Abstract
Background
Genes of the Raf family encode kinases that are regulated by Ras and mediate cellular responses to growth signals. Recently, it was shown that activating mutations of BRaf are found with high frequency in human melanomas. The Ras family member most often mutated in melanoma is NRas.
Methods
The constitutive activation of the Ras/Raf signaling pathway suggests an impact on the clinical course of the tumor. To address this notion, we analyzed tumor DNA from 114 primary cutaneous melanomas and of 86 metastatic lesions obtained from 174 patients for mutations in BRaf (exons 15 and 11) and NRas (exons 1 and 2) by direct sequencing of PCR products and correlated these results with the clinical course.
Results
In 57.5% of the tumors either BRaf or NRas were mutated with a higher incidence in metastatic (66.3%) than in primary lesions (50.9%). Although the majority of BRaf mutations affected codon 599, almost 15% of mutations at this position were different from the well-described exchange from valine to glutamic acid. These mutations (V599R and V599K) also displayed increased kinase and transforming activity. Surprisingly, the additional BRaf variants D593V, G465R and G465E showed a complete loss of activity in the in vitro kinase assay; however, cells overexpressing these mutants displayed increased Erk phosphorylation. The correlation of mutational status and clinical course revealed that the presence of BRaf/NRas mutations in primary tumors did not negatively impact progression free or overall survival. In contrast, however, for metastatic lesions the presence of BRAF/NRAS mutations was associated with a significantly poorer prognosis, i.e. a shortened survival.
Conclusion
We demonstrate a high – albeit lower than initially anticipated – frequency of activating BRaf mutations in melanoma in the largest series of directly analyzed tumors reported to date. Notably, the clinical course of patients harboring activating BRaf mutations in metastatic melanoma was significantly affected by the presence of a constitutive BRaf activation in these.
Background
There are about 133,000 new cases of melanoma worldwide each year, of which almost 80% occur in the Caucasian population of North America, Europe, Australia and New Zealand [1]. The clinical features of melanoma are asymmetry, a coastline border, or multiple colors, i.e. signs reflecting the heterogeneity of the melanoma cells within one tumor. The classification system supersedes histopathological parameters and clinical features, thereby superficial spreading melanoma (SSM), nodular melanoma (NM), acral-lentiginous melanoma (ALM), and lentigo maligna melanoma (LMM) are differentiated. However, prognosis of primary melanoma is essentially determined by vertical tumor thickness and the presence or absence of ulceration [2].
While it is clear that the genetic make-up of the melanoma-prone population is very important, only few melanomas can be ascribed to specific genetic defects. Loss-of-function mutations in MC1-R are associated with red hair, fair skin and the decreased capacity to tan, i.e. a high susceptibility to melanoma [3]. About 20% of melanoma-prone families possess germline mutations in the CDKN2A gene, which encodes p16INK4A, and in rare cases mutations in the gene encoding CDK4 have been reported. Genes identified as having a role in sporadic melanoma include CDKN2A and PTEN, while chromosomal regions 1q, 6q, 7p and 11q may also be involved. Nodular melanoma display amplification of the MYC oncogene and inactivation of p16INK4A is associated with a poorer prognosis [4,5]. Recently, mutations in the BRAF gene, which is localized on chromosome 7, have been reported in melanoma [6,7].
The Raf family of serine/threonine protein kinases consists of three members: A-, B- and C-Raf which differ in their tissue distribution [8] and which have overlapping as well as unique regulatory functions [9,10]. Beside their influence on processes like apoptosis and differentiation the role of Raf proteins in proliferation control is well established. Raf is the first element of the classical cytoplasmic cascade module (Raf-MEK-Erk), which transmits growth signals from activated Ras at the plasma membrane to transcription factors in the nucleus.
Although the oncogenic potential of Raf has been known for a long time [11] only very recently the presence of Raf mutations in human tumors was described. Activating mutations of BRaf are found in melanomas, colorectal cancers, ovarian tumors [6], thyroid [12,13] and lung cancers [14,15] and colangiocarcinomas [16]. Notably, the highest mutational rate of BRaf with up to 68% was described for melanoma [6,7]. The majority of mutations are present in exon 15 within or immediately adjacent to the so-called activation segment. The most frequently occurring amino acid alteration is V599E, which is caused by a T ? A exchange at nucleotide 1796 [6]. In the study of Davies et al. this V599E mutation accounted for 92% of the mutations found in melanoma. Additional mutations were present in exon 11 altering the glycine residues of the highly conserved GXGXXG motif, which is found in most kinases but also in nucleotide binding proteins like Ras.
In the present study we aimed at confirming these observations in a larger series of patients by directly analyzing uncultured tumor biopsies to avoid any culture artifacts and more importantly to correlate the mutational status with the clinical course. We therefore characterized 200 tumor samples, i.e. 114 primary and 86 metastatic melanomas with a well documented clinical follow up for the presence of mutations in either the BRAF or NRAS genes. To this end, our analysis revealed that constitutive activation of the Ras/Raf signaling pathway in melanoma metastases is significantly associated with a poorer prognosis.
Results
Mutations within the BRAF gene
In the study of Davies et al. [6] exon 15 mutations accounted for 94% of BRaf mutations found in melanoma. Therefore we analyzed first exon 15 of 200 tumor samples i.e. 114 primary and 86 metastatic melanomas. It should be noted that the primary tumors were selected to be already in vertical growth phase in order to ensure that the number of subsequent clinical events was sufficient for statistical analysis. This selection further allowed us both to dissect the tumor from neighboring normal tissue and to isolate sufficient amounts of high quality genomic DNA. The mutation data for all200 tumor samples are summarized in table 1.
Table 1.
