Conspectus
Circulating tumor cells (CTCs) are cancer cells that break away from either a primary tumor or a metastatic site and circulate in the peripheral blood as the cellular origin of metastasis. With their role as a “tumor liquid biopsy”, CTCs provide convenient access to all disease sites, including that of the primary tumor and the site of fatal metastases. It is conceivable that detecting and analyzing CTCs will provide insightful information in assessing the disease status without the flaws and limitations encountered in performing conventional tumor biopsies. However, identifying CTCs in patient blood samples is technically challenging due to the extremely low abundance of CTCs among a large number of hematologic cells. To address this unmet need, there have been significant research endeavors, especially in the fields of chemistry, materials science, and bioengineering, devoted to developing CTC detection, isolation, and characterization technologies.
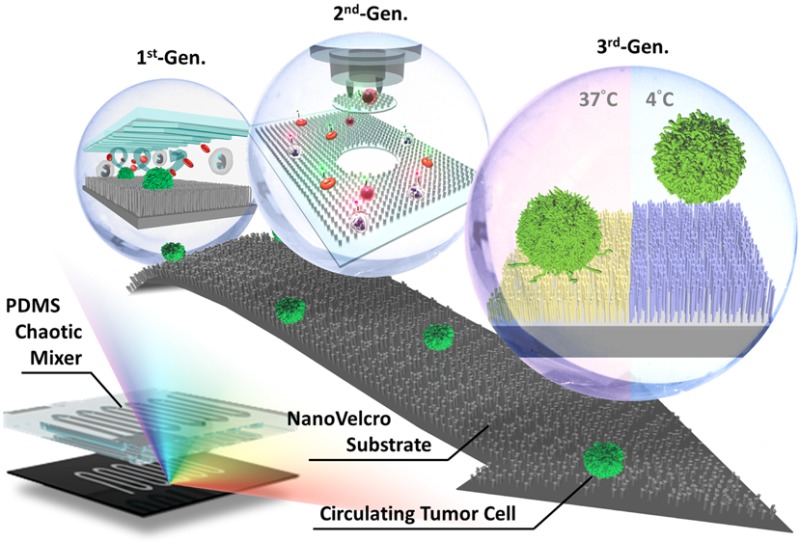
Inspired by the nanoscale interactions observed in the tissue microenvironment, our research team at UCLA pioneered a unique concept of “NanoVelcro” cell-affinity substrates, in which CTC capture agent-coated nanostructured substrates were utilized to immobilize CTCs with high efficiency. The working mechanism of NanoVelcro cell-affinity substrates mimics that of Velcro: when the two fabric strips of a Velcro fastener are pressed together, tangling between the hairy surfaces on two strips leads to strong binding. Through continuous evolution, three generations (gens) of NanoVelcro CTC chips have been established to achieve different clinical utilities. The first-gen NanoVelcro chip, composed of a silicon nanowire substrate (SiNS) and an overlaid microfluidic chaotic mixer, was created for CTC enumeration. Side-by-side analytical validation studies using clinical blood samples suggested that the sensitivity of first-gen NanoVelcro chip outperforms that of FDA-approved CellSearch. In conjunction with the use of the laser microdissection (LMD) technique, second-gen NanoVelcro chips (i.e., NanoVelcro-LMD), based on polymer nanosubstrates, were developed for single-CTC isolation. The individually isolated CTCs can be subjected to single-CTC genotyping (e.g., Sanger sequencing and next-generation sequencing, NGS) to verify the CTC’s role as tumor liquid biopsy. Created by grafting of thermoresponsive polymer brushes onto SiNS, third-gen NanoVelcro chips (i.e., Thermoresponsive NanoVelcro) have demonstrated the capture and release of CTCs at 37 and 4 °C, respectively. The temperature-dependent conformational changes of polymer brushes can effectively alter the accessibility of the capture agent on SiNS, allowing for rapid CTC purification with desired viability and molecular integrity.
This Account summarizes the continuous evolution of NanoVelcro CTC assays from the emergence of the original idea all the way to their applications in cancer research. We envision that NanoVelcro CTC assays will lead the way for powerful and cost-efficient diagnostic platforms for researchers to better understand underlying disease mechanisms and for physicians to monitor real-time disease progression.
1. Introduction
1.1. Circulating Tumor Cells
Metastasis is the most common cause of cancer-related death in patients with solid tumors. A considerable body of evidence indicates that tumor cells are shed from a primary and metastatic tumor mass at different stages of malignant progression. These break-away circulating tumor cells (CTCs)1,2 enter the bloodstream and travel to different tissues of the body as a crucial route for cancer spreading. The current gold standard for diagnosing tumor status requires invasive biopsy, followed by pathological analysis. Unlike tumor biopsies that can be constrained by problems such as sampling bias, CTCs are regarded as the “liquid biopsy”3 of the tumor, providing convenient access to all disease sites, including the primary tumor and fatal metastases. In addition to conventional diagnostic imaging and serum marker detection, detecting and characterizing CTCs in patient blood provides an opportunity for early diagnosis of cancer metastasis. To address this unmet need, there have been significant research endeavors, especially in the fields of chemistry, materials science, and bioengineering, devoted to developing CTC detection, isolation, and characterization technologies. However, identifying CTCs in blood samples has been technically challenging due to the extremely low abundance (a few to hundreds per milliliter) of CTCs among a large number (109 mL–1) of hematologic cells.
1.2. Existing CTC Enrichment Technologies and Their Limitations
The most widely used CTC detection assays are summarized in the following. (i) Immunomagnetic separation: these methods utilize capture agent-labeled magnetic beads to either positively select4,5 CTCs using a cell surface marker, (i.e., anti-epithelial cell adhesion molecule (EpCAM)) or negatively deplete white blood cells (WBCs) using anti-CD45. CellSearch assay4,5 is the only FDA-cleared CTC diagnostic technology for metastatic breast, prostate, and colorectal cancer. Recently, several sophisticated systems (e.g., MagSweeper,6 IsoFlux,7 and VerIFAST8) have been developed to further improve the detection speed and efficiency. In parallel, Massachusetts General Hospital team’s iChip9 applies the negative depletion mechanism and has attracted significant attention. (ii) Flow cytometry: Although flow cytometry10 is one of the most mature technologies for analyzing of subpopulations of cells, the flow-based methodology is unable to provide the CTCs’ morphological information to meet the gold standard set by pathologists. An improved method,11 known as ensemble-decision aliquot ranking, was developed to address this weakness. (iii) Microfluidic chips: Several microfluidic technologies12−14 were developed to achieve higher capture efficiency. However, a majority of these microfluidic CTC technologies suffer from depth of field issues due to the vertical depths of device features (e.g., micropillars12 or herringbones14). Multiple cross-sectional scans are required in order to avoid out-of-focus or superimposed micrographs. (iv) CTC filters: Filter-based approaches15,16 have been established to trap CTCs according to their sizes. Several commercial products, e.g., Clearbridge15 and RareCell,16 are now available on the market. Nevertheless, concerns regarding missing small CTCs have been raised. (v) Other methods: There are review articles17,18 where side-by-side comparisons of a wide collection of CTC detection technologies are presented. Although the existing technologies have demonstrated their capacities for efficient CTC detection, challenges remain in (i) establishing a translational pipeline, where a joint effort between researchers and clinicians can be devoted to clinical validation and FDA approval and (ii) most importantly recovering CTCs with improved purity, viability,19 and molecular integrity20 in order to enable subsequent molecular and functional analyses. It is conceivable that the CTC-derived molecular signatures and functional readouts can provide significant insight into tumor biology during the critical window where treatment intervention could actually make the difference.
