Abstract
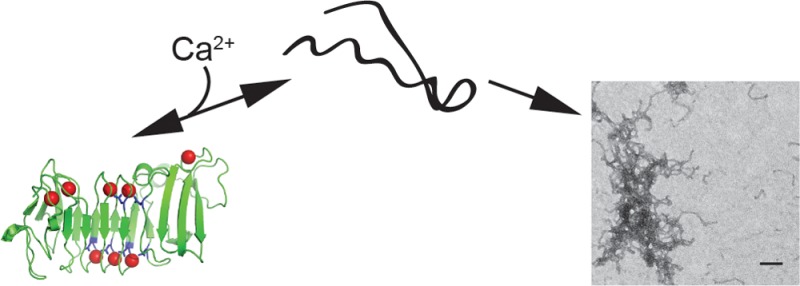
Self-assembling proteins represent potential scaffolds for the organization of enzymatic activities. The alkaline protease repeats-in-toxin (RTX) domain from Pseudomonas aeruginosa undergoes multiple structural transitions in the presence and absence of calcium, a native structural cofactor. In the absence of calcium, this domain is capable of spontaneous, ordered polymerization, producing amyloid-like fibrils and large two-dimensional protein sheets. This polymerization occurs under near-physiological conditions, is rapid, and can be controlled by regulating calcium in solution. Fusion of the RTX domain to a soluble protein results in the incorporation of engineered protein function into these macromolecular assemblies. Applications of this protein sequence in bacterial adherence and colonization and the generation of biomaterials are discussed.
The acquisition of native protein structure is fundamental to the function of newly synthesized polypeptides.1 Failure to obtain the native conformation results in a loss of protein function and is often associated with polypeptide degradation and/or aggregation.2 While non-native proteins have often been considered inactive, their incorporation into large macromolecular structures is being recognized as being biologically important.3−5 In addition, the ability to generate specific protein polymers, including ordered filaments and films, has potential for the production of biomaterial scaffolds, which are increasingly being developed for therapeutic applications.6−8
Many protein sequences have been shown to self-associate and assemble, though these reactions are often induced by nonphysiological conditions, including extreme shifts in buffer composition and pH, salt, temperature, or extended incubation periods.6,9 These harsh treatments are often limiting to the incorporation of native protein functions, as, by design, they usually result in the denaturation of protein structure prior to self-assembly.6 As a result, the identification of protein sequences that can be utilized to facilitate stable polymer assembly under near-physiological conditions would likely facilitate the development and production of biological materials.
The repeats-in-toxin (RTX) family of exoproteins is associated with the virulence of multiple human pathogens, including Escherichia coli, Bordetella pertussis, Serratia marcescens, and Pseudomonas aeruginosa.10−14 The hallmark of this family of secreted proteins is a highly conserved nonapeptide glycine- and aspartate-rich repeat, which is concatenated in C-terminal Ca2+-binding RTX domains. Structural studies of multiple RTX domains indicate that Ca2+ binding facilitates the folding of the RTX domains into compact β-helical structures.15−17 In the absence of Ca2+, these domains are largely disordered, exhibiting limited secondary structure and extended hydrodynamic radii. It is thought that this structural transition serves to regulate the activity and secretion of these proteins from the bacterial cell.
Studies of the P. aeruginosa alkaline protease (AprA) demonstrate that the folding and activation of the enzyme are coupled to Ca2+ binding within the RTX domains.17 These Ca2+ binding events first facilitate the folding of the RTX domain, which, in turn, nucleates the folding of the protease domain. While characterizing the Ca2+-induced folding of the AprA sequence, we previously observed that incubation of the apo RTX domain in near-physiological buffers at both low temperatures and low concentrations resulted in the formation of ordered protein polymers. The characterization of these polymeric structures and their assembly were not explored, nor were the mechanisms by which Ca2+ binding might influence these polymerization events in vitro.
To characterize these structures further and evaluate their ability to form stable assemblies, the RTX domain was purified for in vitro polymerization studies. This polymerization was tightly controlled by Ca2+-induced RTX folding and could be regulated by Ca2+ addition or chelation. Further, deposition of these structures onto solid supports resulted in the formation of large, highly ordered two-dimensional protein sheets. The formation of the fibrillar structures and the protein sheets both depended on the presence of the apo RTX conformations. Saturating Ca2+, which facilitates folding of the RTX domain to its native β-helical conformation, inhibited the production of the polymeric structures. Fusion of the RTX sequences to a soluble, globular protein resulted in the incorporation of specific protein activity into these two-dimensional films. These data suggest that the RTX domain potentially provides a novel scaffold for producing protein-based biomaterials.
Materials and Methods
RTX Domain Purification and Refolding
The RTX domain was purified and stored as previously described.17 Briefly, the RTX domain was cloned by polymerase chain reaction from P. aeruginosa PAO1 genomic DNA (ATCC) into the pET-DUET expression vector (Novagen) for expression in E. coli. The RTX domains contained either an N- or C-terminal six-His tag for purification. Expression cultures were grown at 37 °C, and the insoluble RTX protein was purified from inclusion bodies using guanidinium hydrochloride (GuHCl) as the denaturant. Protein was bound to a Ni-NTA column (GE Health Sciences) in the presence of GuHCl [50 mM Tris, 150 mM NaCl, and 6 M GuHCl (pH 6.8)]. The bound protein was washed [50 mM Tris, 150 mM NaCl, and 40 mM imidazole (pH 6.8)] and eluted [50 mM Tris, 150 mM NaCl, and 400 mM imidazole (pH 6.8)] in the absence of GuHCl. Proteins were further purified by gel filtration chromatography using a Sephacryl S-300 HP 16-60 column (GE Health Sciences) to isolate the monodisperse protein and to remove imidazole from the protein storage buffer [50 mM Tris and 150 mM NaCl (pH 6.8)].
The GFP::RTX and RTX::GFP constructs were synthesized by Genescript and subsequently subcloned into a T7-regulated pET vector for expression in E. coli with an N-terminal six-His tag. The protein was expressed as described for the RTX domain alone. The soluble fraction of the GFP::RTX and RTX::GFP protein was purified under native conditions after lysis by sonication and separation by centrifugation [40000 relative centrifugal force (RCF) for 30 min]. The purification was accomplished using the buffers described for the RTX protein without the GuHCl.
Calcium-Induced RTX Folding
Calcium-induced RTX folding was accomplished as previously described.17 Purified RTX protein was incubated in refolding buffer (50 mM Tris and 150 mM NaCl) in the presence or absence of Ca2+ for 15 min on ice. The refolded protein was separated from any misfolded species by centrifugation (21000 RCF at 4 °C for 10 min) or centrifugal filtration [Millipore Ultracel 100 kDa molecular weight cutoff (MWCO) filters]. The Ca2+-induced RTX folding was confirmed by circular dichroism (CD) spectroscopy and analytical gel filtration chromatography.
Analytical Gel Filtration
Protein samples were injected onto a Tosoh GC-PAK 200 gel filtration column that was pre-equilibrated with buffers in the presence or absence of Ca2+.17 Sample elution was monitored by UV absorbance at 220 and 280 nm using a Shimadzu Prominence high-performance liquid chromatograph. Buffers contained 50 mM Tris and 150 mM NaCl (pH 6.8) and were supplemented with 2 mM Ca2+ or Mg2+.