Mutations in BRaf (exon 11 and 15) and NRas (exon 1 and 2) in melanoma.
| All lesions | Primary tumors | Metastatic lesions | ||||
| Complete data set | 200 | 114 | 86 | |||
| BRaf mutations | 75 | 37.5% | 39 | 34.2% | 36 | 41.9% |
| BRaf or NRas mutations | 115 | 57.5% | 58 | 50.9% | 57 | 66.3% |
| BRaf Exon 15 | ||||||
| wt/wt | 129 | 64.0% | 78 | 67.5% | 51 | 59.3% |
| mut/wt (V599E) | 55 | 27.5% | 30 | 26.3% | 28 | 32.5% |
| mut/mut (V599E) | 2 | 1.0% | 1 | 0.9% | 1 | 1.2% |
| V599R | 5 | 2.5% | 1 | 0.9% | 4 | 4.7% |
| V599K | 6 | 3.0% | 4 | 3.5% | 2 | 2.3% |
| codon 599 mutations | 71 | 35.5% | 36 | 31.6% | 35 | 40.7% |
| % of 599 mutations not V599E | 15% | 14% | 17% | |||
| D593V | 1 | 0.5% | 1 | 0.9% | 0 | 0.0% |
| BRaf Exon 11 (G-loop) | ||||||
| wt/wt | 197 | 98.5% | 112 | 98.2% | 85 | 98.8% |
| G463R | 1 | 0.5% | 1 | 0.9% | 0 | 0.0% |
| G465R | 1 | 0.5% | 0 | 0.0% | 1 | 1.2% |
| G465E | 1 | 0.5% | 1 | 0.9% | 0 | 0.0% |
| G-loop mutations | 3 | 1.5% | 2 | 1.8% | 1 | 1.2% |
| NRas Exon 2 (codon 61) | ||||||
| wt/wt | 163 | 81.5% | 96 | 84.2% | 67 | 77.9% |
| Q61K | 17 | 8.5% | 9 | 7.9% | 8 | 9.3% |
| Q61H | 2 | 1.0% | 1 | 0.9% | 1 | 1.2% |
| Q61R | 18 | 9.0% | 8 | 7.0% | 10 | 11.6% |
| codon 61 mutations | 37 | 18.5% | 18 | 15.8% | 19 | 22.1% |
| NRas Exon 1 (codons 12 and 13) | ||||||
| wt/wt | 196 | 98.0% | 112 | 98.2% | 84 | 97.7% |
| G12S | 2 | 1.0% | 1 | 0.9% | 1 | 1.2% |
| G12A | 1 | 0.5% | 1 | 0.9% | 0 | 0.0% |
| G12V | 1 | 0.5% | 0 | 0.0% | 1 | 1.2% |
| codon 12 mutations | 4 | 2.0% | 2 | 1.8% | 2 | 2.3% |
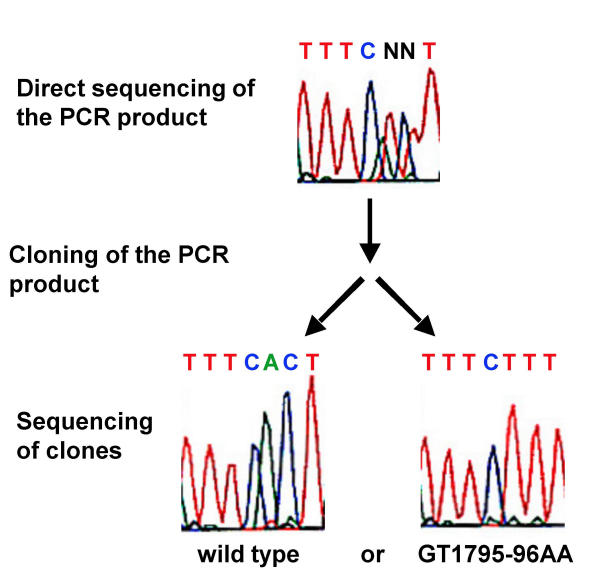
The most frequent alteration was T1796A, which leads to an exchange of valine at position 599 to glutamic acid (V599E) (see table 1). In addition, sequencing of B-Raf exon 15 revealed that in 14 cases two adjoining nucleotides were in a heterozygous state (Figure 1). To answer the question whether both nucleotide alterations are localized on one allele or whether two alleles were affected by independent single nucleotide mutations, the PCR-products were cloned and several of the resulting clones were sequenced. We never found clones in which only one of the two nucleotides was exchanged, but always found either both nucleotides being altered or both in the wild type state (Figure 1). All these 14 mutations affected BRaf at position 599: GT1795-96AA, which leads to Valine599Lysine (V599K), GT1795-96AG, which leads to Valine599Arginine (V599R), and TG1796-97AA which like T1796A leads to Valine599Glutamic acid (V599E) (Table 1). In addition, in one case we observed an A1778T mutation, which leads to an Aspartic Acid 593 Valine (D593V) exchange. Overall the frequency of exon 15 mutations in our cohort of patients is substantially lower than anticipated from the data reported hitherto, i.e. 32,5% for primary and 40,7% for metastatic tumors. Detailed analysis revealed that the presence of exon 15 mutations was dependent on the melanoma subtype, i.e. exon 15 mutations were present in only 2 out of 32 ALM/LMM lesions, whereas the frequency of these mutations in NM or SSM was 42%.
Figure 1.
Mutation of two adjacent nucleotides. Following cloning of the PCR product and sequencing of independent clones we always observed either wild type or double mutants.
In their original report Davies et al. also described, albeit at a lower incidence, mutations in exon 11 of the BRAF gene. These G-loop mutations occurred predominantly in non-melanoma tumors. Nevertheless, we amplified this exon from the 200 tumor samples as well and sequenced the resulting amplicons. This analysis identified 3 additional mutations, i.e. G463R, G465R and G465E, in 3 individual patients, thereby confirming that mutations in the G-loop of BRaf occur at low frequencies in melanoma.