1.3. Evolution of NanoVelcro Cell-Affinity Assays
In contrast to existing CTC detection technologies, our team at UCLA pioneered a unique concept of the “NanoVelcro” cell-affinity assay, in which capture agent-coated nanostructured substrates21−24 were utilized to immobilize CTCs with high efficiency. Through continuous evolution, three generations (gens) of NanoVelcro chips have been established for different clinical utilities. The first-gen NanoVelcro chip,25−27 composed of a silicon nanowire substrate (SiNS) and an overlaid microfluidic chaotic mixer, was created for CTC enumeration. Side-by-side validation studies using clinical blood samples suggested that the sensitivity of first-gen NanoVelcro chip outperforms that of CellSearch assay. In conjunction with the use of laser microdissection (LMD) techniques, the second-gen NanoVelcro chip28,29 with a transparent polymer nanosubstrate was developed for single-CTC isolation. The individually isolated CTCs can be subjected to single-CTC genotyping (e.g., Sanger sequencing and next-generation sequencing, NGS) to verify the CTC’s role as tumor liquid biopsy. Created by grafting thermoresponsive polymer brushes onto SiNS, third-gen NanoVelcro chips30 have demonstrated the capture and release of CTCs at 37 and 4 °C, respectively. The temperature-dependent conformational changes of polymer brushes can effectively alter the accessibility of the capture agent on SiNS, allowing for rapid CTC purification with desired viability and molecular integrity. In this Account, we summarize the continuous development of these NanoVelcro CTC assays and their potential applications in oncology clinics.
1.4. Nanostructured Substrates for Cell Biology
It has been long documented that nanoscale components present in the tissue microenvironment, including the extracellular matrix (ECM) and cell-surface structures (e.g., microvilli) provide structural and biochemical support that regulates cellular behaviors and fates, including adhesion, migration, communication, differentiation, and viability. Inspired by these nanoscale interactions, researchers have been developing nanostructure-embedded substrates, which mimic the features and dimensions of the ECM, in order to understand how different nanosubstrates affect cells and ultimately control cellular behaviors for potential biomedical applications.31−33 Different from the existing studies, the idea behind NanoVelcro CTC assays is to exploit the use of nanostructured substrates for detection, isolation, and characterization of CTCs.
2. Proof-of-Concept Demonstration of NanoVelcro Cell-Affinity Substrates
2.1. Stationary NanoVelcro CTC Assay
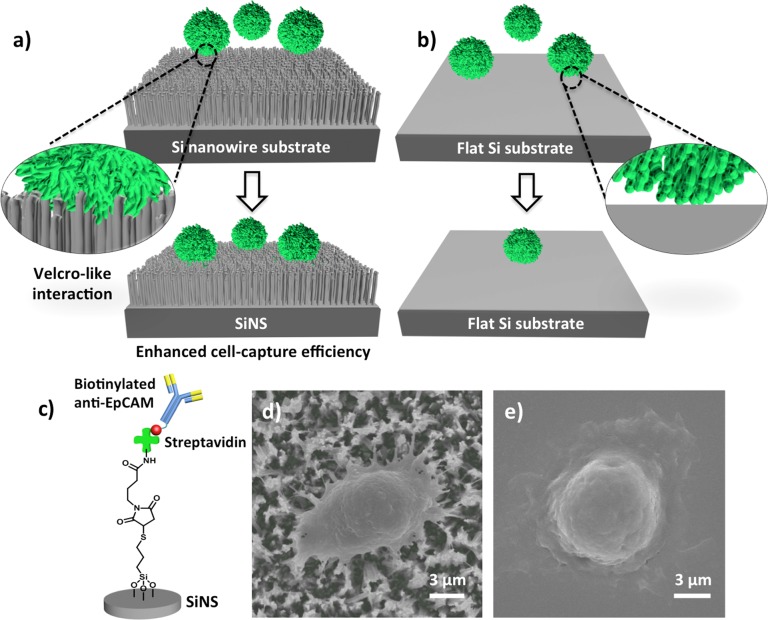
The working mechanism (Figure 1a) of NanoVelcro cell-affinity substrates21−24 mimics that of Velcro: when the two fabric strips of a Velcro fastener are pressed together, tangling between the hairy surfaces on two strips leads to enhanced binding. The proof-of-concept demonstration of the NanoVelcro cell-affinity assay21 lies in the use of a SiNS, which allows for Velcro-like interactions34 between the SiNS and nanoscale cell-surface components. Here, anti-EpCAM was grafted onto the SiNS as the capture agent, conferring specificity to such a cell-affinity assay for recognizing CTCs and resulting in improved cell-capture affinity compared with that of an unstructured (i.e., flat silicon) substrate (Figure 1b). The anti-EpCAM grafted SiNS was fabricated through three continuous steps (Figure 1c): (i) introduction of densely packed silicon nanowires onto a silicon wafer, (ii) silane treatment and covalent conjugation of streptavidin onto the SiNS, and (iii) grafting of biotinylated anti-EpCAM onto the streptavidin-coated SiNS. Through comprehensive optimization, an optimal condition was obtained for performing cell capture on whole blood samples in a stationary device setting. SEM characterization of CTCs on both SiNSs (Figure 1d) and flat Si substrates (Figure 1e) revealed that there were many interdigitated cell-surface components on the SiNS, supporting the proposed NanoVelcro working mechanism. Generally, NanoVelcro cell-affinity substrates are capable of capturing CTCs from artificial blood samples with about 40–70% efficiency.
Figure 1.
Velcro-like working mechanism of NanoVelcro cell-affinity substrates. (a) An anti-EpCAM-coated SiNS was employed to achieve significantly enhanced capture of CTCs in contrast to (b) an anti-EpCAM-coated flat silicon substrate. (c) Anti-EpCAM is grafted onto a SiNS to confer specificity for recognizing CTCs. (d, e) SEM images of a SiNS and a flat Si substrate, on which MCF7 cells were captured.