CD Spectroscopy
RTX folding was monitored as a function of protein secondary structure by circular dichroism spectroscopy using a Jasco 810CD spectrophotometer.17 Spectra were collected with protein concentrations between 4 and 6 μM at 25 °C using a 1 mm path length cuvette. Buffers contained 50 mM Tris and 150 mM NaCl (pH 6.8) with or without 2 mM Ca2+ or Mg2+.
RTX Fibril Formation and Characterization
Amyloid formation occurred spontaneously at protein concentrations above ∼5–10 μM in subsaturating Ca2+. Amyloid formation was followed using thioflavin-T (ThT) fluorescence, which shows a change in emission upon binding amyloid structures. Fluorescence was measured as a function of ThT emission after excitation at 450 nm. For kinetic measurements, emission was monitored at 482 nm; steady-state emission spectra were collected between 460 and 550 nm. For kinetic aggregation experiments, the RTX protein was first concentrated and then cleared by centrifugation or filtration to remove any preformed aggregates that may have appeared during purification or storage. To evaluate the unfolding and subsequent aggregation of the RTX protein, the protein was first diluted into buffers containing 1–2 mM Ca2+ and refolded, as described above, at a final concentration of 10–15 μM. Proteins were then mixed with ThT-containing buffer with Ca2+ or EGTA, and reaction mixtures were transferred to 96-well plates (Costar). The plates were sealed with optical tape for the experiments. Data were collected on a BioTek Synergy 4 multimode plate reader in kinetic mode, with constant shaking between reads. Experiments were read over the course of 24–72 h.
Production of RTX Sheets
RTX sheets were produced by centrifugal concentration of the apo RTX protein in buffers lacking Ca2+ [50 mM Tris and 150 mM NaCl (pH 6.8–7.2)]. Proteins were concentrated using Millipore Ultracel 10 kDa MWCO filters spun at 4000 RCF and 4 °C. Sheets formed spontaneously on the centrifugal concentrator membrane support. Sheets were removed from the support by inversion of the concentrator and/or gentle pipetting and transferred for storage at 4 °C. Alternatively, sheets were produced using an Amicon Ultrafiltration Cell (Millipore) with a 30 kDa MWCO poly(ether sulfone) membrane (Pall) under nitrogen pressure. Sheets formed spontaneously on the membrane filter support and could be displaced by vigorous pipetting.
Analysis of Protein in the Amyloid Structure
RTX polymers were formed in the absence of Ca2+ by incubation overnight at 25 °C and concentration in centrifugal concentrators (Amicon Ultra, 3 kDa MWCO, Millipore). The large polymers were collected from the soluble fraction by brief centrifugation (1000 RCF). This insoluble material was then washed in Ca2+-containing buffer to remove loosely associated or unincorporated RTX protein. The insoluble material from each wash was collected by low-speed centrifugation. After the final wash, the remaining insoluble material was resuspended in buffer and sonicated extensively to break up the protein polymers.18 Samples of the original insoluble material, each of the washes, and the final sonicated material were resuspended in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) sample buffer for analysis by Western blotting. Western blots were performed using the α-RTX antibody described previously.19
Negative Stain Electron Microscopy of Fibrillar Structures
Sample volumes of 3.5 μL were applied to 400 mesh carbon-coated copper grids (EM Sciences) that had been freshly glow-discharged. Grids were lightly blotted, washed in purified water, and stained with 4% uranyl acetate. Grids were visualized with an FEI Tecnai 12 transmission electron microscope operating at 120 kV, and images were recorded on Kodak SO-163 film at 30000× nominal magnification or on a Gatan Ultrascan 1000 CCD camera at 42000× nominal magnification.
Microscopy of RTX Sheets
RTX sheets were separated from soluble protein by centrifugation at 1000 RCF. The sheets were stained with ThT and washed repeatedly in PBS to remove excess dye and soluble RTX protein. The stained sheets were transferred to a MatTek glass-bottom dish for analysis by fluorescence microscopy. Images were acquired using an Olympus Fluoview-1000 inverted microscope on sheets suspended in PBS without fixation. Image stacks were processed using ImageJ.20
Scanning Electron Microscopy (EM) Analysis of RTX Sheets
RTX sheets were fixed in 2.5% glutaraldehyde for 1 h at room temperature and then rinsed several times in 1× PBS. Next, the RTX sheets underwent secondary fixation in 1% OsO4 for 1 h. After several PBS washes, RTX sheets were dehydrated in a graded ethanol series of 15 min washes from 30 to 100%. A final 15 min wash of hexamethyldisilizane (HMDS) was used to further dehydrate the RTX sheets. RTX sheets were then removed from the HMDS, immediately placed on aluminum SEM stubs, and allowed to air-dry. Samples were then sputter-coated with a 3.5 nm gold/palladium coating (Cressington Auto 108) and viewed in a JEOL JSM-6335F scanning electron microscope at 3 kV.
Results
Folding and Misfolding of the RTX Domain of Alkaline Protease
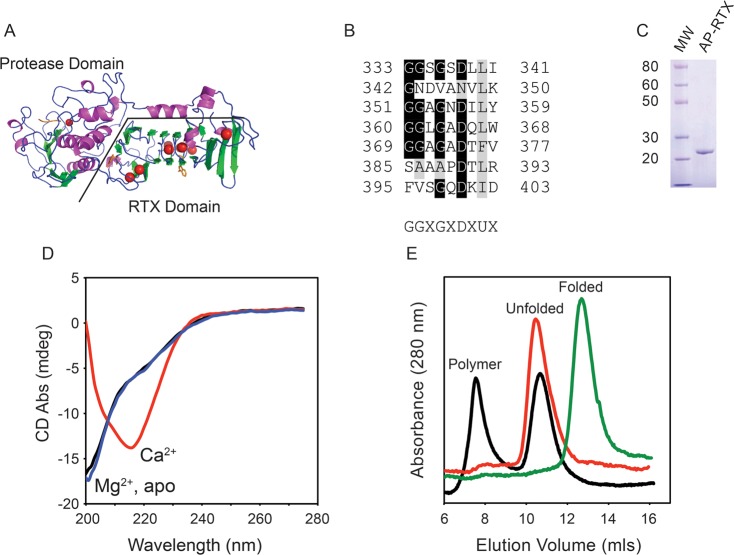
Alkaline protease is a two-domain protein containing an N-terminal proteolytic domain and a C-terminal Ca2+-binding RTX domain (Figure 1A).21 Previous studies have shown that Ca2+ binding in the conserved RTX domain is coupled to the folding and activation of the protease domain, putatively providing a means of protease regulation during biosynthesis and secretion.17 This structural regulation is controlled by multiple Ca2+ ions that are coordinated by a nonapeptide repeat that is the hallmark of the RTX family of proteins. These repeats are enriched with glycine and aspartate residues, which serve to coordinate the bound Ca2+ ions (Figure 1B).21,22
Figure 1.