Constitutive activation of BRaf by the exchange of Valine 599 to Lysine or Arginine
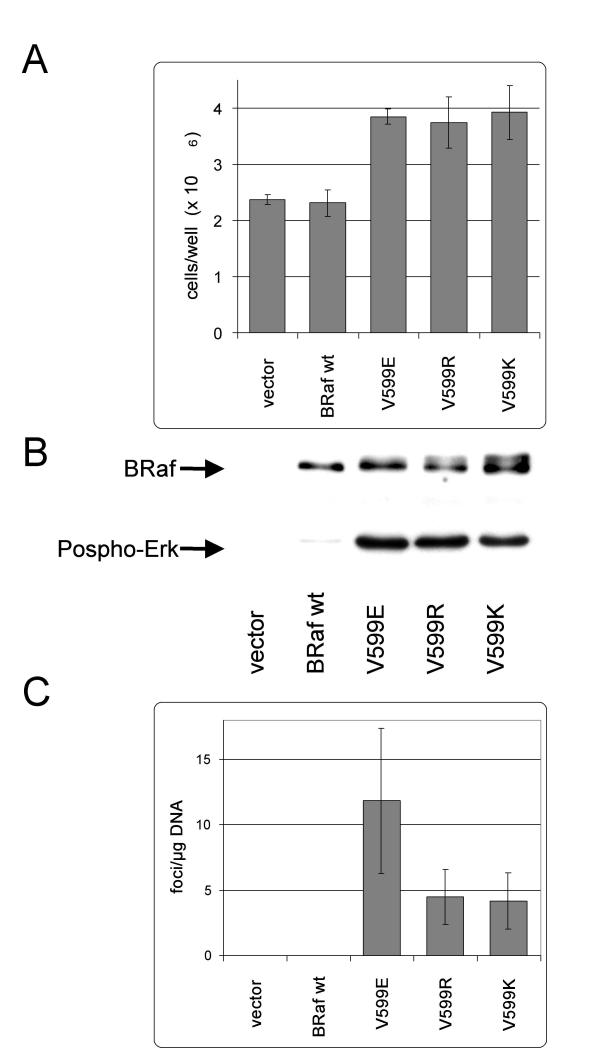
Almost 20% of the BRaf mutations in our patient cohort differed from the V599E mutation. This observation prompted us to test whether these amino acid changes would also increase kinase and transforming activity of BRaf. To this end, expression constructs were generated by in vitro mutagenesis covering these mutations and these were transiently expressed in NIH3T3 cells. Two days post transfection all 599 variants caused a change in morphology towards a more refractile, transformed phenotype. After only 3 additional days of culture it was obvious that the 599 mutants reached a 1.6 to 1,7 fold higher cell density compared to cells expressing either wild type BRaf or an empty vector (figure 2A).
Figure 2.
Mutations V599R and V599K like V599E increase BRaf kinase and transforming activity. NIH3T3 cells were transiently transfected with expression constructs harbouring a wild type BRaf cDNA or the indicated mutant. A) Five days post transfection the cells per well were counted. Shown are the mean values (±SD) of 3 wells each. B) In vitro Raf kinase assay: Immunoprecipitated BRaf was subjected to a kinase reaction with purified MEK and ERK proteins as series connected substrates. B-Raf and Phospho-ERK were detected on a western blot. A representative experiment is shown. C) Focus assay. The number of transformed foci overgrowing the cell monolayer was counted 18 days post transfection. For this assay the cells were split 2 days post transfection from a well of a 6 well plate to a 10 cm dish. Shown are the mean values (±SD) of 6 plates each.
The next step was to measure the intrinsic kinase activity of these BRaf variants. To this end, BRaf protein was immunoprecipated two days post transfection of NIH3T3 cells with the respective expression constructs. The protein was then subjected to a coupled in vitro kinase assay in which the kinase cascade is reconstituted by addition of bacterially expressed and purified GST tagged MEK and His tagged Erk protein. On the western blot all 599 mutants showed up as double band, consistent with autophosphorylation by mutant BRaf whereas BRafwt appeared as one band (figure 2B). Furthermore, there was a strong increase in Erk phosphorylation by all four 599 variants (figure 2B) indicating that these mutants have an increased ability to activate MEK. The oncogenic potential of the exchange of valine599 to arginine or lysine was further confirmed by the ability to transform NIH3T3 cells. Indeed, as already observed with BRafV599E, transfection of expression vectors coding for these mutations resulted in the development of typical foci, which were detectable after a two week culture of confluent cells (figure 2C). Transfection of DNA containing the respective BRafwt cDNA or the empty vector did not yield any foci.
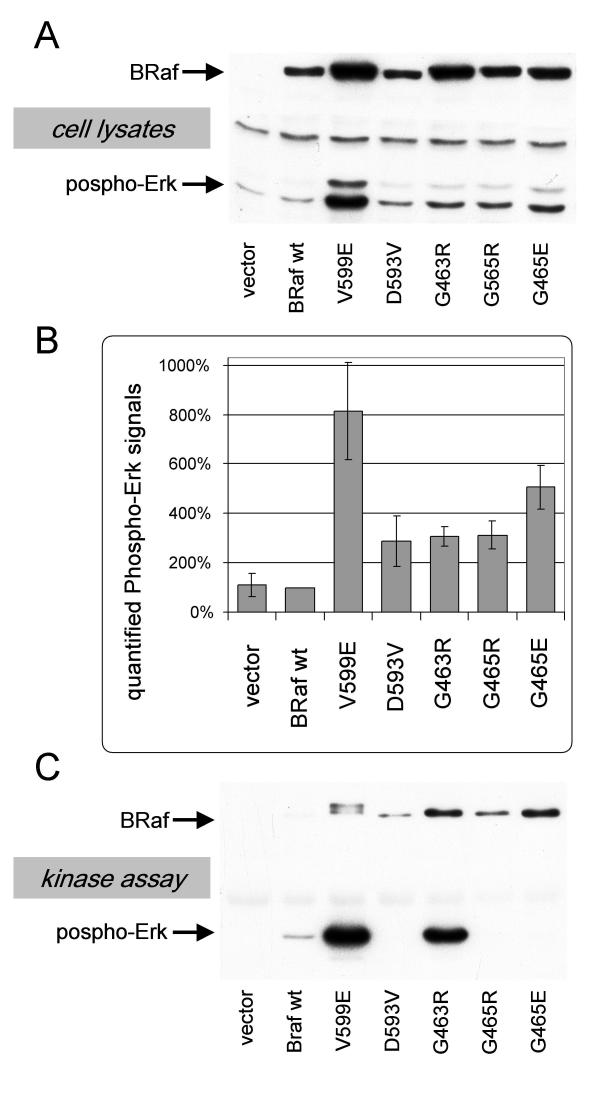
Sequencing of BRaf exons 15 and 11 in our melanoma samples had led to the identification of four mutations, which do not affect codon 599. Analysis of the activating potential of these mutations, however, did not yield consistent data. Following transfection of NIH3T3 cells with expression vectors encoding the BRaf mutants D593V, G463R, G465R or G465E did not show any visible changes in morphology, no higher cell densities were reached and there was no significant increase in focus formation in the transformation assay (data not shown). Still, the phosphorylation status of Erk was increased when the mutants D593V, G463, G465R or G465E were expressed although this increase was somewhat weaker in comparison toV599E (figure 3A and 3B). Surprisingly, when these BRaf mutants were immunoprecipitated and subjected to the in vitro kinase assay the mutants D593V, G465R and G465E showed decreased rather than increased kinase activity (figure 3C). Only BrafG463R did not lose its kinase function following immunoprecipitation and showed slightly higher kinase activity.
Figure 3.