2.2. General Applicability NanoVelcro CTC Substrates
In addition to SiNS-based nanosubstrates, we have also adopted different fabrication approaches to incorporate various nanomaterials into NanoVelcro substrates. By an eletrospinning process, horizontally oriented TiO2 nanofibers were deposited onto glass slides. After anti-EpCAM conjugation, enhanced CTC capture efficiency was observed for the resulting TiO2 nanofiber-embedded NanoVelcro substrates,23 where the densities of TiO2 nanofibers affected their CTC-capture performance. Alternatively, an electrochemical method was employed to deposit organic conducting polymer (i.e., PEDOT) nanodots22 onto ITO-coated glass substrates. Carboxylic groups on PEDOT backbones allow for convenient conjugation with anti-EpCAM, and an enhanced capture performance observed for the nanodot-embedded NanoVelcro substrates was determined by the sizes and densities of the nanodots. Recently, a new approach24 combining chemical oxidative polymerization and a modified PDMS transfer printing technique was established for introducing highly regular PEDOT nanorods onto glass substrates for CTC capture. Given the outstanding electrical transport properties, inherent biocompatibility, and manufacturing flexibility of PEDOT, we foresee that these PEDOT-based NanoVelcro substrates will be integrated with downstream electrical sensing and phenotyping after capturing CTCs.
Besides our own attempts, there has been extensive research devoted to testing different nanostructured materials for capturing CTCs and other types of rare cells. Since our proof-of-concept publication21 in 2009, nanostructured materials, for example, layer-by-layer (LbL)-assembled nanostructures,35 gold clusters on silicon nanowires,36 Fe3O4 nanoparticles,37 polymer nanotubes,38 TiO2 nanoparticles,39 dendrimers,40 and other nanomaterials,41,42 were deposited on cell-capture substrates to show enhanced cell-capture performance. Both our work and that of others summarized above provide very solid support for the general applicability of NanoVelcro cell-affinity assays and their potential for cell-sorting applications.
3. First-Generation NanoVelcro Chips for CTC Enumeration
3.1. Device Configuration of First-Gen NanoVelcro Chips
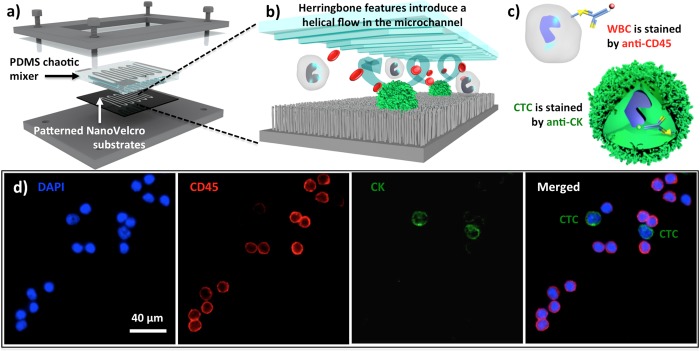
On the basis of the stationary NanoVelcro cell-affinity substrates,21−24 we foresaw that further improvement on capture performance could be achieved by increasing the contact frequency between CTCs and nanosubstrates. With the incorporation of an overlaid PDMS chaotic mixer43 onto a lithographically patterned NanoVelcro substrate (Figure 2a,b), we developed first-gen NanoVelcro chips.25 When a blood sample containing CTCs flows through the device, the herringbone microstructures on the roof of the chaotic mixer induce43 vertical flows in the microchannel, resulting in an enhanced cell–substrate contact frequency. Validation studies using artificial blood samples (i.e., cancer cell-spiked blood) reveal that this CTC assay exhibits >85% cell-capture performance. In parallel our laboratory has also established a three-color immunocytochemistry (ICC) protocol44 for parallel staining of 4′,6-diamidino-2-phenylindole (DAPI), anti-CD45, and anti-cytokeratin (CK) to identify nanosubstrate-immobilized CTCs. Single-cell image cytometry data (Figure 2c,d) covering CK/CD45 expression and CTC footprint sizes can be used to identify CTCs from nonspecifically captured WBCs and cellular debris.
Figure 2.
(a, b) Configuration of 1st-gen NanoVelcro CTC chip. The device is composed of a patterned NanoVelcro substrate and an overlaid PDMS chaotic mixer. (c) CTCs (DAPI+/CK+/CD45–, sizes >6 μm) can be clearly distinguished from nonspecifically captured WBCs (DAPI+/CK–/CD45+, sizes <12 μm) by a three-color ICC protocol. (d) Fluorescent images of two prostate cancer CTCs captured on the substrate along with nonspecifically captured WBCs.
3.2. Clinical Utility of First-Gen NanoVelcro Chips
To assess the performance of first-gen NanoVelcro chips in a clinical setting, we conducted side-by-side analytical validation studies between NanoVelcro CTC chips and CellSearch assay. The blood samples were collected from prostate cancer patients with different disease severities. Figure 2d shows fluorescence micrographs of two CTCs isolated from a prostate cancer patient’s blood (1.0 mL). In 17 out of 26 clinical blood samples (Figure 3a), first-gen chips exhibited25 significantly greater sensitivity and superior dynamic range in CTC enumeration. It is known that the clinical utility of CellSearch assay is constrained by its high blood consumption (7.5 mL), poor sensitivity, and dynamic range. We envision that the great performance observed for first-gen NanoVelcro chips will open up opportunities for (i) better monitoring of disease progression and treatment response and (ii) detecting CTCs at a relatively early stage of disease.
Figure 3.
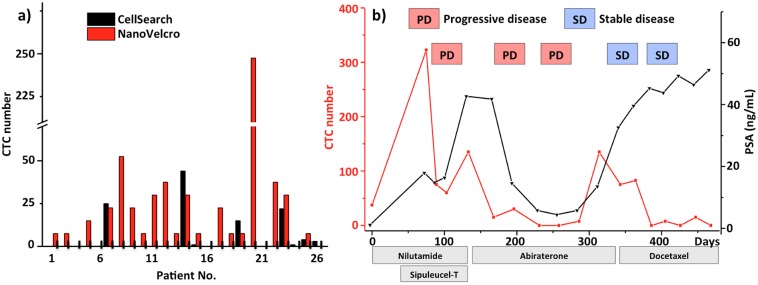
(a) CTC enumeration results obtained from 1st-gen NanoVelcro chips and a CellSearch assay on matched samples from 26 prostate cancer patients. (b) Serial CTC and PSA changes of a prostate cancer patient are plotted over his multiple treatment responses and progressions.