Calcium-regulated conformation of the RTX domain. The calcium-dependent conformational changes of the alkaline protease domain from P. aeruginosa were evaluated in vitro. (A) Cartoon of the determined structure of alkaline protease. The N-terminal protease domain and a C-terminal, β-helix RTX domain are indicated. Calcium ions are shown as red spheres. (B) Individual RTX nonapeptide repeats from the Pseudomonas alkaline protease after their alignment. The nonapetide consensus sequence, G-G-X-G-X-(D/N)-X-U-X, is shown below, where X is any amino acid and U is a hydrophobic amino acid. (C) Representative Coomassie blue-stained SDS–PAGE gel of the purified RTX protein showing a single predominant species consistent with an unfolded monomer of RTX. (D) Representative CD spectra of the RTX protein refolded in the presence (red) and absence (black) of Ca2+ and in the presence of Mg2+ (blue). (E) Representative chromatogram from analytical gel filtration of the Ca2+-free (red) and Ca2+-bound (green) RTX protein immediately after refolding. Data for the RTX protein in the absence of Ca2+ after incubation for 24 h are colored black. The elution peaks for the folded, unfolded, and polymer states are labeled.
Our previous work suggested that the RTX domain could form amyloid-like structures that bound ThT, though the nature of these structures and their assembly were not characterized.17 To further explore the Ca2+-free properties of the RTX domain, these polymeric structures, and their modes of assembly, the RTX protein was expressed in E. coli and purified for in vitro structural studies. The RTX protein was expressed abundantly and could be purified to homogeneity as judged by Coomassie blue staining of SDS–PAGE gels (Figure 1C). Typical yields were 5–8 mg of purified protein/L of expression culture. Elution from preparative sizing columns in the absence of Ca2+ showed the protein eluted in an unfolded conformation with a predicted hydrodynamic radius larger than that expected for the RTX sequence, consistent with previous reports of Ca2+-dependent RTX folding.15,17 Notably, during purification, little to no RTX protein was found in the column void (>650 kDa), suggesting wide-scale aggregation was not associated with these altered hydrodynamic properties under purification conditions.
To characterize the unfolded state and confirm the conformational regulation by Ca2+ binding, the RTX protein was evaluated by circular dichroism spectroscopy. Previous work has shown that, in the absence of Ca2+, the purified soluble RTX protein adopts a predominantly random coil conformation, consistent with that reported for one of the CyaA RTX domains.15−17 For refolding experiments, the RTX protein was rapidly diluted into buffer in the presence or absence of divalent cations. The refolding reaction mixture was then cleared by centrifugation to remove misfolded species. Previous studies have demonstrated that this refolding procedure is highly efficient and yields >85–90% folded protein.17
When diluted into buffers without Ca2+, the RTX protein appeared to adopt a predominantly random coil conformation, with a strong absorbance minimum near 200 nm (Figure 1D). A small shoulder of absorbance was routinely observed near 220 nm, suggesting some limited residual structure was likely present in the sample. When saturating (>500 μM) Ca2+ was present in the buffer, the CD spectrum showed a single predominant absorbance minimum at 217 nm, consistent with the RTX domain folding into a β-rich structure. This shift in CD absorbance is also consistent with secondary structures of the RTX domains seen in the crystal structure of alkaline protease and related RTX proteases and the Ca2+-regulated folding of other RTX domains.15,17,23 To evaluate the specificity of the Ca2+-induced secondary structural changes, protein was refolded in the presence of Mg2+. When samples were diluted into buffers with 2 mM Mg2+, the CD spectra appeared to be unchanged with both the 200 nm minimum and the 220 nm shoulder being observed. This suggested that the observed changes in secondary structure were the result of specific conformational changes resulting from Ca2+ binding.
Additional conformational changes were evident with the RTX domain in the absence of Ca2+. After overnight incubation at 4 °C or after concentration beyond ∼5–10 μM, the RTX domain appeared to form polymers in solution. Overnight incubation of the RTX domain in the absence of Ca2+ showed a time-dependent formation of protein polymers by analytical gel filtration chromatography. Refolding in the presence and absence of Ca2+ was initially accomplished after preclearing the apo RTX protein solution by centrifugation to remove protein aggregates formed during purification or storage (Figure 1E, red and green traces). After centrifugation and in the presence of Ca2+, the RTX protein eluted as a single predominant peak at 13.2 mL, consistent with a compact, folded protein with an apparent molecular weight of roughly 20–25 kDa, as determined by known molecular weight standards. This elution profile was consistent with previous measurements of Ca2+-bound RTX protein.17 The apoprotein eluted at approximately 10.5 mL as a single symmetrical peak. This change in elution volume was consistent with our previous reports of the RTX domain folding and the extended, random coil conformation seen in the CD spectra.17 Analytical gel filtration of the RTX protein refolded in the presence of 2 mM Mg2+ showed no differences from that of the apoprotein (data not shown). These data were consistent with the Ca2+ specific changes in secondary structure measured by CD.
Analytical gel filtration of the apo RTX protein, after overnight incubation at 4 °C, indicated a population of the protein was spontaneously forming oligomers or polymers. When the uncleared apoprotein was evaluated in the absence of Ca2+, two predominant peaks were seen by analytical gel filtration (Figure 1E, black trace). The Ca2+-free, extended conformation of the RTX domain was seen at ∼10.5 mL, consistent with the peak seen after centrifugation. In addition, a population of the protein eluted in the column void volume at ∼7.8 mL, consistent with RTX polymerization. The appearance and stability of the void peak occurred spontaneously and did not require heating, freeze–thaw cycles, or other perturbations after preparation. This peak was not observed immediately after purification or refolding, as the monodisperse RTX monomer was isolated by preparative gel filtration chromatography and was the initial starting material. Additionally, no large-scale flocculent precipitation was seen under these experimental conditions. No changes in the Ca2+-bound RTX protein were observed after similar overnight incubation at 4 °C (data not shown).
Predicted Self-Assembly Sequences within the RTX Domain
β-Rich structures are often associated with protein aggregation.24 Given the β-content of the RTX sequence and the observation of spontaneous association in the absence of flocculent precipitation, computational evaluation of the RTX sequence was performed to assess the amyloid forming tendencies of the sequence. The RTX sequence was evaluated using a consensus web-based tool, AmylPred, which utilizes multiple parallel prediction algorithms to define regions of sequence that may contribute to amyloid or amyloid-like packing and association.25 Evaluation of the RTX primary sequence suggests that multiple regions within the domain show potential to form amyloid or amyloid-like structures (Figure 2). Eight distinct peptide sequences of five or more amino acids are predicted to potentially form amyloid structure (Figure 2, saw teeth). These regions were found throughout the RTX domain and did not appear to be fully correlated with the specific β-structures formed in the native RTX domain, though significant overlap is seen with predicted amyloid-forming sequences and determined regions of β-structure (Figure 2, boxes). Similarly, the predicted amyloid-forming regions did not fully correlate with the nonapeptide repeats (Figure 2, asterisks). Four of the predicted amyloid-forming sequences were located within the C-terminal “capping region” of the RTX domain, which is thought to help stabilize the Ca2+-bound fold but does not directly bind Ca2+ (spanning residues 410–421, 429–435, 442–447, and 455–461). The lengths of the putative amyloid-forming sequences ranged from 6 to 12 amino acids.
Figure 2.
Secondary structure and aggregation propensities of the RTX domain. The sequence of the RTX domain from P. aeruginosa alkaline protease is shown. Secondary structure elements are shown above the sequence with β-structure indicated by bars and α-structure indicated by curves. Sequences of five or more amino acids predicted to form amyloid structure are indicated as saw teeth below the protein sequence. Asterisks indicate the diglycine motifs present in the RTX nonapeptide repeats. The indicated residue numbering and secondary structure assignment are consistent with the X-ray structures from Protein Data Bank entries 1KAP and 1JIW.