Analysis of the BRaf mutants D593V, G463R, G465R and G465E. NIH3T3 cells were transiently transfected with expression constructs harbouring a wild type BRaf cDNA or the indicated mutant. A) Cells were lysed two days post transfection and the lysates were analysed for BRaf and Phospho-Erk on a western blot. B) Quantified Phospho Erk signals from blots shown in A. Given are the mean values (±SD) from two experiments. C) In vitro Raf kinase assay: Immunoprecipitated BRaf was subjected to a kinase reaction with purified MEK and Erk proteins as series connected substrates. B-Raf and Phospho-Erk were detected on a western blot. A representative experiment is shown.
Mutations in NRas and BRaf V599 are exclusive
Previous reports suggested that BRAF V599 mutation uncouple tumor cells from their proliferation requirements for RAS. Hence, coincidence of activating RAS and BRAF mutations would not result in an additional growth advantage for the tumor cell. Indeed, Davies et al. reported that the presence of NRAS and BRAF mutations were mutually exclusive in the limited series of melanoma samples they analyzed similar to our earlier finding regarding Ras versus C-Raf mutations in a mouse model for carcinogen induced lung tumor formation [17,18]. The analysis of the 200 melanoma lesions for mutations within exon 1 and 2 of the RAS gene demonstrated that in 20.5% codon 12 (2%) or codon 61 (18,5%) (see table 1) were mutated and with only one exception mutations in NRas and BRaf were exclusive. This one exception showed an NRasQ61R and a BRafD593V mutation. Notably, for the D593V mutation we could not demonstrate any transforming activity (data not shown).
NRAS and BRAF mutations do not necessarily persist during disease progression
For 24 patients we were able to analyze the mutational status in a longitudinal fashion, i.e. in the primary tumor and subsequently evolving metastasis. In the majority of cases the genotype regarding BRAF (exon 11 and 15) and NRAS (exon 1 and 2) in the different tumor lesions was identical. However, in seven cases the genotype showed changes (table 2). In four cases mutations in NRAS (n = 1) or BRAF (n = 3) could only be detected in the metastatic lesion, but not in the primary tumor. In two cases there was a change in the way the Ras/Raf signalling pathway was activated, i.e. the BRafV599 mutation in the primary tumor was replaced by an NRasQ61K mutation in the metastasis. In one case an NRasQ61K mutation was present in the primary tumor, but not detectable in the metastatic lesion. Notably, this metastatic lesion also did not harbour any other activating mutation in the above described regions of either RAS or BRAF.
Table 2.
Longitudinal analysis of BRaf and NRas genotypes
| primary tumor | Metastasis | |||
| Nr. | NRas61 | BRaf 599 | NRas61 | BRaf 599 |
| 1 | wt | wt | wt | wt |
| 2 | wt | wt | wt | wt |
| 3 | Q61H | wt | Q61H | wt |
| 4 | wt | wt | wt | V599R |
| 5 | Q61K | wt | Q61K | wt |
| 6 | wt | wt | wt | wt |
| 7 | Q61K | wt | Q61K | wt |
| 8 | wt | wt | wt | wt |
| 9 | Q61R | wt | Q61R | wt |
| 10 | wt | V599E | wt | V599E |
| 11 | wt | wt | wt | wt |
| 12 | wt | wt | wt | wt |
| wt | V599E | |||
| 13 | wt | wt | wt | wt |
| 14 | Q61K | wt | wt | wt |
| 15 | wt | V599R | wt | V599R |
| 16 | wt | V599E | wt | V599E |
| Q61K | wt | |||
| 17 | Q61K | wt | Q61K | wt |
| 18 | wt | V599E | Q61K | wt |
| 19 | wt | V599E | wt | V599E |
| 20 | wt | wt | Q61R | wt |
| 21 | wt | V599E | wt | V599E |
| 22 | wt | wt | wt | wt |
| 23 | wt | wt | wt | wt |
| 24 | wt | wt | wt | V599K |
None of the samples showed mutations in BRaf exon 11 or Nras exon 1. In two cases (12 and 16) samples of two different metastasis excised from one patient were analyzed
Constitutive activation of the Ras/Raf signaling pathway in metastatic melanoma correlates with the clinical course
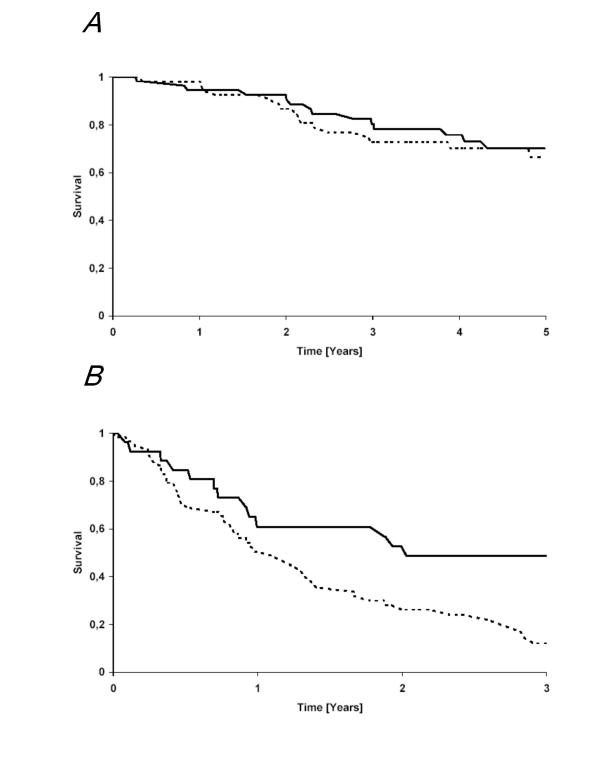
Since the Ras/Raf signaling pathway effectuates several hallmarks of cancer, its constitutive activation may influence the clinical course of tumors. However, the recent detection of constitutive activation of this pathway in the majority of benign melanocytic lesions argues against this hypothesis in melanoma [7]. To address this notion, we correlated the mutational status of NRAS and BRAF with the clinical course. Multivariate analysis revealed that well established and validated prognostic parameters for primary cutaneous melanoma, i.e. tumor thickness and the presence or absence of ulceration, did not correlate with either the presence of any NRas or BRaf mutation (data not shown). However, it should be noted that we only analyzed primary tumors, which were in the vertical growth phase. Hence, no statement concerning pre-invasive tumors in the radial growth phase can be extracted from our study. The strategy was actually chosen in order to allow enough events with respect to disease progression within the clinical follow up of approximately 5 years. To this end, the presence of BRaf/NRas mutations in the primary tumor did not negatively impact progression free (log-rank p = 0,74) or overall survival (log-rank p = 0,28) (figure 4A).