Continuous improvement of first-gen NanoVelcro CTC chips has led to a device configuration26 with a smaller device footprint, paving the way for a cost-efficient CTC enumeration assay that can benefit more cancer patients. The validation studies26 were jointly conducted by the Uro-Oncology teams at both UCLA Hospital and Cedars-Sinai Medical Center. Forty prostate cancer patients (32 with metastatic disease and 8 with localized disease) were recruited, and CTCs were identified in all 40 patients. We further performed follow-up measurements in these patients over the courses of different treatments. As a result of the NanoVelcro chips’ high sensitivity, we found that patients responding to the given therapies have a significant negative change in their CTC numbers. In the index patient, we performed serial measurements for up to 460 days, during which multiple therapies were given with variable responses (Figure 3b). CTC counts faithfully represented the initial response and subsequent failures. However, we observed a CTC–PSA disagreement when his disease was stabilized by docetaxel therapy. The patient’s CTC numbers remained low despite PSA progression when bone scan confirmed his stable disease. This case indicates that CTC number measured by NanoVelcro CTC chip may be a more reliable biomarker in the clinical assessment of prostate cancer. In addition to prostate cancer, our studies toward applying NanoVelcro chips for different types of cancer (e.g., breast, lung, and pancreatic cancer, as well as melanoma) are making continuous progress.
3.3. An Alternative Capture Agent, Aptamer
Anti-EpCAM remains the most commonly used CTC capture agent for the majority of epithelial origin solid tumors. Due to the poor stability and high cost of antibody, the dissemination and translation of CTC-based diagnostics can be constrained especially in a low-resource environment. This limitation can be addressed by replacing anti-EpCAM with aptamers, which were generated through an in vitro cell-SELEX (systematic evolution of ligands by exponential enrichment) process targeting a specific type of cancer cells. Recently, we were able to produce two single-stranded DNA aptamers via cell-SELEX45 processes in the presence of A549 non-small-cell lung cancer (NSCLC) cells. We demonstrated27 that the aptamer-grafted NanoVelcro chip (Figure 4) is capable of not only capturing NSCLC CTCs from blood with high efficiency but also recovering the nanosubstrate-immobilized CTCs upon enzymatic treatment.
Figure 4.
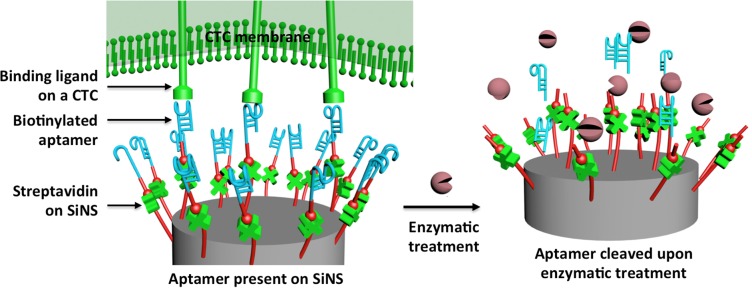
Molecular mechanism governing the capture and enzymatic release of NSCLC CTCs from the aptamer-coated SiNS.
Recently, an interesting phenomenon46 that capture-agent free nanosubstrates on glass slides exhibit a differential affinity to cancer cells rather than WBCs was reported. Although this differential affinity is potentially applicable for CTC detection, concerns have been raised for the lack of understanding of the molecular and cellular mechanism that governs such a differential affinity. Further experimental data supporting their utility in clinical setting remain to be provided.
4. Second-Generation NanoVelcro-LMD Technology for Single-CTC Isolation
The development of first-gen NanoVelcro chips has led to a highly sensitive CTC enumeration technology that has demonstrated its clinical utility for monitoring disease progression and reporting treatment responses. To pave the way toward molecular analysis of CTCs, we developed second-gen NanoVelcro-LMD technology28,29 (Figure 5a) by coupling LMD techniques with a transparent nanosubstrate covered with poly(lactic-co-glycolic acid) (PLGA) nanofibers. Unlike the SiNS used in the first-gen chips, the transparent PLGA nanosubstrates within NanoVelcro-LMD technology28,29 allows single-CTC identification and isolation, followed by a wide range of molecular analyses (e.g., RT-PCR, Sanger sequencing, and NGS).
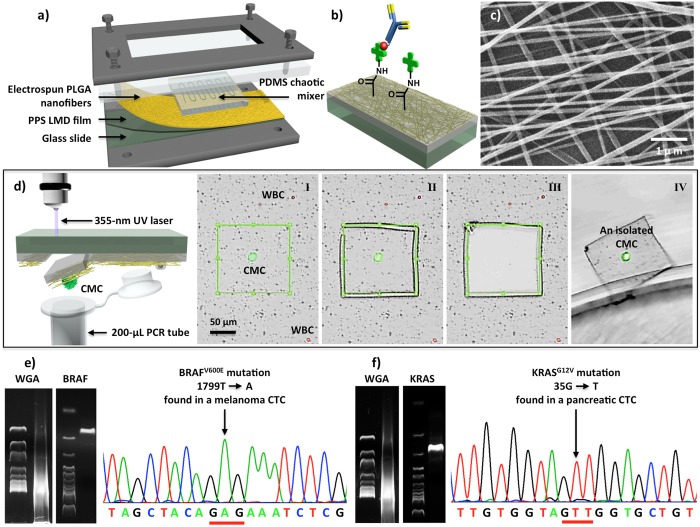
Figure 5.
(a) A chip holder assembles PLGA nanofiber-embedded NanoVelcro chips and PDMS chaotic mixer in a sandwiched configuration. (b) Streptavidin is covalently attached onto the PLGA nanofibers for conjugation of the biotinylated capture agents (i.e., anti-CD146 for melanoma or anti-EpCAM for prostate and pancreatic cancer). (c) SEM image of the electrospun PLGA nanofibers. (d) Process of LMD-based single CTC isolation includes (I) CTC identification, (II) laser-dissection of the identified CTC, (III, IV) release of the CTC from the substrate into a 200-μL PCR tube. (e) Results of single-CTC WGA and gel electrophoresis after PCR amplification using a BRAF-specific primer. Confirmatory Sanger sequencing showed individual melanoma CTCs carrying distinct BRAFV600E mutation. (f) Detecting KRASG12V mutation in single pancreatic CTCs.
4.1. Preparation of PLGA NanoVelcro Chips
The PLGA NanoVelcro substrates28,29 were prepared by depositing electrospun PLGA nanofibers onto a commercial LMD slide (i.e., a regular microscope slide covered with a 1.2-μm-thick polymer membrane, Figure 5a). After covalently attaching streptavidin onto the PLGA nanofibers, conjugation of biotinylated antibodies confers capture specificity to PLGA NanoVelcro substrates (Figure 5b,c) to identify CTCs from whole blood samples.
