGTT A-3', and to the complementary strand downstream of Bgl II, oD4-3r, 5'-TGTCTAGACG AACGCCAAAG AT-3' were used to screen cDNAs derived from human cell lines to identify PCR products that are ~300 bp shorter (due to removal of a putative intron sequence) than the product (~1050 bp) when using pBSK-D4 as the template. A PCR product of ~700 bp was detected from cDNA template derived from CA-HPV-10 cells. The ~700 bp PCR product was then cleaved with Hpa I and Bgl II to generate a ~670 bp fragment for ligating into the vector pBSK-D43X that had been cleaved with Hpa I and Bgl II for the generation of the plasmid pBSK-D43X1. DNA sequence analysis shows the cDNA insert in pBSK-D43X contains extensive homology to the rat PLCδ4 sequence.
Construction of vectors for expression of human hPLCδ4 in mammalian cells
To assemble a full-length human PLCδ4a cDNA for expression study, a ~600 bp Nhe I - Eco RI fragment was isolated from digestion with Nhe I + Eco RI of the ~2200 bp PCR product generated by amplification from the template pBSK-D4 using the primers oD4-1f, 5'-GTGATCTGGT GCTAGCTGGT GGAAC-3' and oD4-1r, 5'-ACACCAATGC ATTCCCGTGA AATGCCCACC-3'; and a ~1700 bp Eco RI - Nsi I fragment isolated from digestion with Eco RI + Nsi I of the ~1770 bp PCR product generated by amplification from the template pBSK-D43X1 using the primers oD4-2f and oD4-1r were combined and inserted into the pCivIrPu vector that had been cleaved with Nhe I and Nsi I via a three-part ligation for the generation of the plasmid pPuroD4F. pCivIrPu is derived from the plasmids pIRESpuro (Clonetech, Palo Alto, CA) and pCIneo (Promega, Madison, WI). Specifically, the 980 bp Spe I - Not I fragment from pCIneo was inserted into the cloning vector pSL1180 (Amersham Pharmacia, Piscataway, NJ) between the restriction sites Spe I and Not I to generate pSLCI. The 1000 bp Spe I - Nsi I fragment from pSLCI was then isolated for insertion into the vector pIRESpuro between the restriction sites Spe I and Nsi I to generate pCivIrPu.
To determine whether the human hPLCδ4a cDNA sequence encodes a protein with phospholipase c (PLC) activity, the plasmid pPuroD4F was stably transfected into MCF-7 cells (American Type Culture Collection, Manassas, VA) to generate puromycin-resistant clones. Specifically, pPuroD4F was digested with BspH I before electroporating into these cell lines with a Cell-Porator™ (Life Technologies, Gaithersburg, MD) using conditions described previously [38]. After adherence of the transfected cells 48 hours later, the cells were grown in the presence of 1.0 μg/ml puromycin to select for cells that had incorporated the plasmid.
Construction of vectors for expression of GFP-PH/PLCδ4 fusion proteins in mammalian cells
Coding region corresponding to the PH domain of PLCδ4 splice variant were amplified with PCR primers oPHd4_1f, 5'-GCGCAGCGAA TTCATGGCGT CCCTGCTGCA AGAC-3' and o oPHd4_1r, 5'-GATCTAGTGT CGACCCAAGA TCCACCAACA GCTGGAG-3' to generate a 400 bp fragment. The fragment was then cleaved with EcoR I and Sal I to generate a 390 bp fragment for insertion between the EcoR I and Sal I sites of pEGFP-n1 (Clontech, Palo Alto, CA) for generation of pPHd4GFP, an expression plasmid for fusion protein containing the PH domain of PLCδ4 upstream of GFP.