Figure 4.
Influence of BRaf or NRas mutation on overall survival. Kaplan-Meier plots for overall survival for (A) primary tumors and (B) metastases stratified for absence (full line) or presence of either a BRAF or a NRAS mutation (dotted line). A), i.e., as well as B) Overall survival was measured from removal of primary tumor time or the respective metastasis to time of death.
In contrast, however, for metastatic lesions the presence of BRAF/NRAS mutations was associated with a significantly poorer prognosis (log-rank test p = 0,02, t-test p = 0,0013), i.e. a shortened overall survival from the removal of the respective metastases (Figure 4B). In this respect it is important to note, that the majority of analyzed samples were cutaneous or soft tissue metastases and that the distribution of locations of metastases between the groups harboring a BRAF/NRAS mutation or not was not significantly shifted to metastatic lesions with an impaired prognosis.
Discussion
Genes of the Raf family encode kinases that are regulated by Ras and mediate cellular responses to growth signals. Recently, it was shown that activating mutations of BRaf could be found with high frequency in melanomas. In a panel of 200 melanoma samples in 37.5% a BRaf and 57.5% BRaf or NRas mutations were present. These frequencies are low compared to those, i.e. up to and 68% for BRaf and 90% for BRaf or NRas mutations, initially reported [6,7]. However, this lower incidence is consistent with other more recent publications which also describe lower frequencies for these mutations in melanoma: (i) Lang et al. detected only 6 cases with BRafV599E in 24 samples of metastatic melanoma lesions obtained from Scottish patients [19], (ii) Gorden et al. reported a frequency of BRafV599 mutations of 40% in a cohort of 77 metastatic melanoma lesions [20], and (iii) only 2 out of 20 primary melanoma in radial growth phase harbored either a NRas or BRaf mutation [21]. Hence, it seems likely that with increasing sample number the frequency of NRAS and BRAF mutations in melanoma will level at a high albeit lower number than initially anticipated. In addition, it has become evident from this and other reports that in some subtypes of cutaneous melanoma, such as ALM and LMM, BRAF mutations appear at much lower frequency. To this end, in our patient cohort in only 3 out of 32 ALM/LMM BRAF was mutated.
Phosphorylation of threonine 598 and serine 601 within the BRaf activation segment is necessary for the activation of the kinase [22]. It was suggested that the introduction of a negative charge by the exchange of valine to glutamic acid at position 599 might mimic these phosphorylation events thereby leading to increased BRaf activity [6,23]. However, as shown in the present report the introduction of positively charged residues (lysine or arginine) at position 599 also leads to activation. Thus, the constitutive activation of BRaf is not necessarily due to the negative charge but the switch from a small hydrophobic to a large hydrophilic residue seems to be sufficient. In the inactive Raf kinase the activation segment is hidden in a hydrophobic pocket. Upon phosphorylation of threonine 598 and serine 601, or upon exchange of valine to a hydrophilic residue the activation segment may become exposed and accessible for interaction.
We have further demonstrated that all three G-loop and the 593 mutation seem to render BRaf more active, as the Phospho-Erk level is raised in transfected cells. The activation by these alterations, however, is substantially weaker than that induced by the 599 mutations, since cells experimentally expressing these BRaf variants do not show the refractile phenotype, nor do they reach higher cell densities and only a few foci overgrow the monolayer of cells. The discrepancy that the D593V, G465R and G465E variants lead to increased Erk phosphorylation if expressed in the cell, but not if tested in an in vitro kinase assay may be due to the fact that these mutations lead to destabilization of the protein conformation which is functionally relevant under in vitro conditions. Another possible explanation would be that these mutants need a cellular partner, which would transmit the signal to Erk. A candidate for such a partner would be CRaf. It should be interesting to determine the constellation of cancer genes in this subset of melanomas in which these weakly in vivo Erk activity mutations of BRaf have been selected.
The major finding of the present study, however, is that the presence of BRaf mutations in a metastatic melanoma lesion is associated with a poor prognosis, i.e. shortened survival from either the removal of the respective metastases or clinical diagnosis of stage IV disease. For primary tumors, however, the presence of BRaf mutations did not impact the prognosis. A notion further substantiated by the results of multivariate analysis which revealed that for this patient cohort mutations in the BRAF gene were not correlated with established markers for prognosis such as tumor thickness or ulceration.
How might this discrepancy be explained? Considering carcinogenesis as a multistage process one can assume that in the step from primary melanoma to metastasis the cancer cells have to undergo additional genetic or epigenetic alterations. Our data show that in the absence of these alteration(s) the poor prognosis properties of activated BRaf are masked. This might happen if activated BRaf itself would counteract the development of these alterations. In this respect it should be noted that Dong et al. could demonstrate that oncogenic BRAF mutations rather correlate with progression than initiation of human melanoma [21].
There are several lines of evidence suggesting that activated Raf proteins can suppress genetic instability. For example, if mice with lung-specific expression of activated C-Raf (BxB) are crossed to p53-/- mice we find only an acceleration of the development of multiple lung tumors, but no metastatic phenotype [24]. This observation was surprising because p53 is well known to be necessary for the maintenance of a stable genome and in cancer cells loss of p53 function gives rise to clones with greater malignant potential [25]. The preliminary analysis by comparative genomic hybridization has indeed shown that the genome of these C-Raf BxB/p53-/- tumors is as stable as the genome in C-Raf BxB tumors or in normal lung tissue (Balmain and Rapp, unpublished observation). Additionally, oncogenically activated RAS or RAF have been shown to induce growth arrest or senescence rather than unrestricted proliferation in normal fibroblasts via up-regulation of p53, p21Cip1, or p16 [26,27]. On the other hand the Ras/Raf/Mek/Erk pathway is also clearly involved in the oncogenic process. Raf Kinase Inhibitor Protein (RKIP) was identified as a metastasis suppressor in prostate cancer [28] placing Raf in a pro-metastatic pathway. Moreover, high ratios of Erk/p38 signaling determine the human head and neck carcinoma cell line (Hep3) towards tumor growth and metastasis whereas low Erk/p38 ratios are associated with dormancy [29,30]. The sequential increase in the level of activated Smad2 and HRas has been shown to drive the progression from a differentiated squamous carcinoma to an invasive spindle cell carcinoma [31]. The higher incidence of NRas or BRaf mutations in metastasic (66.3%) than in primary melanoma (50.9%) in this study together with the observation that BRAF mutations are more frequent in advanced than in early primary melanoma suggests that activation of this pathway favors the metastatic process [21].