4.2. NanoVelcro-LMD Technology, Followed by Mutational Analysis
We first applied NanoVelcro-LMD technology to isolate28 single melanoma CTCs (Figure 5d) for detecting a signature oncogenic mutation (i.e., BRAFV600E), which is present in 60% of melanomas and has been targeted by specific inhibitors (e.g., vemurafenib). Here, a melanoma-specific anti-CD146 was used as the capture agent. To validate the clinical utility of NanoVelcro-LMD technology, we then performed single-CTC isolation and genotyping using peripheral blood samples collected from several stage-IV melanoma patients, whose melanomas have been previously characterized to contain BRAFV600E. The individually isolated melanoma CTCs were subjected to whole genome amplification (WGA) and PCR targeting BRAF, and Sanger sequencing was then employed to detect the BRAFV600E mutation in the melanoma CTCs. Notably, the Sanger sequencing data obtained for the BRAFV600E mutation in single melanoma CTCs (Figure 5e) displayed a strong signal-to-noise ratio. In contrast, varying levels of sequencing noise and BRAFV600E signals were often encountered when biopsied melanoma tissues were sequenced.
The same single-CTC genotyping approach has also been used for different solid tumors with specific signature oncogenic mutations. For example, using anti-EpCAM as capture agent, we were able to isolate single CTCs from artificial and clinical blood samples. Genotyping of the individually isolated pancreatic CTCs disclosed a KRASG12V mutation (Figure 5f), consistent with that found in patients’ tumors.
4.3. NanoVelcro-LMD Technology, Followed by Whole Exome Sequencing
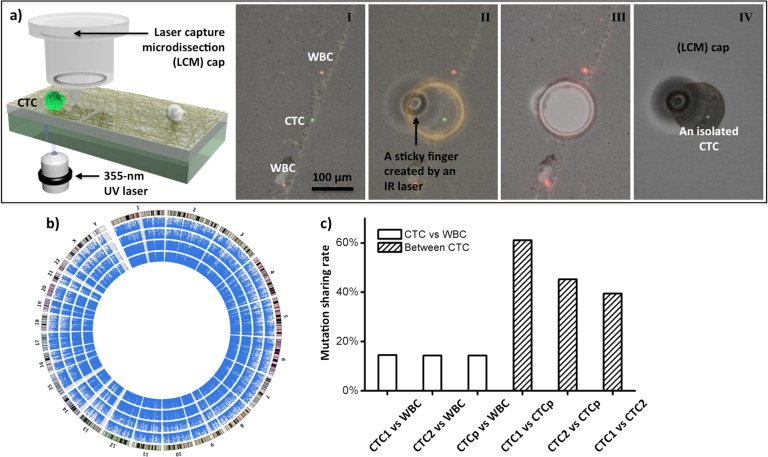
Due to frequent cell loss caused28 by the static charge during collection processes, we replaced29 the LMD technology with a modified version, that is, laser capture microdissection (LCM). The LCM technology prevented the cell loss by employing an IR-laser to melt the LCM cap, dropping down a “sticky finger” for adhering onto laser-dissected NanoVelcro substrate (Figure 6a). This system allows for isolation of single CTCs from patients with advanced prostate cancer suitable for NGS to detect mutations in a broader mutational landscape (Figure 6b,c). Prostate cancer is known for its long natural course of up to 10–15 years in most cases. The combined use of single-CTC isolation and NGS can be used to monitor the evolution of tumor heterogeneity20 over time. Beyond single-CTC exome sequencing, our continuous efforts will be devoted to exploring the second-gen NanoVelcro-LCM/LMD technology in conjunction with advanced NGS including whole genome sequencing, transcriptomic analysis by RNA sequencing, and epigenetic studies. The information will help to understand the tumor heterogeneity and clonal evolution, as well as provide real-time monitoring of patients’ disease progression and response to specific treatments.
Figure 6.
(a) Process of single-CTC isolation using a LCM system includes (I) CTC identification; (II) determination of IR sticky finger positions and UV dissection route; (III) UV laser dissection; (IV) collection of the identified CTC onto the LCM cap. (b) Circos plots representing the coverage areas of Exome-Seq. The rings from the inside out represent WBCs, pooled CTCs (CTCp), CTC-1, and CTC-2. The outermost ring represents the karyotype of the human reference genome. (c) The shared mutations between CTCs and WBC are compared with the shared mutations among CTCs. Adapted with permission from ref (29). Copyright 2013 John Wiley and Sons.
5. Third-Generation Thermoresponsive NanoVelcro Chips
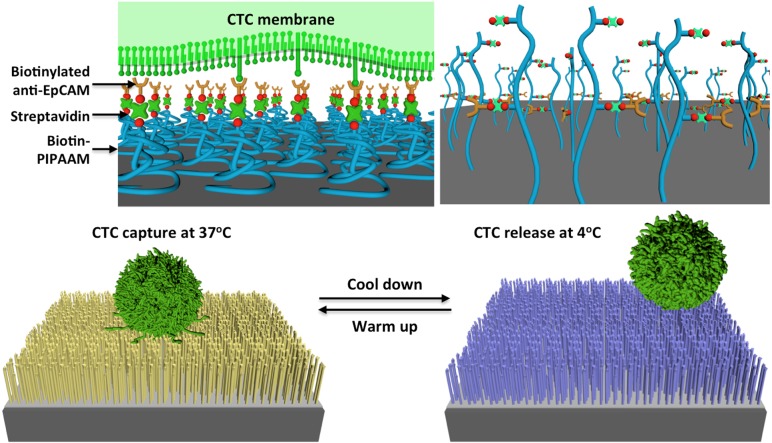
As the molecular characterization20 and functional analysis19 of CTCs are increasingly conducted, there is an urgent need to isolate CTCs with higher efficiency, higher cell quality, and less technical demand. Although the second-gen NanoVelcro-LCM approach32,33 possesses a great precision in single-CTC isolation, it suffers from a labor-consuming process and poor viabilities of the isolated CTCs. The third-gen thermoresponsive NanoVelcro chip30 was developed to address these issues. Created by grafting thermoresponsive polymer brushes47 (i.e., poly(N-isopropylacrylamide, PIPAAm) onto SiNS, thermoresponsive NanoVelcro chips (Figure 7) can capture and release CTCs at 37 and 4 °C, respectively. The uniqueness of our idea is that the temperature-dependent conformational changes of polymer brushes can effectively alter the accessibility of the capture agent on the NanoVelcro, allowing for rapid CTC purification with desired CTC viability and molecular integrity. More specifically, we strategically introduced biotin groups onto the polymer brushes for conjugation of a CTC-capture agent, anti-EpCAM. At 37 °C, anti-EpCAM and hydrophobic domains of the polymer brushes are present on the surfaces of NanoVelcro substrates, enabling CTC capture. When the temperature is reduced to 4 °C, the conformational change of the polymer brushes induces an internalization of anti-EpCAM, leading to CTC release. The thermoresponsive NanoVelcro chip exhibited enhanced CTC-capture efficiency, and the majority of the captured cells were successfully released at 4 °C with approximately 90% viability.
Figure 7.
Operational mechanism of the thermoresponsive NanoVelcro CTC substrate for capturing and releasing CTCs at 37 and 4 °C, respectively. The temperature-dependent conformational changes of polymer brushes effectively alter the accessibility of anti-EpCAM on NanoVelcro substrates.