Cell culture
Cell culture reagents were from Invitrogen (Carlsbad, CA). Human tumor cell lines were from the American Type Culture Collection (Manassas, VA). MCF-7, MCF-7 cells stably overexpressing PLCδ4, or MCF-7 cells transfected with control vector were grown in RPMI supplemented with 10% FBS. Spheroids were grown on Costar® Ultra-Low Cluster Plates (Corning, Acton, MA) in RPMI supplemented with 10% FBS. Cell proliferation assays were performed using the CellTiter 96® AQueous One Solution (Promega, Madison, WI). For colonies grow in soft agar, cells in soft agar colony assay were cultured in 6-well plates containing 104 or 105 cells/well in RPMI 1640 with 0.35% agar, 5% FBS, 10% tryptose phosphate broth, 100 nM 17-β-estradiol. The plates were incubated for 10 days, after which the cultures were inspected and photographed.
Antibodies
Phospho-PKC (pan), phospho-PKCα/βII (Thr638/641), phospho-PKCφ (Thr538), phosphoPKCζ/λ (Thr410/403), phospho-MEK1/2 (Ser217/221), phospho-Erk1/2 (Thr202/183+Tyr204/185), total Erk1/2, phospho-p90RSK (Thr359+Ser363), phospho-SAPK p54/p46 (Thr183+Tyr185), phospho-p38 MAPK (Thr180+Tyr182), phospho-Akt (Ser473), total ER-α, phospho-ER-α (Ser167), total EGFR, phospho-EGFR (Tyr992), phospho-EGFR (Tyr1068), phospho-(Ser) PKC substrate and Lamin A/C antibodies were from Cell Signaling Technology (Beverly, MA). Hsp90 antibodies were from Biosource International (Camarillo, CA). HER-2 antibodies were from Upstate Cell Signaling Solutions (Charlottsville, VA). β-actin antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Polyclonal antibodies raised in rabbits immunized with a synthetic peptide WEQQQTMARHLTEI corresponding to amino acids 390–402 of human PLCδ4 were obtained from Covance Research Products (Berkeley, CA). Western blot analyses were performed using the ECL Plus™ detection reagents from Amersham Biosciences (Piscataway, NJ).
Assay of phospholipase C activity in PLCδ4 overexpressing cells
Puromycin-resistant clones were randomly selected to check for PLC activity in cell extracts using a assay that measures the conversion of radiolabeled PIP2 or PI to DAG and IP3 or inositol-1-phosphate (Ins(1)P). To prepare cell extracts for assay, cells from plates were scraped into PBS + protease inhibitors. These cells were sonicated 3 times for 20 seconds using a probe sonicator. Protein concentrations were determined and the lysates were used for the assays. The assay of hydrolysis of PIP2 or PI was based on methods described previously [39,40]. Specifically, each PI hydrolysis assay was in a total volumn of 200 μl composed of 250 μM PI (50 nmol), 20,000 cpm (0.030 μCi) of [3H]PI, 100 mM NaCl, 1 mM EGTA, 4 mM CaCl2, 0.1 mM DTT, 50 mM HEPES buffer, pH 7.0, 1 mg/ml sodium deoxycholate. A substrate solution enough for 50 assays was prepared by drying 5 ml of 500 μM PI + 1.5 μCi [3H]PI under N2 before resuspending in 5 ml of 2 mg/ml sodium deoxycholate by sonication. A buffer solution (10×) was made up of 250 mM HEPES pH 7.0, 1 M NaCl, 1 mM DTT. A Ca2+/EGTA solution (4×) was made up of 4 mM EGTA, 12 mM CaCl2, 100 mM HEPES pH 7.0 (free 3 mM Ca2+ in assay). After mixing 20 μl buffer solution, 50 μl Ca2+/EGTA, and 30 μl enzyme extract, the reaction was started by addition of 100 μl substrate solution. After incubating for 10 min at 37°C, the reaction was terminated with 0.75 ml chloroform/methanol/HCl (100:200:0.6, by volume), followed by 0.25 ml of chloroform (water saturated) and then 0.25 ml 1 N HCL containing 5 mM EGTA. After centrifugation to separate the phases, 300 μl of aqueous phase was then counted in scintillation counter.