Therefore, activated BRaf may have two-edged features during oncogenesis, i.e. good prognosis properties by stabilizing the genome and poor prognosis properties by contributing to several other hallmarks of cancer [23]. This is reminiscent of transforming growth factor β, which has been implicated in both tumor suppression and progression [32]. However at the metastatic stage of the disease when the full set of genetic alterations might have taken place the good prognosis properties become irrelevant and the bad prognosis properties become dominant. A report by Kumar et al. describing the mutational status of 38 metastatic melanomas further demonstrate the complexity of the clinical consequences of constitutive BRaf activation [33]. In their patient cohort the presence of BRAF mutations was associated with a favorable response to therapy, i.e. a longer progression free survival as compared to cases without mutation. In our patient cohort we did not analyze for any correlation of mutational status and response to therapy due to the fact that the number of objective responses induced by our therapeutic measures (12%) was substantially lower than that observed by Kumar et al. (55%). Hence, the number of patients eligible for this analysis was too low [34].
Is activation of the MAP kinase pathway a necessary event for the development of melanoma? Constitutive activation of Erk is a general feature in melanoma [35] which was attributed to the constitutive activity of BRaf and NRas. However, in 43% of the cases we did not detect any BRaf or NRas alterations. This is in agreement with other studies, although the reported gap was smaller [6,7]. A candidate to fill this gap is the lipid and protein phosphatase PTEN. This tumor suppressor can negatively regulate the MAP kinase pathway [36]. Like for BRaf/NRas a reciprocal mutational status has been reported for NRas and PTEN in human melanoma cell lines [37]. It will be interesting to see whether also PTEN alterations and BRaf/NRas mutations in melanoma are exclusive. Furthermore, autocrine secretion of growth factors may also contribute to increased Erk activity in melanoma cells [35]. Our observation that NRAS and BRAF mutation are not necessarily concordant in the primary tumor and the subsequently evolving metastases does not only illustrate the plasticity of tumor cells, but also that different oncogenic mechanisms resulting in activation of Erk may be engaged in one individual patient. In a recent report of Schmidt-Kittler et al. it was described that genomic aberrations in tumor cells disseminated in the bone marrow generally do not resemble the aberrations in the primary tumor from which they arose [38]. Hence, tumor cells seem to disseminate very early and evolve to metastatic disease largely independent from the primary tumor.
Conclusions
In conclusion, we demonstrate a high frequency of activating BRaf mutations, which are not restricted to the V599E variant, in cutaneous melanoma with an increased incidence in metastatic lesions. Strikingly, the constitutive activation of the Ras/Raf signaling pathway was associated with an impaired prognosis if present in metastatic but not in primary melanoma. This indicates the intricacy of the outcome of a constitutive activation of this signaling pathway in the oncogenesis of melanoma.
Material and methods
Tumor material
Paraffin embedded tumor samples from 114 primary and 86 metastatic melanomas were obtained by surgical excision. All tumors have undergone routine histology for diagnosis. The three immediately following slides from the blocks were used for DNA extraction and the forth slide was used to confirm the presence of the lesion. Prior to DNA extraction adjacent normal tissue was macroscopically dissected. Informed consent was obtained from all patients prior to any of these measures.
Polymerase chain reaction
Genomic DNA was isolated from paraffin embedded tumor samples using a DNA Isolation Kit (Qiagen). Applying conventional PCR for BRaf exon 15 on these templates we had frequent drop outs not yielding the desired product. We therefore switched to nested PCR protocols with an initial PCR reaction (22 cycles, total volume of 30 µl) followed by a second PCR reaction (35 cycles, total volume of 55 µl) containing 1 µl of the first reaction as template. Primers and conditions for the different amplicons were as follows:
BRAF exon 15
First PCR reaction: 2 min at 95°C, 22 cycles (25 sec at 95°C, 25 sec at 55°C, 30 sec at 72°C) and 5 min at 72°C; Primers: cataatgcttgctctgatagg/ggccaaaaatttaatcagtgga
Second PCR reaction: 2 min at 95°C, 35 cycles (20 sec at 95°C, 20 sec at 55°C, 30 sec at 72°C) and 5 min at 72°C; Primers: cataatgcttgctctgatagg/tagcctcaattcttaccatc
BRAF exon 11
First PCR reaction: 2 min at 95°C, 22 cycles (30 sec at 95°C, 30 sec at 58°C, 30 sec at 72°C) and 5 min at 72°C; Primers: gtcccgactgctgtgaa/gtttggcttgacttgacttttt
Second PCR reaction: 2 min at 95°C, 35 cycles (30 sec at 95°C, 30 sec at 58°C, 30 sec at 72°C) and 5 min at 72°C; Primers: gtcccgactgctgtgaac/acgggactcgagtgatgatt
NRAS exon 1
First PCR reaction: 2 min at 95°C, 22 cycles (30 sec at 95°C, 30 sec at 58°C, 30 sec at 72°C) and 5 min at 72°C; Primers: ttatccggtgtttttgcgttctc/gctaccactgggcctcacctcta
Second PCR reaction: 2 min at 95°C, 35 cycles (30 sec at 95°C, 30 sec at 58°C, 30 sec at 72°C) and 5 min at 72°C; Primers: agtactgtagatgtggctcgc/actgggcctcacctctatg
NRAS exon 2
First PCR reaction: 2 min at 95°C, 22 cycles (30 sec at 95°C, 30 sec at 60°C, 30 sec at 72°C) and 5 min at 72°C; Primers: cccccttaccctccacacc/gaggttaatatccgcaaatgactt
Second PCR reaction: 2 min at 95°C, 35 cycles (30 sec at 95°C, 30 sec at 53°C, 30 sec at 72°C) and 5 min at 72°C; Primers: gattcttacagaaaacaagtg/atgacttgctattattgatgg
Sequencing
Samples were sequenced using BigDye™ Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) according to the manufacturer's manual, and analyzed on an ABI PRISM 3100 Avant Genetic Analyzer.
Plasmid construction
For transfection into NIH3T3 cells the plasmid pCMV5-BRaf containing a full length human BRaf cDNA was used. The different mutations were introduced by PCR with Pfu Polymerase (Stratagene) using mutation specific primers. Following restriction digestion with Dpn I the PCR product was transformed into competent DH5α E-coli cells. All clones were verified by sequencing.