Our continuous research efforts will be devoted to building a user-interface of third-gen thermoresponsive NanoVelcro chips. The goal is to enable rapid purification of CTCs from whole blood samples, thus paving the way for downstream CTC characterization. In addition to performing CTC molecular analysis, obtaining viable CTCs for ex vivo expansion (i.e., culture) will set the stage for a wide range of applications, e.g., generating CTC-derived cancer lines and in vitro screening for potential therapeutics for individual cancer patients, a promising opportunity for bringing personalized medicine to fruition.
6. Conclusions and Future Perspectives
Over the past decade, detection, isolation, and characterization of CTCs have been a research focal point that attracts significant attention from broad research disciplines, including oncology, cancer biology, bioengineering, materials science, chemistry, and other related fields. Although a variety of CTC detection technologies have been demonstrated, continuous efforts are needed to acquire translational data in order to benefit cancer patients. Going beyond CTC detection, it is crucial to further develop highly efficient CTC isolation and purification platforms in order to pave the way for the subsequent molecular and functional analyses. Ultimately, the CTC-derived molecular signatures and functional readouts will be able to provide significant insight into tumor biology during the critical window where treatment intervention could actually make the difference.
Based on the unique NanoVelcro working mechanism, our team at UCLA has successfully demonstrated three generations of NanoVelcro CTC chips capable of detecting, isolating, and purifying CTCs from blood samples with high efficiency. In the presence of different capture agents, NanoVelcro CTC chips were used to capture CTCs shed from several types of solid tumors, including prostate, breast, lung, and pancreatic cancer, as well as melanoma. We were able to subject the CTCs isolated by NanoVelcro CTC chips to subsequent molecular analyses, for example, Sanger sequencing and NGS to verify CTC’s role as tumor liquid biopsy. Meanwhile, we have been testing the feasibility to culture the purified CTCs in order to pave the way for a wide range of applications that will impact realization of personalized medicine. Our continuous research endeavors will be devoted to translating our discovery in the research laboratory to oncology clinics.
Biographies
Millicent Lin obtained her B.S. (2009) at University of California, Santa Cruz. She studied Clinical Medicine at Peking University from 2009 to 2011 and is currently working in Prof. Tseng’s lab. Her research interests are mainly on CTC isolation and characterization.
Jie-Fu Chen received his M.D. (2013) at National Taiwan University. He is currently a postdoctoral fellow at UCLA under Prof. Tseng’s supervision, focusing on clinical application of NanoVelcro assays and molecular characterization of CTCs.
Yi-Tsung Lu received his M.D. (2011) at National Taiwan University. His research interest has always been on the development of diagnostic devices for cancer since he was a medical student.
Yang Zhang is a Ph.D. student at University of Texas, El Paso. He received his B.S. (2009) at Harbin Institute of Technology in Biotechnology and his M.S. (2012) in Electrical & Computer Engineering at University of Texas El Paso. He is currently a visiting student in Prof. Tseng’s lab, working on CTC imaging.
Jinzhao Song obtained his Ph.D. (2013) from Institute of Chemistry, Chinese Academy of Science. He is currently a postdoctoral fellow in Prof. Tseng’s lab, focusing on the development of NanoVelcro CTC Assays and their downstream molecular analyses.
Shuang Hou obtained his Ph.D. (2008) in chemistry from Institute of Chemistry, Chinese Academy of Sciences. After his first postdoctoral study at Max-Planck Institute, he joined Prof. Tseng’s lab. Currently, he is an assistant researcher, instrumental behind the development of NanoVelcro CTC assays.
Zunfu Ke is an associate professor in the Department of Pathology at Sun Yat-Sen University (SYSU), China. He also serves as a molecular pathologist in the 1st Affiliated Hospital of SYSU. Dr. Ke received his M.D. (2005) at Wuhan University and Ph.D. (2008) in pathology at SYSU. He is currently a visiting physician in Prof. Tseng’s lab, working on exploiting NanoVelcro Assays for different types of cancers.
Hsian-Rong Tseng is a professor in the Department of Molecular & Medical Pharmacology at UCLA. He also holds memberships in the California NanoSystems Institute, Crump Institute for Molecular Imaging, and Jonsson Comprehensive Cancer Center on the UCLA campus. Prof. Tseng obtained his Ph.D. (1998) in Organometallic Chemistry from National Taiwan University. After joining UCLA Medical School in 2003, his research interest has been to develop nanostructured materials and microfluidic platforms as enabling technologies for facilitating the advancement of cancer diagnosis and treatment.
Supporting Information Available
This work was supported by National Institutes of Health (Grants R21-CA151159, R33-CA157396, P50-CA92131, R33-CA174562, and PO1-CA168585), Prostate Cancer Foundation (Creativity Award), UCLA Jonsson Comprehensive Cancer Center (Impact Award), and National Natural Science Foundation of China (Grants 30900650/H1615 and 81372501/H1615). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): Dr. Tseng has financial interest in CytoLumina Technologies Corp., a company licensed the intellectural properties associated with the “NanoVelcro diagnostic platforms” described in the manuscript. In addition, Dr. Tseng’s home institution, UCLA, has taken equity in CytoLumina Technologies Corp. as part of the licensing transaction.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kaiser J. Cancer’s circulation problem. Science 2010, 327, 1072–1074. [DOI] [PubMed] [Google Scholar]
- Criscitiello C.; Sotiriou C.; Ignatiadis M. Circulating tumor cells and emerging blood biomarkers in breast cancer. Curr. Opin. Oncol. 2010, 22, 552–558. [DOI] [PubMed] [Google Scholar]
- van de Stolpe A.; Pantel K.; Sleijfer S.; Terstappen L. W.; den Toonder J. M. Circulating tumor cell isolation and diagnostics: Toward routine clinical use. Cancer Res. 2011, 71, 5955–5960. [DOI] [PubMed] [Google Scholar]