For the assay of PLC activity using PIP2 as the substrate, each PI hydrolysis assay was in a total volumn of 200 μl composed of 40 μM PI (8 nmol), 200 μM PE (40 nmol), 20,000 cpm (0.030 μCi) of [3H]PIP2, 3 mM MgCl2, 100 mM NaCl, 1 mM EGTA, 1 mM CaCl2 (free 45 μM Ca2+ in assay), 0.1 mM DTT, 50 mM HEPES buffer, pH 7.0, 1 mg/ml sodium deoxycholate. A substrate solution enough for 50 assays was prepared by drying 80 μM PIP2 (5 ml) + 400 μM PE + 1.5 μCi [3H]PIP2 under N2 before resuspending in 5 ml of 65 mM HEPES pH 7.0, 100 mM NaCl, 2 mg/ml sodium deoxycholate by sonication. A MgCl2 solution was made up of 12 mM MgCl2, 50 mM HEPES pH 7.0, 200 mM NaCl, 0.4 mM DTT. A Ca2+/EGTA solution was made up of 10 mM EGTA, 10 mM CaCl2, 50 mM HEPES pH 7.0. After mixing 50 μl MgCl2 solution, 20 μl Ca2+/EGTA, and 30 μl enzyme extract, the reaction was started by addition of 100 μl substrate solution. After incubating for 10 min at 37°C, the reaction was terminated with 0.75 ml chloroform/methanol/HCl (100:200:0.6, by volume), followed by 0.25 ml of chloroform (water saturated) and then 0.25 ml 1 N HCL containing 5 mM EGTA. After centrifugation to separate phases, 300 μl of aqueous phase was then counted in scintillation counter.
Preparation of nuclear extracts
MCF-7 cells were harvested with 0.2% trypsin and washed two times with cold PBS. The cells were suspended in buffer A (10 mM Tris/HCl, pH 7.8), 1% Nonidet P-40, 10 mM β-mercaptoethanol, 0.5 mM PMSF, 1 μg/ml aprotinin and leupeptin) for 2 min on ice. An equal volume of distilled H2O was added, and the cells were allowed to swell for 2 min. The cells were sheared by ten passages through a 22-gauge needle. The nuclei were recovered by centrifugation at 400 × g for 6 min and washed once with buffer B (10 mM Tris/HCl (pH 7.4), 2 mM MgCl2, 0.5 mM PMSF, 1 μg/ml aprotinin and leupeptin). These nuclei were sonicated 3 times for 20 seconds at an output of 1 using a probe sonicator prior to PI hydrolysis assay.
Northern blot, PCR and tissue array analysis
Northern hybridization was carried out to detect the expression of PLCδ4 mRNA in different normal tissues. Specifically, nylon membranes containing mRNA isolated from various human tissues and tumor cell lines (Clontech, Palo Alto, CA) and cDNAs derived from various matched tumor and normal tissues on Cancer Profiling Array I and II (Clontech, Palo Alto, CA) were hybridized in ULTRAhyb™ (Ambion, Austin, TX) with PLCδ4 (2000 bp Nhe I-Bgl II fragment from pPuroD4F) coding region cDNA fragment labeled with [α-32P]dCTP using a Strip-Ez labeling kit (Ambion, Austin, TX). To evaluate the relative abundance of full length PLCδ4 and splice variant PLCδ4b, primers, oD4_2f, 5'-AGACACGTCC CAGTCTGGAA CC-3', and oD4_2r, 5'-CTGCTTCCTC TTCCTCATAT TC-3', located upstream and downstream of the coding region for exons 7, 8, and 9 were used to detect PCR products in a Rapid-Scan™ Gene Expression Panel (OriGene, Rockville, MD) generated from cDNA samples derived from various normal human tissues. The full length PLCδ4 would generate a 800 bp product; whereas the splice variant PLCδ4b with exons 7, 8, and 9 deleted, a 300 bp product.