Regulated Assembly of the Apo RTX Domain
Visual inspection of tubes containing the apo RTX domain incubated overnight suggested that the protein was polymerizing in solution, as small deposits could been seen on the walls and base of the plastic storage tubes. However, large-scale flocculent precipitation was not evident after prolonged incubation in the absence of Ca2+ at all concentrations tested (∼1–30 μM). However, after inversion or vortexing, noticeable deposits adhered to the sides of tubes containing the apo RTX protein. These deposits were not seen when a millimolar Ca2+ concentration was included in the refolding buffer after prolonged incubation times (>7–10 days), suggesting that these conformational changes were arising from the apo state (data not shown). Given the natural β-forming propensities of the RTX sequences and the prediction of amyloid-forming sequences within the RTX domain, we sought to characterize these RTX polymers further using thioflavin-T (ThT) binding. Thioflavin-T has previously been used to identify and characterize the presence of amyloid structures with a variety of unrelated proteins.26−28 Spectral and quantal shifts in ThT fluorescence occur when the dye binds amyloid structures.
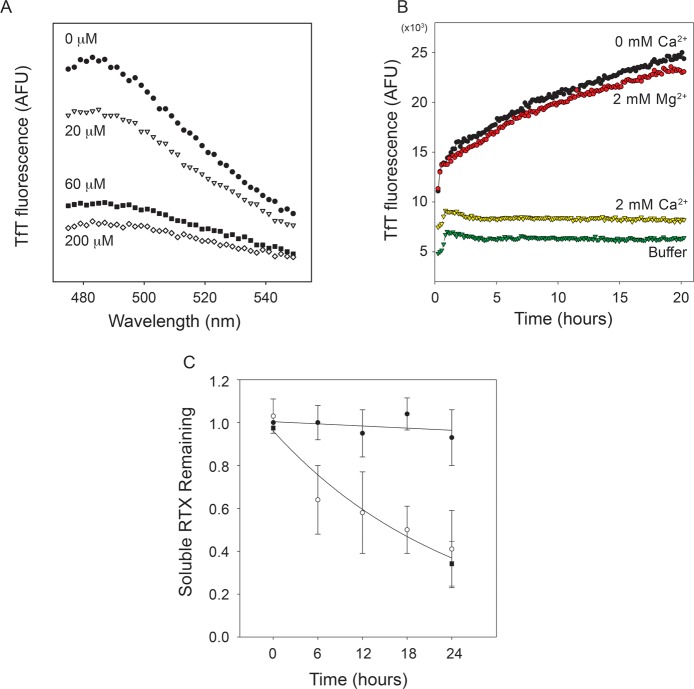
To test whether amyloid or amyloid-like structures were being formed by the apo RTX domain, ThT was included in refolding reactions with multiple Ca2+ concentrations. For these experiments, the RTX protein was refolded at 10 μM, a concentration that promoted competing reactions of folding into a soluble monomer and association into polymers. The proteins were incubated in Ca2+, and emission spectra were collected to evaluate ThT incorporation and alterations in the fluorescence of the dye (Figure 3A). Our previous work has demonstrated that the RTX domain cooperatively binds Ca2+ with an apparent affinity of 55 μM and a Hill coefficient of at least 2.8.17 In the presence of 200 μM Ca2+, a concentration sufficient to fold the RTX domain, the RTX protein showed minimal ThT fluorescence at 485 nm, the reported spectral peak for amyloid-bound ThT.26 As Ca2+ concentrations were reduced, the peak intensity at 485 nm increased, consistent with the formation of amyloid or amyloid-like structures in subsaturating Ca2+. The coincident increase in the level of RTX folding and the decrease in ThT fluorescence were consistent with the model of competing RTX folding pathways. These data suggested that the conformation that had amyloid-like character and bound ThT originated from and/or was dependent on the presence of apo RTX. Inclusion of Mg2+ in refolding buffers did not decrease ThT fluorescence (data not shown), suggesting the alterations in RTX polymerization and ThT binding were Ca2+ specific.
Figure 3.
Characterization of RTX polymers. The Ca2+-dependent aggregation of the RTX domain was evaluated using ThT fluorescence. (A) The purified RTX domain was refolded into buffers containing increasing concentrations of Ca2+, and emission spectra of ThT fluorescence were collected after overnight incubation. Increasing Ca2+ concentrations resulted in decreasing maximal emission intensities [0 (●), 20 (▽), 60 (■), and 200 μM Ca2+ (◇)]. Background fluorescence of buffer without RTX protein was subtracted from each of the emission spectra. (B) The kinetics of ThT fluorescence were monitored by emission at 482 nm as a function of time in the presence or absence of Ca2+ and Mg2+. RTX protein in the absence of Ca2+ showed an increase in ThT fluorescence (black circles), as did the protein incubated in Mg2+ (red circles). This increase in TfT fluorescence was not observed with the Ca2+-bound protein (yellow triangles) or in buffer controls (green triangles). (C) Analytical gel filtration was used to assess the soluble, monomeric RTX protein in solution through the time course of aggregation. The level of soluble RTX protein in the absence of Ca2+ decreased as a function of incubation time (○), as did the level of protein incubated in 2 mM Mg2+ (■). Calcium-bound (2 mM) RTX protein remained soluble and monomeric over the incubation times tested (●).
To evaluate the kinetics of the RTX-induced ThT fluorescence, the purified RTX proteins were incubated in buffers containing ThT in the presence or absence of Ca2+ and Mg2+ and fluorescence was measured kinetically. In the presence of 2 mM Ca2+, the RTX protein showed ThT fluorescence similar to that of buffer controls (Figure 3B, green and yellow triangles). After transient changes in fluorescence associated with thermal equilibration and mixing, no observable changes in ThT fluorescence were seen over the time course of 20–24 h. In the absence of Ca2+, ThT fluorescence showed a dramatic increase in fluorescence intensity (Figure 3B, black circles). An initial, rapid rise in fluorescence slowed with time, suggesting depletion of some component in the reaction. This apparent rise to maximum was seen in all kinetic experiments. The increase in fluorescence intensity was dependent on the presence of Ca2+-free protein, as the Ca2+-saturated protein showed little to no change in ThT fluorescence over the time courses and protein concentrations evaluated. Inclusion of 2 mM Mg2+ showed negligible effects on ThT fluorescence compared to that of the Ca2+-free sample (Figure 3B, red circles). These data provide additional evidence that the Ca2+-induced folding seen by CD and gel filtration protected the RTX from polymerization and was Ca2+ specific.