- Cristofanilli M.; Budd G. T.; Ellis M. J.; Stopeck A.; Matera J.; Miller M. C.; Reuben J. M.; Doyle G. V.; Allard W. J.; Terstappen L. W.; Hayes D. F. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [DOI] [PubMed] [Google Scholar]
- Shaffer D. R.; Leversha M. A.; Danila D. C.; Lin O.; Gonzalez-Espinoza R.; Gu B.; Anand A.; Smith K.; Maslak P.; Doyle G. V.; Terstappen L. W.; Lilja H.; Heller G.; Fleisher M.; Scher H. I. Circulating tumor cell analysis in patients with progressive castration-resistant prostate cancer. Clin. Cancer Res. 2007, 13, 2023–2029. [DOI] [PubMed] [Google Scholar]
- Talasaz A. H.; Powell A. A.; Huber D. E.; Berbee J. G.; Roh K. H.; Yu W.; Xiao W. Z.; Davis M. M.; Pease R. F.; Mindrinos M. N.; Jeffrey S. S.; Davis R. W. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 3970–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb W.; Fan A.; Tran T.; Danila D. C.; Keys D.; Schwartz M.; Ionescu-Zanetti C. Mutational analysis of circulating tumor cells using a novel microfluidic collection device and qPCR assay. Transl. Oncol. 2013, 6, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casavant B. P.; Guckenberger D. J.; Berry S. M.; Tokar J. T.; Lang J. M.; Beebe D. J. The VerIFAST: An integrated method for cell isolation and extracellular/intracellular staining. Lab Chip 2013, 13, 391–396. [DOI] [PubMed] [Google Scholar]
- Ozkumur E.; Shah A. M.; Ciciliano J. C.; Emmink B. L.; Miyamoto D. T.; Brachtel E.; Yu M.; Chen P. I.; Morgan B.; Trautwein J.; Kimura A.; Sengupta S.; Stott S. L.; Karabacak N. M.; Barber T. A.; Walsh J. R.; Smith K.; Spuhler P. S.; Sullivan J. P.; Lee R. J.; Ting D. T.; Luo X.; Shaw A. T.; Bardia A.; Sequist L. V.; Louis D. N.; Maheswaran S.; Kapur R.; Haber D. A.; Toner M. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 2013, 5, 179ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.; Wang H.; Hartmann L. C.; Cheng J. X.; Low P. S. In vivo quantitation of rare circulating tumor cells by multiphoton intravital flow cytometry. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 11760–11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.; Schiro P. G.; Kuo J. S.; Koehler K. M.; Sabath D. E.; Popov V.; Feng Q.; Chiu D. T. An automated high-throughput counting method for screening circulating tumor cells in peripheral blood. Anal. Chem. 2013, 85, 2465–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagrath S.; Sequist L. V.; Maheswaran S.; Bell D. W.; Irimia D.; Ulkus L.; Smith M. R.; Kwak E. L.; Digumarthy S.; Muzikansky A.; Ryan P.; Balis U. J.; Tompkins R. G.; Haber D. A.; Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. A.; Okagbare P. I.; Feng J.; Hupert M. L.; Patterson D.; Gottert J.; McCarley R. L.; Nikitopoulos D.; Murphy M. C.; Soper S. A. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J. Am. Chem. Soc. 2008, 130, 8633–8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott S. L.; Hsu C. H.; Tsukrov D. I.; Yu M.; Miyamoto D. T.; Waltman B. A.; Rothenberg S. M.; Shah A. M.; Smas M. E.; Korir G. K.; Floyd F. P. Jr.; Gilman A. J.; Lord J. B.; Winokur D.; Springer S.; Irimia D.; Nagrath S.; Sequist L. V.; Lee R. J.; Isselbacher K. J.; Maheswaran S.; Haber D. A.; Toner M. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 18392–18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. J.; Yobas L.; Lee G. Y.; Ong C. N.; Lim C. T. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed. Microdevices 2009, 11, 883–892. [DOI] [PubMed] [Google Scholar]
- Lecharpentier A.; Vielh P.; Perez-Moreno P.; Planchard D.; Soria J. C.; Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br. J. Cancer 2011, 105, 1338–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel K.; Brakenhoff R. H.; Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer 2008, 8, 329–340. [DOI] [PubMed] [Google Scholar]
- Fan Z. H.; Beebe D. J. Lab on a chip and circulating tumor cells. Lab Chip 2014, 14, 12–13. [DOI] [PubMed] [Google Scholar]
- Yu M.; Bardia A.; Aceto N.; Bersani F.; Madden M. W.; Donaldson M. C.; Desai R.; Zhu H.; Comaills V.; Zheng Z.; Wittner B. S.; Stojanov P.; Brachtel E.; Sgroi D.; Kapur R.; Shioda T.; Ting D. T.; Ramaswamy S.; Getz G.; Iafrate A. J.; Benes C.; Toner M.; Maheswaran S.; Haber D. A. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014, 345, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr J. G.; Adalsteinsson V. A.; Cibulskis K.; Choudhury A. D.; Rosenberg M.; Cruz-Gordillo P.; Francis J. M.; Zhang C. Z.; Shalek A. K.; Satija R.; Trombetta J. J.; Lu D.; Tallapragada N.; Tahirova N.; Kim S.; Blumenstiel B.; Sougnez C.; Lowe A.; Wong B.; Auclair D.; Van Allen E. M.; Nakabayashi M.; Lis R. T.; Lee G. S.; Li T.; Chabot M. S.; Ly A.; Taplin M. E.; Clancy T. E.; Loda M.; Regev A.; Meyerson M.; Hahn W. C.; Kantoff P. W.; Golub T. R.; Getz G.; Boehm J. S.; Love J. C. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 2014, 32, 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Wang H.; Jiao J.; Chen K. J.; Owens G. E.; Kamei K.; Sun J.; Sherman D. J.; Behrenbruch C. P.; Wu H.; Tseng H. R. Three-dimensional nanostructured substrates toward efficient capture of circulating tumor cells. Angew. Chem., Int. Ed. 2009, 48, 8970–8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine J.; Luo S. C.; Wang S.; Zhu B.; Tseng H. R.; Yu H. H. Functionalized conducting polymer nanodots for enhanced cell capturing: the synergistic effect of capture agents and nanostructures. Adv. Mater. 2011, 23, 4788–4792. [DOI] [PubMed] [Google Scholar]
- Zhang N.; Deng Y.; Tai Q.; Cheng B.; Zhao L.; Shen Q.; He R.; Hong L.; Liu W.; Guo S.; Liu K.; Tseng H. R.; Xiong B.; Zhao X. Z. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv. Mater. 2012, 24, 2756–2760. [DOI] [PubMed] [Google Scholar]