Preparation of siRNAs and transfections
Several siRNAs targeting PLC-δ4 (accession number BC006355) were synthesized by Dharmacon Research (Lafayette, CO). siRNAs were annealed essentially as described [41]. The sense strands for each siRNA duplex were as follows. PLC-δ4 specific siRNA #1 beginning at nt 1612, 5'-GAGCAGAACCTTCAGAATAdTdT-3', #2 beginning at nt 457, 5'-GAGCAGGGCTTCACCATTGdTdT-3', #3 beginning at nt, 765, 5'-GGAAGGAGAAGAATTCGTAdTdT-3', #4 beginning at nt 1752, 5'-GATATCATCTTTCTCTGAAdTdT-3' and "Smart Pool" siRNAs which combined the above PLC-δ4 siRNAs #1–4. The following siRNAs were obtained from Dharmacon Research (sense strands); Lamin A/C 5'-CUGGACUUCCAGAAGAACAdTdT-3' and Non-specific Control I 5'-NNATGAACGTGAATTGCTCAA-3'. Human MCF7-PLCd4c16 breast cancer cells were grown in RPMI1640, supplemented with 10% fetal bovine serum. Cells were seeded at 60,000 cells/well in a six-well dish. These seed density resulted in 15% confluency and was optimal for transfection efficiencies and growth by 72 hrs. Cells were transfected with 27 nM siRNA in serum free Opti-MEM I using Oligofectamine (Invitrogen, Carlsbad, California). Following 4 hr incubation with siRNAs, fetal bovine serum to 2% v/v final was added. Cells were harvested and extracts prepared for Western analysis 72 hr after transfection.
Authors' contributions
DWL conceived of the study, participated in its design and coordination, and drafted the manuscript. DWL and CT carried out the molecular cloning and expression. CT and JB carried out the signaling and cell growth studies. AB and MC carried out the siRNA studies. VM and DW carried out the biochemical assays. JS oversaw the project. All authors read and approved the final manuscript.
Contributor Information
David W Leung, Email: dwleung1@msn.com.
Chris Tompkins, Email: ctompkins@acceleratorcorp.com.
Jim Brewer, Email: jbrewer@dynamac.com.
Alexey Ball, Email: aball@ctiseattle.com.
Mike Coon, Email: mcoon@ctiseattle.com.
Valerie Morris, Email: valmorr@yahoo.com.
David Waggoner, Email: wags@u.washington.edu.
Jack W Singer, Email: jsinger@ctiseattle.com.
References
- Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol Rev. 2000;80:1291–1335. [Google Scholar]
- Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Annu Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RL. Mammalian phosphoinositide-specific phospholipase C. Biochim Biophys Acta. 1999;1441:255–267. doi: 10.1016/S1388-1981(99)00150-X. [DOI] [PubMed] [Google Scholar]
- Lopez I, Mak EC, Ding J, Hamm HE, Lomasney JW. A novel bifunctional phospholipase c that is regulated by Galpha 12 and stimulates the Ras/mitogen-activated protein kinase pathway. J Biol Chem. 2001;276:2758–2765. doi: 10.1074/jbc.M008119200. [DOI] [PubMed] [Google Scholar]
- Kelley GG, Reks SE, Ondrako JM, Smrcka AV. Phospholipase C(epsilon): a novel Ras effector. EMBO J. 2001;20:743–754. doi: 10.1093/emboj/20.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Hu CD, Masago M, Kariyai K, Yamawaki-Kataoka Y, Shibatohge M, Wu D, Satoh T, Kataoka T. Regulation of a novel human phospholipase C, PLCepsilon, through membrane targeting by Ras. J Biol Chem. 2001;276:2752–2757. doi: 10.1074/jbc.M008324200. [DOI] [PubMed] [Google Scholar]
- Saunders CM, Larman MG, Parrington J, Cox LJ, Royse J, Blayney LM, Swann K, Lai FA. PLC zeta: a sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development. 2002;129:3533–3544. doi: 10.1242/dev.129.15.3533. [DOI] [PubMed] [Google Scholar]