Cell culture
NIH 3T3 cells were grown under standard conditions (37°C, 5% CO2) in DMEM (Life Technologies, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone), 168 mM L-Glutamin (Life Technologies, Inc.) and 100 units/ml streptomycin and penicillin (Life Technologies, Inc.).
Focus assay
2 × 105 NIH 3T3 cells per well were seeded in 6 well culture plates 1 day prior to transfection. Transfections were carried out using the Lipofectamine method (Life Technologies, Inc.) according to the manufacturers manual. 2 days post transfection cells were split into 10 cm culture plates. Focus formation was scored 14 – 18 days later.
In vitro kinase assay
The in vitro kinase assay procedure was similar to that described by Weber et al. [39]. Transfected NIH3T3 cells were harvested 2 days post transfection and washed twice with PBS. From now on all steps were carried out on ice or at 4°C. Cells were lysed in 1 ml NP40-buffer (25 mM Tris, 150 mM NaCl, 10 mM Na4P2O7, 25 mM β-glycerophosphate, 25 mM NaF, 10% glycerine, 0.75% NP-40, 20 µg/ml Leupeptin, 20 µg/ml Aprotenin, 1 mM Benzamidine, 1 mM PMSF). For the purpose of preclearing the lysate was incubated with 15 µl protein agarose A (Amersham) for 30 min. Following centrifugation the supernatant was incubated for 2 hours with 30 µl protein agarose A beads and 5 µl of a polyclonal α-BRaf antiserum. The beads carrying the immunoprecipitated BRaf protein were then washed twice with NP-40 buffer and once with kinase buffer (25 mM HEPES pH 7.5, 150 mM NaCl, 25 mM β-glycerophosphate, 10 mM MgCl2, 1 mM DTT). The kinase reaction was carried out for 25 minutes in a shaker at 26°C in 60 µl kinase buffer containing 15 µM vanadate, 170 µM ATP and the bacterially expressed purified proteins GST-MEK (3 µg) [40] and His-Erk-2 (400 ng) [41]. After adding 4x Laemmli buffer the samples were boiled and the proteins were separated on a 9% SDS-Polyacrylamidgel. Following blotting Phospho-Erk and BRaf were detected using the monoclonal antibody α-Phospho-p44/42 MAP kinase (Thr202/Tyr204) (Cell Signaling) and the monoclonal α-BRaf antibody S12 [42] respectively.
Statistics
Time to progression, i.e. progression free survival, was measured from the day of removal of the tumor lesion to the first onset of tumor progression. Overall survival was measured either from the first day of removal of the respective tumor lesion or diagnosis of stage IV disease to the last documented staging examination, i.e. the date last seen. The Kaplan-Meier technique was used to calculate survival data. The log-rank test was used to analyse survival differences among subgroups of patients. The survival analysis was performed in June 2003.
Bivariate analysis of the relationships between prognostic factors and NRAS and BRAF genotypes were assessed according to the U-test following Mann and Whitney or the student's t-test, depending on the nature and distribution of the data. Independent prognostic factors and NRAS and BRAF genotypes should be identified in multivariate analysis using configuration-frequency-method. A p-value of <0.05 was considered to be significant. All calculations were performed using MEDAS statistical software (Grund EDV-Systeme, Margetshöchheim, Germany).
Authors' contributions
RH and JCB contributed equally to this research. RH performed most of the mutation analysis and did the cell culture experiments and the kinase assays. RH and JCB wrote the manuscript. JCB and EB provided the gDNA from the tumor samples and the corresponding clinical data. PT performed the statistical analyses. AK and RG contributed to mutation analysis. UR was the supervisor of the project and together with JCB designed the study.
All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We are grateful to Katrin Mueller-Blech, Svetlana Hilz and Tamara Potapenko for excellent technical assistance. Special thanks to Reinhold Krug who did all the sequencing work and to Ludmilla Wixler for giving an introduction into the Raf kinase assay. This study was supported by grants from the Deutsche Forschungsgemeinschaft (DFG Ra 642/6-2), the Deutsche Krebshilfe (10-1793-Ra-7) and the Sander Stiftung (2000.033.2).
Contributor Information
Roland Houben, Email: roland.houben@mail.uni-wuerzburg.de.
Jürgen C Becker, Email: becker_JC@klinik.uni-wuerzburg.de.
Andreas Kappel, Email: akappel@recognomics.com.
Patrick Terheyden, Email: terheyden_P@klinik.uni-wuerzburg.de.
Eva-B Bröcker, Email: broecker_e@klinik.uni-wuerzburg.de.
Rudolf Goetz, Email: goetz@mail.uni-wuerzburg.de.
Ulf R Rapp, Email: rappur@mail.uni-wuerzburg.de.