- Hsiao Y.-S.; Luo S.-C.; Hou S.; Zhu B.; Sekine J.; Kuo C.-W.; Chueh D.-Y.; Yu H.-h.; Tseng H.-R.; Chen P. 3D bioelectronic interface: Capturing circulating tumor cells onto conducting polymer-based micro/nanorod arrays with chemical and topographical control. Small 2014, 10, 3012–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Liu K.; Liu J.; Yu Z. T.; Xu X.; Zhao L.; Lee T.; Lee E. K.; Reiss J.; Lee Y. K.; Chung L. W.; Huang J.; Rettig M.; Seligson D.; Duraiswamy K. N.; Shen C. K.; Tseng H. R. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew. Chem., Int. Ed. 2011, 50, 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. T.; Zhao L.; Shen Q.; Garcia M. A.; Wu D.; Hou S.; Song M.; Xu X.; Ouyang W. H.; Ouyang W. W.; Lichterman J.; Luo Z.; Xuan X.; Huang J.; Chung L. W.; Rettig M.; Tseng H. R.; Shao C.; Posadas E. M. NanoVelcro chip for CTC enumeration in prostate cancer patients. Methods 2013, 64, 144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.; Xu L.; Zhao L.; Wu D.; Fan Y.; Zhou Y.; Ouyang W. H.; Xu X.; Zhang Z.; Song M.; Lee T.; Garcia M. A.; Xiong B.; Hou S.; Tseng H. R.; Fang X. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv. Mater. 2013, 25, 2368–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S.; Zhao L.; Shen Q.; Yu J.; Ng C.; Kong X.; Wu D.; Song M.; Shi X.; Xu X.; OuYang W. H.; He R.; Zhao X. Z.; Lee T.; Brunicardi F. C.; Garcia M. A.; Ribas A.; Lo R. S.; Tseng H. R. Polymer nanofiber-embedded microchips for detection, isolation, and molecular analysis of single circulating melanoma cells. Angew. Chem., Int. Ed. 2013, 52, 3379–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.; Lu Y. T.; Li F.; Wu K.; Hou S.; Yu J.; Shen Q.; Wu D.; Song M.; Ouyang W. H.; Luo Z.; Lee T.; Fang X.; Shao C.; Xu X.; Garcia M. A.; Chung L. W.; Rettig M.; Tseng H. R.; Posadas E. M. High-purity prostate circulating tumor cell isolation by a polymer nanofiber-embedded microchip for whole exome sequencing. Adv. Mater. 2013, 25, 2897–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S.; Zhao H.; Zhao L.; Shen Q.; Wei K. S.; Suh D. Y.; Nakao A.; Garcia M. A.; Song M.; Lee T.; Xiong B.; Luo S. C.; Tseng H. R.; Yu H. H. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv. Mater. 2013, 25, 1547–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettinger C. J.; Langer R.; Borenstein J. T. Engineering substrate topography at the micro- and nanoscale to control cell function. Angew. Chem., Int. Ed. 2009, 48, 5406–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Wang S. Three-dimensional nano-biointerface as a new platform for guiding cell fate. Chem. Soc. Rev. 2014, 43, 2385–2401. [DOI] [PubMed] [Google Scholar]
- Saracino G. A. A.; Cigognini D.; Silva D.; Caprini A.; Gelain F. Nanomaterials design and tests for neural tissue engineering. Chem. Soc. Rev. 2013, 42, 225–262. [DOI] [PubMed] [Google Scholar]
- Fischer K. E.; Aleman B. J.; Tao S. L.; Hugh Daniels R.; Li E. M.; Bunger M. D.; Nagaraj G.; Singh P.; Zettl A.; Desai T. A. Biomimetic nanowire coatings for next generation adhesive drug delivery systems. Nano Lett. 2009, 9, 716–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.; Jang Y.; Seo J.; Nam J.-M.; Char K. Nanoparticle-functionalized polymer platform for controlling metastatic cancer cell adhesion, shape, and motility. ACS Nano 2011, 5, 5444–5456. [DOI] [PubMed] [Google Scholar]
- Park G.-S.; Kwon H.; Kwak D. W.; Park S. Y.; Kim M.; Lee J.-H.; Han H.; Heo S.; Li X. S.; Lee J. H.; Kim Y. H.; Lee J.-G.; Yang W.; Cho H. Y.; Kim S. K.; Kim K. Full surface embedding of gold clusters on silicon nanowires for efficient capture and photothermal therapy of circulating tumor cells. Nano Lett. 2012, 12, 1638–1642. [DOI] [PubMed] [Google Scholar]
- Banerjee S. S.; Paul D.; Bhansali S. G.; Aher N. D.; Jalota-Badhwar A.; Khandare J. Enhancing surface interactions with colon cancer cells on a transferrin-conjugated 3D nanostructured substrate. Small 2012, 8, 1657–1663. [DOI] [PubMed] [Google Scholar]
- Liu X.; Chen L.; Liu H.; Yang G.; Zhang P.; Han D.; Wang S.; Jiang L. Bio-inspired soft polystyrene nanotube substrate for rapid and highly efficient breast cancer-cell capture. NPG Asia Mater. 2013, 5, e63. [Google Scholar]
- He R.; Zhao L.; Liu Y.; Zhang N.; Cheng B.; He Z.; Cai B.; Li S.; Liu W.; Guo S.; Chen Y.; Xiong B.; Zhao X.-Z. Biocompatible TiO2 nanoparticle-based cell immunoassay for circulating tumor cells capture and identification from cancer patients. Biomed. Microdevices 2013, 15, 617–626. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Chen L.; Xu T.; Liu H.; Liu X.; Meng J.; Yang G.; Jiang L.; Wang S. Programmable fractal nanostructured interfaces for specific recognition and electrochemical release of cancer cells. Adv. Mater. 2013, 25, 3566–3570. [DOI] [PubMed] [Google Scholar]
- Benson K.; Prasetyanto E. A.; Galla H.-J.; Kehr N. S. Self-assembled monolayers of bifunctional periodic mesoporous organosilicas for cell adhesion and cellular patterning. Soft Matter 2012, 8, 10845–10852. [Google Scholar]
- Kumeria T.; Kurkuri M. D.; Diener K. R.; Parkinson L.; Losic D. Label-free reflectometric interference microchip biosensor based on nanoporous alumina for detection of circulating tumour cells. Biosens. Bioelectron. 2012, 35, 167–173. [DOI] [PubMed] [Google Scholar]
- Stroock A. D.; Dertinger S. K.; Ajdari A.; Mezic I.; Stone H. A.; Whitesides G. M. Chaotic mixer for microchannels. Science 2002, 295, 647–651. [DOI] [PubMed] [Google Scholar]
- Sun J.; Masterman-Smith M. D.; Graham N. A.; Jiao J.; Mottahedeh J.; Laks D. R.; Ohashi M.; DeJesus J.; Kamei K.; Lee K. B.; Wang H.; Yu Z. T.; Lu Y. T.; Hou S.; Li K.; Liu M.; Zhang N.; Wang S.; Angenieux B.; Panosyan E.; Samuels E. R.; Park J.; Williams D.; Konkankit V.; Nathanson D.; van Dam R. M.; Phelps M. E.; Wu H.; Liau L. M.; Mischel P. S.; Lazareff J. A.; Kornblum H. I.; Yong W. H.; Graeber T. G.; Tseng H. R. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Res. 2010, 70, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X.; Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc. Chem. Res. 2010, 43, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.; Weng S.; Zhang F.; Allen S.; Li X.; Bao L.; Lam R. H.; Macoska J. A.; Merajver S. D.; Fu J. Nanoroughened surfaces for efficient capture of circulating tumor cells without using capture antibodies. ACS Nano 2013, 7, 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano T.; Yamada N.; Okuhara M.; Sakai H.; Sakurai Y. Mechanism of cell detechment from temperature-modulated hydrophilic-hydrophobic polymer surfaces. Biomaterials 1995, 16, 297–303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.