To confirm that the RTX domains were associating and that this assembly was correlated with the changes in ThT fluorescence, analytical gel filtration and UV absorbance were used to assess changes in the concentrations of soluble RTX protein during aggregation experiments. Samples from aggregation experiments were removed at regular intervals and cleared by high-speed centrifugation to remove the polymeric structures. The soluble, monomeric RTX protein was then evaluated by analytical gel filtration (Figure 3C). In the presence of Ca2+, the relative quantity of soluble RTX protein showed little aggregation, with the peak for the monomeric RTX showing minimal changes in area. In contrast, the soluble, Ca2+-free RTX protein decreased as a function of incubation time. At 6 h, approximately 65% of the Ca2+-free RTX protein remained in solution, as measured by the area of the elution peak. The decrease in the level of RTX protein continued throughout the incubation period, with the peak area decreasing to roughly 40% of the starting area at 24 h. Similarly, protein incubated at millimolar Mg2+ concentrations showed a similar decrease in soluble protein as a function of time, consistent with incorporation of RTX protein into the insoluble fraction. These data demonstrated that the RTX protein was polymerizing into higher-order structures and that these association events correlated with the measured changes in ThT fluorescence.
The observed polymerization and ThT binding experiments suggested that the non-native amyloid-like polymer could be produced from the random coil, apo state of the RTX protein. The ability of Ca2+ to inhibit the formation of this species suggested that these processes might be controllable, in vitro, by the addition or removal of Ca2+. To evaluate the control of RTX polymerization, a series of kinetic experiments in which Ca2+ was added or removed by chelation and ThT binding was evaluated fluorescently were performed. The RTX protein was first refolded in the presence of 1 mM Ca2+, and the evolution of ThT fluorescence was followed as a function of time. As before, the presence of Ca2+ inhibited the formation of ThT-binding structures for extended periods of time (Figure 4A, yellow and magenta lines). After incubation, EGTA was added to chelate free Ca2+ from the RTX buffers. As the Ca2+-induced RTX folding is reversible, complete chelation of free Ca2+ should rapidly facilitate the unfolding of the RTX domain and lead to its association, as measured by ThT binding.17 Addition of 1 mM EGTA at multiple time points in the experiment resulted in a rapid increase in ThT fluorescence (Figure 4A, arrowheads). These data confirmed that ThT-binding conformations of the RTX domain arose from the apo state and could be induced by Ca2+ chelation from the folded, Ca2+-bound conformation.
Figure 4.
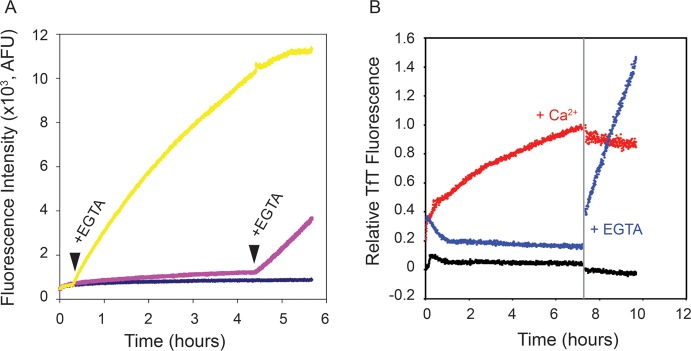
Conformational dependence of RTX aggregation. (A) Ca2+-bound RTX protein was evaluated by ThT binding kinetically, and free Ca2+ was chelated by addition of EGTA. Points of EGTA addition are labeled and indicated by arrowheads. (B) The stability of the ThT-bound, Ca2+-free species was assessed by addition of Ca2+. The vertical gray bar marks the addition of Ca2+ (red data set) or EGTA (blue data set). Data for native RTX protein in 2 mM Ca2+ (black data set) are shown as a control.
The reversibility of the apparent amyloid formation and the increase in ThT fluorescence were evaluated by the addition of Ca2+ to kinetic reaction mixtures in which ThT-binding structures were preformed. After a period of incubation, either Ca2+ or EGTA was added to alter the conformation of the RTX domains in solution (Figure 4B). Incubation of the RTX domain in the absence of Ca2+ resulted in increased ThT fluorescence (Figure 4B, red circles). Addition of 1 mM Ca2+ to this reaction rapidly arrested the increase in ThT fluorescence. Prolonged incubation of this reaction mixture showed minimal changes in fluorescence despite the high concentrations of free Ca2+. As a control, the Ca2+-bound protein was treated with EGTA, as in Figure 4A, and a rapid increase in ThT fluorescence was seen (Figure 4B, blue circles). Changes in buffer fluorescence were not significant after addition of either Ca2+ or EGTA (data not shown). These data provided further evidence that the changes in ThT fluorescence were associated with protein conformations arising from the apo state. The Ca2+-induced folding of the RTX domain effectively limits the formation of these ThT-binding conformations. Further, once formed, the ThT-bound structures appeared to be highly stable in the presence of Ca2+, as the addition of Ca2+ to the preformed polymers did not appear to alter the ThT fluorescence over the time courses evaluated.
Multimodal Assembly of Non-native RTX Protein
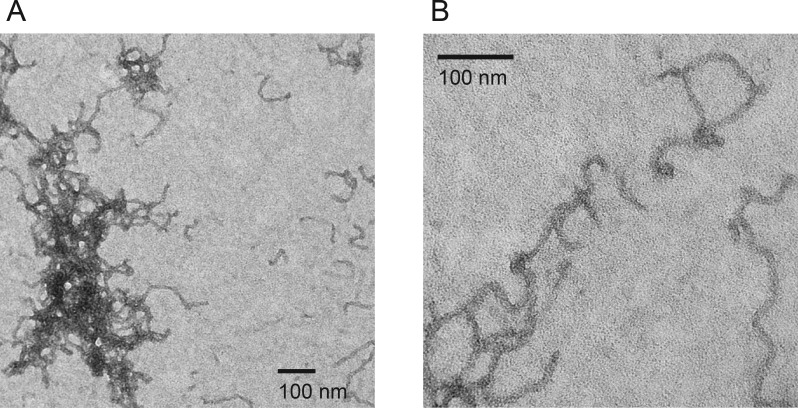
To further evaluate the non-native RTX structures that bound ThT, the protein was incubated in the absence of Ca2+ and cleared by centrifugation. The insoluble material was collected, washed, and imaged by negative stain transmission electron microscopy (Figure 5A,B), revealing structures resembling amyloid fibrils and protofibrils. The fibrillar structures grew in length to 300–500 nm and beyond and showed a relatively uniform diameter of ∼11 nm. The structures appeared to be unbranched, but many were kinked or bent. While individual fibrillar structures could be observed, a majority of the material appeared to be intertwined or tangled.
Figure 5.
Electron micrographs of RTX fibrils. Negative stain electron microscopy was used to visualize the structures formed by the Ca2+-free RTX domain. (A) Electron micrograph of the apo RTX domain fibrils. The RTX protein appears as regular fibrillar structures with a uniform width, but varying lengths and shapes. Nets or tangles of fibrils were most often seen, though individual fibrils were also visualized. (B) Electron micrograph of individual fibrillar structures. Fibrils appeared to be unbranched but often tangled and kinked.
In addition to the fibrillar structures observed by EM, large protein structures were observed forming while the RTX protein was being concentrated in the absence of Ca2+ using centrifugal concentrators. In the absence of Ca2+ and with protein concentrations above ∼5–8 μM, translucent sheetlike structures as large as ∼5 cm2 formed, though they were routinely smaller and could be readily observed by eye in solution. The apparent upper bound of sheet size was limited by the size of the solid support upon which the RTX protein was deposited.