References
- Ferlay J, Bray F, Parkin D, Pisani P. IARC Cancer Bases. Vol. 5. Lyon: IARCPress; 2001. Cancer Incidence and Mortality Worldwide. [Google Scholar]
- Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, Cascinelli N, Urist M, McMasters KM, Ross MI, Kirkwood JM, et al. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19:3622–3634. doi: 10.1200/JCO.2001.19.16.3622. [DOI] [PubMed] [Google Scholar]
- Schaffer JV, Bolognia JL. The melanocortin-1 receptor: red hair and beyond. Arch Dermatol. 2001;137:1477–1485. doi: 10.1001/archderm.137.11.1477. [DOI] [PubMed] [Google Scholar]
- Halachmi S, Gilchrest BA. Update on genetic events in the pathogenesis of melanoma. Curr Opin Oncol. 2001;13:129–136. doi: 10.1097/00001622-200103000-00008. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Trent JM. The genetics of cutaneous melanoma. Clin Lab Med. 2000;20:667–690. [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- Naumann U, Eisenmann-Tappe I, Rapp UR. The role of Raf kinases in development and growth of tumors. Recent Results Cancer Res. 1997;143:237–244. doi: 10.1007/978-3-642-60393-8_16. [DOI] [PubMed] [Google Scholar]
- Hagemann C, Rapp UR. Isotype-specific functions of Raf kinases. Exp Cell Res. 1999;253:34–46. doi: 10.1006/excr.1999.4689. [DOI] [PubMed] [Google Scholar]
- Busca R, Abbe P, Mantoux F, Aberdam E, Peyssonnaux C, Eychene A, Ortonne JP, Ballotti R. Ras mediates the cAMP-dependent activation of extracellular signal-regulated kinases (ERKs) in melanocytes. Embo J. 2000;19:2900–2910. doi: 10.1093/emboj/19.12.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp UR, Goldsborough MD, Mark GE, Bonner TI, Groffen J, Reynolds FH, Jr, Stephenson JR. Structure and biological activity of v-raf, a unique oncogene transduced by a retrovirus. Proc Natl Acad Sci U S A. 1983;80:4218–4222. doi: 10.1073/pnas.80.14.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–1457. [PubMed] [Google Scholar]
- Cohen Y, Xing M, Mambo E, Guo Z, Wu G, Trink B, Beller U, Westra WH, Ladenson PW, Sidransky D. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- Naoki K, Chen TH, Richards WG, Sugarbaker DJ, Meyerson M. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62:7001–7003. [PubMed] [Google Scholar]
- Tannapfel A, Sommerer F, Benicke M, Katalinic A, Uhlmann D, Witzigmann H, Hauss J, Wittekind C. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut. 2003;52:706–712. doi: 10.1136/gut.52.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm SM, Rapp UR. Oncogene activation: c-raf-1 gene mutations in experimental and naturally occurring tumors. Toxicol Lett. 1993;67:201–210. doi: 10.1016/0378-4274(93)90056-4. [DOI] [PubMed] [Google Scholar]
- Rapp UR, Fensterle J, Albert S, Gotz R. Raf kinases in lung tumor development. Adv Enzyme Regul. 2003;43:183–195. doi: 10.1016/S0065-2571(03)00002-5. [DOI] [PubMed] [Google Scholar]
- Lang J, Boxer M, MacKie R. Absence of exon 15 BRAF germline mutations in familial melanoma. Hum Mutat. 2003;21:327–330. doi: 10.1002/humu.10188. [DOI] [PubMed] [Google Scholar]
- Gorden A, Osman I, Gai W, He D, Huang W, Davidson A, Houghton AN, Busam K, Polsky D. Analysis of BRAF and N-RAS Mutations in Metastatic Melanoma Tissues. Cancer Res. 2003;63:3955–3957. [PubMed] [Google Scholar]
- Dong J, Phelps RG, Qiao R, Yao S, Benard O, Ronai Z, Aaronson SA. BRAF Oncogenic Mutations Correlate with Progression rather than Initiation of Human Melanoma. Cancer Res. 2003;63:3883–3885. [PubMed] [Google Scholar]
- Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. Embo J. 2000;19:5429–5439. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer KE, Pritchard CA. Raf proteins and cancer: B-Raf is identified as a mutational target. Biochim Biophys Acta. 2003;1653:25–40. doi: 10.1016/s0304-419x(03)00016-7. [DOI] [PubMed] [Google Scholar]
- Fedorov LM, Papadopoulos T, Tyrsin OY, Twardzik T, Gotz R, Rapp UR. Loss of p53 in craf-induced transgenic lung adenoma leads to tumor acceleration and phenotypic switch. Cancer Res. 2003;63:2268–2277. [PubMed] [Google Scholar]
- Soussi T. The p53 tumor suppressor gene: from molecular biology to clinical investigation. Ann N Y Acad Sci. 2000;910:121–137. doi: 10.1111/j.1749-6632.2000.tb06705.x. discussion 137–129. [DOI] [PubMed] [Google Scholar]
- Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, Lowe SW. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkhoff E, Rapp UR. High-intensity Raf signals convert mitotic cell cycling into cellular growth. Cancer Res. 1998;58:1636–1640. [PubMed] [Google Scholar]
- Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, Keller ET. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst. 2003;95:878–889. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- Aguirre-Ghiso JA, Estrada Y, Liu D, Ossowski L. ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK) Cancer Res. 2003;63:1684–1695. [PubMed] [Google Scholar]
- Aguirre-Ghiso JA, Liu D, Mignatti A, Kovalski K, Ossowski L. Urokinase receptor and fibronectin regulate the ERK(MAPK) to p38(MAPK) activity ratios that determine carcinoma cell proliferation or dormancy in vivo. Mol Biol Cell. 2001;12:863–879. doi: 10.1091/mbc.12.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oft M, Akhurst RJ, Balmain A. Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat Cell Biol. 2002;4:487–494. doi: 10.1038/ncb807. [DOI] [PubMed] [Google Scholar]
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- Kumar R, Angelini S, Czene K, Sauroja I, Hahka-Kemppinen M, Pyrhonen S, Hemminki K. BRAF mutations in metastatic melanoma: a possible association with clinical outcome. Clin Cancer Res. 2003;9:3362–3368. [PubMed] [Google Scholar]
- Becker JC, Kampgen E, Brocker E. Classical chemotherapy for metastatic melanoma. Clin Exp Dermatol. 2000;25:503–508. doi: 10.1046/j.1365-2230.2000.00690.x. [DOI] [PubMed] [Google Scholar]
- Smalley KS. A pivotal role for ERK in the oncogenic behaviour of malignant melanoma? Int J Cancer. 2003;104:527–532. doi: 10.1002/ijc.10978. [DOI] [PubMed] [Google Scholar]
- Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene. 2003;22:3113–3122. doi: 10.1038/sj.onc.1206451. [DOI] [PubMed] [Google Scholar]
- Tsao H, Zhang X, Fowlkes K, Haluska FG. Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. Cancer Res. 2000;60:1800–1804. [PubMed] [Google Scholar]
- Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, Kaufmann M, Diebold J, Arnholdt H, Muller P, et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:7737–7742. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CK, Slupsky JR, Herrmann C, Schuler M, Rapp UR, Block C. Mitogenic signaling of Ras is regulated by differential interaction with Raf isozymes. Oncogene. 2000;19:169–176. doi: 10.1038/sj.onc.1203261. [DOI] [PubMed] [Google Scholar]
- Alessi DR, Saito Y, Campbell DG, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall CJ, Cowley S. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. Embo J. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins DJ, Zhen E, Cheng M, Xu S, Vanderbilt CA, Ebert D, Garcia C, Dang A, Cobb MH. Regulation and properties of extracellular signal-regulated protein kinases 1, 2, and 3. J Am Soc Nephrol. 1993;4:1104–1110. doi: 10.1681/ASN.V451104. [DOI] [PubMed] [Google Scholar]
- Sithanandam G, Kolch W, Duh FM, Rapp UR. Complete coding sequence of a human B-raf cDNA and detection of B-raf protein kinase with isozyme specific antibodies. Oncogene. 1990;5:1775–1780. [PubMed] [Google Scholar]