- Baek KJ, Kang S, Damron D, Im M. Phospholipase Cdelta1 is a guanine nucleotide exchanging factor for transglutaminase II (Galpha h) and promotes alpha 1B-adrenoreceptor-mediated GTP binding and intracellular calcium release. J Biol Chem. 2001;276:5591–5597. doi: 10.1074/jbc.M008252200. [DOI] [PubMed] [Google Scholar]
- Murthy SN, Lomasney JW, Mak EC, Lorand L. Interactions of G(h)/transglutaminase with phospholipase Cdelta1 and with GTP. Proc Natl Acad Sci U S A. 1999;96:11815–11819. doi: 10.1073/pnas.96.21.11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Fukami K, Yu H, Takenawa T. A new phospholipase C delta 4 is induced at S-phase of the cell cycle and appears in the nucleus. J Biol Chem. 1996;271:355–360. doi: 10.1074/jbc.271.1.355. [DOI] [PubMed] [Google Scholar]
- Santi P, Solimando L, Zini N, Santi S, Riccio M, Guidotti L. Inositol-specific phospholipase C in low and fast proliferating hepatoma cell lines. Int J Oncol. 2003;22:1147–1153. doi: 10.3892/ijo.22.5.1147. [DOI] [PubMed] [Google Scholar]
- Lee SB, Rhee SG. Molecular cloning, splice variants, expression, and purification of phospholipase C-delta 4. J Biol Chem. 1996;271:25–31. doi: 10.1074/jbc.271.1.25. [DOI] [PubMed] [Google Scholar]
- Nagano K, Fukami K, Minagawa T, Watanabe Y, Ozaki C, Takenawa T. A novel phospholipase C delta4 (PLCdelta4) splice variant as a negative regulator of PLC. J Biol Chem. 1999;274:2872–2879. doi: 10.1074/jbc.274.5.2872. [DOI] [PubMed] [Google Scholar]
- Fukami K, Takenaka K, Nagano K, Takenawa T. Growth factor-induced promoter activation of murine phospholipase C delta4 gene. Eur J Biochem. 2000;267:28–36. doi: 10.1046/j.1432-1327.2000.00943.x. [DOI] [PubMed] [Google Scholar]
- Fukami K, Yoshida M, Inoue T, Kurokawa M, Fissore RA, Yoshida N, Mikoshiba K, Takenawa T. Phospholipase Cdelta4 is required for Ca2+ mobilization essential for acrosome reaction in sperm. J Cell Biol. 2003;161:79–88. doi: 10.1083/jcb.200210057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–353. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- Lee CW, Park DJ, Lee KH, Kim CG, Rhee SG. Purification, molecular cloning, and sequencing of phospholipase C-beta 4. J Biol Chem. 1993;268:21318–21327. [PubMed] [Google Scholar]
- Varnai P, Lin X, Lee SB, Tuymetova G, Bondeva T, Spat A, Rhee SG, Hajnoczky G, Balla T. Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J Biol Chem. 2002;277:27412–27422. doi: 10.1074/jbc.M109672200. [DOI] [PubMed] [Google Scholar]
- Razzini G, Brancaccio A, Lemmon MA, Guarnieri S, Falasca M. The role of the pleckstrin homology domain in membrane targeting and activation of phospholipase Cbeta(1) J Biol Chem. 2000;275:14873–14881. doi: 10.1074/jbc.275.20.14873. [DOI] [PubMed] [Google Scholar]
- Oh AS, Lorant LA, Holloway JN, Miller DL, Kern FG, El Ashry D. Hyperactivation of MAPK induces loss of ERalpha expression in breast cancer cells. Mol Endocrinol. 2001;15:1344–1359. doi: 10.1210/me.15.8.1344. [DOI] [PubMed] [Google Scholar]
- Lee CS, deFazio A, Ormandy CJ, Sutherland RL. Inverse regulation of oestrogen receptor and epidermal growth factor receptor gene expression in MCF-7 breast cancer cells treated with phorbol ester. J Steroid Biochem Mol Biol. 1996;58:267–275. doi: 10.1016/0960-0760(96)00039-8. [DOI] [PubMed] [Google Scholar]
- Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284:99–110. doi: 10.1016/S0014-4827(02)00099-X. [DOI] [PubMed] [Google Scholar]
- Tsuji N, Kamagata C, Furuya M, Kobayashi D, Yagihashi A, Morita T, Horita S, Watanabe N. Selection of an internal control gene for quantitation of mRNA in colonic tissues. Anticancer Res. 2002;22:4173–4178. [PubMed] [Google Scholar]