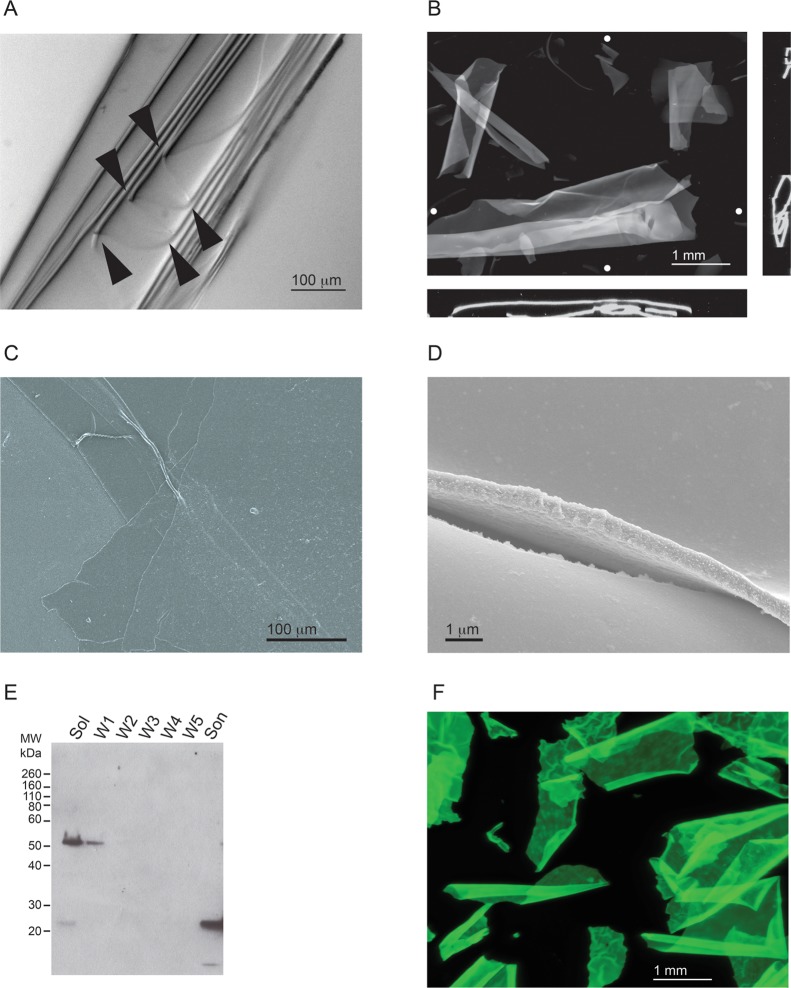
The sheets were imaged by bright-field and fluorescence microscopy after staining with ThT and by scanning electron microscopy (SEM) (Figure 6). The sheets appeared to be highly regular and ordered by both bright-field (Figure 6A) and fluorescence microscopy (Figure 6B) and were observed to curl or fold upon themselves in solution along a single axis. Microscopy allowed the identification of discrete edges and folds, suggesting that the sheets were flexible. Similarly, fluorescence microscopy provided further evidence of large, sheetlike structures that tended to wrap or coil in solution. The natural tendency of the sheets to wrap or fold appeared to be dynamic and reversible with agitation of the solution. Orthogonal projections of image stacks showed the sheets often wrapped loosely with a relatively uniform sheet thickness across all cross sections analyzed. Low-magnification SEM also showed the sheets to be uniform and regular with folds, wrinkles, and other undulations consistent with the light microscopy images (Figure 6C). At higher magnifications, the surface of the sheets appeared to be highly regular and uniform with few small protrusions and irregularities (Figure 6D). The thickness was estimated to be between 500 and 600 nm on the basis of edge analyses of sheets imaged at high magnifications.
Figure 6.
Characterization of RTX-containing protein sheets. The RTX protein formed ordered polymers, which appeared as protein sheets and were visible in solution. (A) Representative image of a RTX sheet folded upon itself. Multiple folds of the sheet can be seen along an edge and are indicated by arrowheads. (B) Representative fluorescence microscopy image of sheets stained with ThT. Image stacks of RTX sheets in suspension were collected and analyzed. The RTX sheets were usually wrapped or folded along a single dimension. Orthogonal views (x–z, bottom; y–z, side) indicate the coiling or wrapping of the RTX sheets. The Z projection is shown as the sum of all stacks. White circles show the approximate positions of the vertical and horizontal orthogonal slices. (C) Scanning electron micrograph of RTX sheets at low magnification with visible folds and creases. (D) Representative image of the edge and surface of the RTX sheets at high magnification. The sheets appear to be uniform in thickness and along the edge of visualized surfaces. (E) Western blot of the material found in the RTX sheets that confirms the presence of the RTX protein. The starting material was pelleted by low-speed centrifugation and washed with refolding buffer. The supernatants were analyzed by Western blotting after each step in the procedure. Lanes are labeled as follows: Sol, soluble material after the first centrifugation step; W1–W5, successive washes of the centrifuged pellet; Son, final material after sonication. The blot was probed with a polyclonal α-RTX antibody. (F) RTX::GFP sheets were formed in the absence of Ca2+ and visualized by fluorescence microscopy. A Z projection representing the sum of all stack images is shown visualized using the intrinsic fluorescence of the GFP fusion.
To confirm that these polymeric sheets contained the RTX protein, the sheets were isolated by low-speed centrifugation and washed multiple times in reaction buffer containing Ca2+ before being sonicated. The sheets were first pelleted, and the supernatant was taken for Western blot analysis (Figure 6E, Sol). Multiple reactive bands were detected, with the prominent band having an apparent molecular weight of ∼52 kDa, roughly consistent with a dimer of RTX proteins and suggesting an SDS-resistant intermediate species may be forming along the polymerization pathway. A fraction of the protein was also detected at ∼25 kDa, consistent with the presence of a small quantity of RTX monomer. The high-molecular weight band, as well as the monomeric species, was washed from the sheets after centrifugation and exchange into reaction buffer with Ca2+ (Figure 6E, lanes W1–W5). After multiple successive washes to remove loosely associated and soluble RTX protein, the final pellet containing visible RTX sheets was resuspended and sonicated to break up the protein polymers. Analysis of the sonicated samples revealed the presence of monomeric RTX species by Western blotting, consistent with its presence in the polymeric material (Figure 6E, Son). Absorbance measurements of the sonicated material showed peaks consistent with peptide bonds and aromatic side chains (data not shown). No other significant absorbance peaks were observed.
The controllable formation of both amyloid-like fibrils and ordered sheets suggested that the RTX domain might be useful for the generation of biomaterials. To evaluate the potential that the RTX sequence could serve as a scaffold for the generation of large macromolecular structures, GFP chimeras were produced with both N- and C-terminal RTX fusions. GFP was chosen as it is a soluble protein that is easily monitored by the measurement of its intrinsic fluorescence. GFP::RTX and RTX::GFP chimeras were generated using the RTX domain and a short linker (GGSSG) fused to the N- or C-terminus of GFP. The entire domain was fused in frame to allow for the Ca2+-controllable transition from monomeric to polymeric species. The GFP::RTX protein was expressed poorly under the expression conditions tested. In contrast, the RTX::GFP protein was expressed robustly in E. coli and was subsequently purified by Ni-NTA and gel filtration chromatography. The soluble fraction of the RTX::GFP protein was purified in the presence of Ca2+ to promote the folding of the RTX domain and inhibit potential aggregation events during the purification process. Purified RTX::GFP protein yields were approximately 1.5–2 mg/L of culture.
Following purification, assembly of the amyloid-like fibrils and protein sheets was accomplished by dilution into Ca2+-free buffers or buffers containing EGTA. The proteins were then incubated in the absence of Ca2+ and deposited onto solid supports as described for the generation of RTX sheets. In the absence of Ca2+, the RTX::GFP protein formed large macromolecular polymers, similar to those of the RTX domain (Figure 6F). The RTX::GFP sheets appeared to be uniform in thickness on the basis of fluorescence microscopy analyses. The surfaces similarly appeared uniform, though slight irregularities could be seen in the chimeric sheets (Figure 6F). The surface irregularities were observed as small folds or puckering in the sheets. The polymeric structures were not observed in the presence of Ca2+, suggesting that the conformational dependence of assembly seen with the RTX domain was likely important for the assembly of the RTX::GFP chimera. As with the isolated RTX protein, the inclusion of Mg2+ had no discernible effects on the RTX::GFP protein behavior, suggesting the conformational regulation of polymerization was Ca2+ specific.
Discussion
The structures of RTX domains are highly dynamic and tightly regulated by Ca2+ binding to the canonical glycine- and aspartate-rich repeats.16,17,29,30 Previous work has focused on the structural transitions between the denatured and native conformations of RTX proteins from multiple virulence factors. Here we show that the association of apo RTX domains leads to the formation of polymeric molecular structures that share physical properties with amyloid fibrils. Deposition of these apo domains onto a solid support facilitates the generation of highly structured two-dimensional protein sheets. The assembly into fibrils or films was regulated by Ca2+-induced changes in protein conformation. Finally, fusion of the RTX domain to a soluble, globular protein facilitated the generation of protein fibrils and sheets that incorporated the biological function of the globular protein (GFP) into the protein assembly.
Consistent with previous reports, specific binding of Ca2+ to the RTX domain results in its folding and is not seen with other divalent cations (Figure 1).16,17 In the absence of Ca2+, the RTX domain is capable of adopting non-native conformations and self-assembles into polymeric species that resemble amyloid-like structures. These assemblies are predicted to be associated with multiple regions of the denatured RTX domain (Figure 2). These structures share similar ThT binding characteristics with amyloids (Figures 3 and 4) and grow as amyloid-like fibrils that are unbranched and exhibit a regular width (Figure 5). The apo RTX domain is also capable of forming large uniform sheets with a smooth surface and uniform thickness both alone and when fused N-terminally to a globular, soluble protein (Figure 6).
Self-assembly was previously reported for a synthetic β-helical peptide that was engineered using consensus RTX nonapeptide repeats.29 Unlike the RTX domain in this study, these peptides contained only the core Ca2+-binding nonapeptide repeats and lacked N- and C-terminal “capping” sequences. Further, the peptides were synthesized with non-native tryptophan residues that served as intrinsic structural probes. The folding and assembly of these synthetic RTX sequences showed key differences with the native domains reported here. The engineered peptides did not bind Ca2+ and did not undergo the disorder-to-order structural transition of the natural RTX domains, nor did they respond significantly to other divalent cations. In contrast, the presence of La3+ led to their self-assembly and the reversible formation of protein fibrils. The fibrillar structures were also unbranched and showed modest ThT binding properties. It is not clear how La3+ participates in the formation of the fibrillar structures in these engineered proteins and from what precursor structures these polymers are formed.
In contrast, we found that self-association of the AprA RTX domain was specifically inhibited by the presence of Ca2+. As such, assembly into either fibrils or sheets likely represents intermolecular interactions occurring from unsatisfied protein–protein or protein–ligand interactions in the unfolded ensemble. The mechanisms by which assembly is initiated in a single dimension (fibril) or in two dimensions (sheet) are yet to be established and warrant additional investigation. It is possible that the sheets represent a further assembly of laterally packed fibrils. Alternatively, the sheets may represent a fundamentally different polymer structure of the RTX domains. It is also not clear whether these pathways are mutually exclusive or if they are linked. However, it is clear that the macromolecular polymers emerge from the apo, unfolded ensemble that is tightly regulated and rapidly controlled, in vitro, by the addition and chelation of Ca2+ under mild ionic and pH conditions.
Similar polymerization under mild conditions has been reported for ultrabithorax (Ubx), a transcription factor from Drosophila that spontaneously forms films at an air–liquid interface.6 The ability to generate such films provides a potential template for a variety of biomaterials.31 The assembly of Ubx does not require harsh chemical treatments and/or prolonged incubation at high protein concentrations. Like that of Ubx, the polymerization of AprA RTX occurs rapidly and spontaneously under near-physiological conditions. Moreover, the demonstration that these scaffolds can incorporate soluble, folded proteins suggests that both scaffolds could be used to incorporate enzymatic or other native biological functions into large biomaterial polymers.31
The AprA RTX domain provides several unique properties for studying the structural transitions into the observed amyloid-like conformations. First, the association and assembly of RTX domains occur under very mild biochemical conditions. The assembly into both amyloid fibrils and protein sheets can be accomplished at near-neutral pH and under physiological salt and buffer conditions. In contrast, the formation of many amyloid structures, often, but not always, requires nonphysiological pH, nonphysiological salt conditions, and/or organic solvents.6,28 Second, this assembly occurs rapidly, occurring over the time course of tens of minutes to hours. Finally, the generation of these macromolecular assemblies can be tightly controlled by the addition and chelation of Ca2+. This Ca2+-regulated switch between structural states allows for the purification of monomeric proteins followed by the removal of Ca2+ and the initiation of assembly. The regulation of these structural transitions may also occur in biological systems and facilitate bacterial infection.
From a biological perspective, protein self-association and the formation of functional amyloids contribute to a variety of bacterial adhesion and biofilm properties.3,4,32 The transition from a soluble monomeric protein to a “sticky” protein polymer may serve to facilitate bacterial adhesion to both biotic and abiotic surfaces. In addition, the formation of these large macromolecular structures may contribute to the protein components of the biofilm matrix. Given the dynamic structural properties of the RTX domains and the involvement of the RTX proteins in bacterial virulence, it is possible that these alternative conformations may also serve a biological function and contribute to previously unappreciated modes of pathogen virulence. The self-association of these domains may serve as a scaffold for native “biomaterials” that were evolved to facilitate pathogen virulence but could similarly be leveraged for other novel biomedical and biological applications.
Conclusions
The RTX domain from P. aeruginosa is conformationally dynamic and regulated by binding to Ca2+ cofactors. The apo-state domain is capable of self-assembly and forms highly ordered protein structures, including fibrillar amyloid-like structures and two-dimensional sheets. These assembly events can be tightly controlled by the addition and chelation of Ca2+ and can be transferred to fusion proteins that incorporate novel biological function into these macromolecular assemblies under very mild conditions. This approach demonstrates that the RTX domain can be used as a scaffold for the production of protein-based biomaterials that include enzymatic or other native-state protein properties.