- Tilli CM, Ramaekers FC, Broers JL, Hutchison CJ, Neumann HA. Lamin expression in normal human skin, actinic keratosis, squamous cell carcinoma and basal cell carcinoma. Br J Dermatol. 2003;148:102–109. doi: 10.1046/j.1365-2133.2003.05026.x. [DOI] [PubMed] [Google Scholar]
- Lewis Benjamin P., Green Richard E., Brenner Steven E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. PNAS. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Vonderheide RH. Universal tumor antigens as targets for immunotherapy. Cytotherapy. 2002;4:317–327. doi: 10.1080/146532402760271091. [DOI] [PubMed] [Google Scholar]
- French Kevin J., Schrecengost Randy S., Lee Brian D., Zhuang Yan, Smith Staci N., Eberly Justin L., Yun Jong K., Smith Charles D. Discovery and Evaluation of Inhibitors of Human Sphingosine Kinase. Cancer Res. 2003;63:5962–5969. [PubMed] [Google Scholar]
- Chung CH, Bernard PS, Perou CM. Molecular portraits and the family tree of cancer. Nat Genet. 2002;32 Suppl:533–540. doi: 10.1038/ng1038. [DOI] [PubMed] [Google Scholar]
- Cohen BD, Kiener PA, Green JM, Foy L, Fell HP, Zhang K. The relationship between human epidermal growth-like factor receptor expression and cellular transformation in NIH3T3 cells. J Biol Chem. 1996;271:30897–30903. doi: 10.1074/jbc.271.48.30897. [DOI] [PubMed] [Google Scholar]
- Haley JD, Waterfield MD. Contributory effects of de novo transcription and premature transcript termination in the regulation of human epidermal growth factor receptor proto-oncogene RNA synthesis. J Biol Chem. 1991;266:1746–1753. [PubMed] [Google Scholar]
- Xian W, Rosenberg MP, DiGiovanni J. Activation of erbB2 and c-src in phorbol ester-treated mouse epidermis: possible role in mouse skin tumor promotion. Oncogene. 1997;14:1435–1444. doi: 10.1038/sj.onc.1200980. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci U S A. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Shiquan, Liu Wenjing, Jakubczak John L., Erexson Gregory L., Tindall Kenneth R., Chan Richard, Muller William J., Adhya Sankar, Garges Susan, Merlino Glenn. Genetic instability favoring transversions associated with ErbB2-induced mammary tumorigenesis. PNAS. 2002;99:3770–3775. doi: 10.1073/pnas.052710299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono JS, Rowinsky EK. The ErbB receptor family: a therapeutic target for cancer. Trends Mol Med. 2002;8:S19–S26. doi: 10.1016/S1471-4914(02)02306-7. [DOI] [PubMed] [Google Scholar]
- Herrera R, Sebolt-Leopold JS. Unraveling the complexities of the Raf/MAP kinase pathway for pharmacological intervention. Trends Mol Med. 2002;8:S27–S31. doi: 10.1016/S1471-4914(02)02307-9. [DOI] [PubMed] [Google Scholar]
- Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/S1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Tolstoshev CM, Bassett D.E.,Jr. Gene discovery in dbEST. Science. 1994;265:1993–1994. doi: 10.1126/science.8091218. [DOI] [PubMed] [Google Scholar]
- Cachianes G, Ho C, Weber RF, Williams SR, Goeddel DV, Leung DW. Epstein-Barr virus-derived vectors for transient and stable expression of recombinant proteins. Biotechniques. 1993;15:255–259. [PubMed] [Google Scholar]
- Nozawa Y, Banno Y. Phosphatidylinositol-specific phospholipase C from human platelets. Methods Enzymol. 1991;197:518–526. doi: 10.1016/0076-6879(91)97178-2. [DOI] [PubMed] [Google Scholar]
- Rhee SG, Ryu SH, Lee KY, Cho KS. Assays of phosphoinositide-specific phospholipase C and purification of isozymes from bovine brains. Methods Enzymol. 1991;197:502–511. doi: 10.1016/0076-6879(91)97176-Y. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]