This work was supported, in part, by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK083284 to P.H.T. and Faculty Development Funding from The University of Pittsburgh School of Medicine.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Sela M.; Anfinsen C. B.; Harrington W. F. (1957) The correlation of ribonuclease activity with specific aspects of tertiary structure. Biochim. Biophys. Acta 26(3), 502–512. [DOI] [PubMed] [Google Scholar]
- King J.; Haase-Pettingell C.; Robinson A. S.; Speed M.; Mitraki A. (1996) Thermolabile folding intermediates: Inclusion body precursors and chaperonin substrates. FASEB J. 10(1), 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M. R.; Robinson L. S.; Pinkner J. S.; Roth R.; Heuser J.; Hammar M.; Normark S.; Hultgren S. J. (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295(5556), 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P.; Nielsen J. L.; Dueholm M. S.; Wetzel R.; Otzen D.; Nielsen P. H. (2007) Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9(12), 3077–3090. [DOI] [PubMed] [Google Scholar]
- McGlinchey R. P.; Gruschus J. M.; Nagy A.; Lee J. C. (2011) Probing fibril dissolution of the repeat domain of a functional amyloid, Pmel17, on the microscopic and residue level. Biochemistry 50(49), 10567–10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer A. M.; Huang Z.; Oriakhi A.; Lu Y.; Lou J.; Matthews K. S.; Bondos S. E. (2009) The Drosophila transcription factor ultrabithorax self-assembles into protein-based biomaterials with multiple morphologies. Biomacromolecules 10(4), 829–837. [DOI] [PubMed] [Google Scholar]
- Kasotakis E.; Mitraki A. (2013) Designed self-assembling peptides as templates for the synthesis of metal nanoparticles. Methods Mol. Biol. 996, 195–202. [DOI] [PubMed] [Google Scholar]
- Woolfson D. N. (2010) Building fibrous biomaterials from α-helical and collagen-like coiled-coil peptides. Biopolymers 94(1), 118–127. [DOI] [PubMed] [Google Scholar]
- Jahn T. R.; Radford S. E. (2008) Folding versus aggregation: Polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 469(1), 100–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo J.; Murgier M.; Filloux A.; Lazdunski A. (1990) Cloning of the Pseudomonas aeruginosa alkaline protease gene and secretion of the protease into the medium by Escherichia coli. J. Bacteriol. 172(2), 942–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepelaire P.; Wandersman C. (1990) Protein secretion in Gram-negative bacteria. The extracellular metalloprotease B from Erwinia chrysanthemi contains a C-terminal secretion signal analogous to that of Escherichia coli α-hemolysin. J. Biol. Chem. 265(28), 17118–17125. [PubMed] [Google Scholar]
- Letoffe S.; Delepelaire P.; Wandersman C. (1991) Cloning and expression in Escherichia coli of the Serratia marcescens metalloprotease gene: Secretion of the protease from E. coli in the presence of the Erwinia chrysanthemi protease secretion functions. J. Bacteriol. 173(7), 2160–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch R. A.; Forestier C.; Lobo A.; Pellett S.; Thomas W. Jr.; Rowe G. (1992) The synthesis and function of the Escherichia coli hemolysin and related RTX exotoxins. FEMS Microbiol. Immunol. 5(1–3), 29–36. [DOI] [PubMed] [Google Scholar]
- Butterworth M. B.; Zhang L.; Heidrich E. M.; Myerburg M. M.; Thibodeau P. H. (2012) Activation of the epithelial sodium channel (ENaC) by the alkaline protease from Pseudomonas aeruginosa. J. Biol. Chem. 287(39), 32556–32665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez A. C.; Karst J. C.; Davi M.; Guijarro J. I.; Ladant D.; Chenal A. (2010) Characterization of the regions involved in the calcium-induced folding of the intrinsically disordered RTX motifs from the Bordetella pertussis adenylate cyclase toxin. J. Mol. Biol. 397(2), 534–549. [DOI] [PubMed] [Google Scholar]
- Chenal A.; Guijarro J. I.; Raynal B.; Delepierre M.; Ladant D. (2009) RTX calcium binding motifs are intrinsically disordered in the absence of calcium: Implication for protein secretion. J. Biol. Chem. 284(3), 1781–1789. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Conway J. F.; Thibodeau P. H. (2012) Calcium-induced Folding and Stabilization of the Pseudomonas aeruginosa Alkaline Protease. J. Biol. Chem. 287(6), 4311–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saborio G. P.; Permanne B.; Soto C. (2001) Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411(6839), 810–813. [DOI] [PubMed] [Google Scholar]
- Butterworth M. B.; Zhang L.; Liu X.; Shanks R. M.; Thibodeau P. H. (2014) Modulation of the Epithelial Sodium Channel (ENaC) by Bacterial Metalloproteases and Protease Inhibitors. PLoS One 9, e100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A.; Rasband W. S.; Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9(7), 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann U.; Wu S.; Flaherty K. M.; McKay D. B. (1993) Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: A two-domain protein with a calcium binding parallel β roll motif. EMBO J. 12(9), 3357–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann U. (1994) Crystal structure of the 50 kDa metallo protease from Serratia marcescens. J. Mol. Biol. 242(3), 244–251. [DOI] [PubMed] [Google Scholar]
- Angkawidjaja C.; You D. J.; Matsumura H.; Kuwahara K.; Koga Y.; Takano K.; Kanaya S. (2007) Crystal structure of a family I.3 lipase from Pseudomonas sp. MIS38 in a closed conformation. FEBS Lett. 581(26), 5060–5064. [DOI] [PubMed] [Google Scholar]
- Richardson J. S.; Richardson D. C. (2002) Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. U.S.A. 99(5), 2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frousios K. K.; Iconomidou V. A.; Karletidi C. M.; Hamodrakas S. J. (2009) Amyloidogenic determinants are usually not buried. BMC Struct. Biol. 9, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVine H. III (1999) Quantification of β-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 309, 274–284. [DOI] [PubMed] [Google Scholar]
- Liu C. W.; Giasson B. I.; Lewis K. A.; Lee V. M.; Demartino G. N.; Thomas P. J. (2005) A precipitating role for truncated α-synuclein and the proteasome in α-synuclein aggregation: Implications for pathogenesis of Parkinson disease. J. Biol. Chem. 280(24), 22670–22678. [DOI] [PubMed] [Google Scholar]
- Chiti F.; Dobson C. M. (2009) Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 5(1), 15–22. [DOI] [PubMed] [Google Scholar]
- Lilie H.; Haehnel W.; Rudolph R.; Baumann U. (2000) Folding of a synthetic parallel β-roll protein. FEBS Lett. 470(2), 173–177. [DOI] [PubMed] [Google Scholar]
- Linhartova I.; Bumba L.; Masin J.; Basler M.; Osicka R.; Kamanova J.; Prochazkova K.; Adkins I.; Hejnova-Holubova J.; Sadilkova L.; Morova J.; Sebo P. (2010) RTX proteins: A highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 34(6), 1076–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.; Salim T.; Brawley A.; Patterson J.; Matthews K. S.; Bondos S. E. (2011) Functionalization and patterning of protein-based materials using active Ultrabithorax. Adv. Funct. Mater. 21(14), 2633–2640. [Google Scholar]
- Dueholm M. S.; Petersen S. V.; Sonderkaer M.; Larsen P.; Christiansen G.; Hein K. L.; Enghild J. J.; Nielsen J. L.; Nielsen K. L.; Nielsen P. H.; Otzen D. E. (2010) Functional amyloid in Pseudomonas. Mol. Microbiol. 77(4), 1009–1020. [DOI] [PubMed] [Google Scholar]