Abstract
Dollar spot is the most economically important disease of amenity turfgrasses in the United States, yet little is known about the source of primary inoculum for this disease. With the exception of a few isolates from the United Kingdom, Sclerotinia homoeocarpa, the causal agent of dollar spot, does not produce spores. Consequently, it was assumed that overwintering of this organism in soil, thatch, and plant debris provides primary inoculum for dollar spot epidemics. Overwintering of S. homoeocarpa in roots and shoots of symptomatic and asymptomatic creeping bentgrass turfgrass was quantified over the course of a three-year field experiment. Roots did not consistently harbor S. homoeocarpa, whereas S. homoeocarpa was isolated from 30% of symptomatic shoots and 10% of asymptomatic shoots in the spring of two out of three years. The presence of stroma-like pathogen material on leaf blades was associated with an increase in S. homoeocarpa isolation and colony diameter at 48 hpi. Commercial seed has also been hypothesized to be a potential source of initial inoculum for S. homoeocarpa. Two or more commercial seed lots of six creeping bentgrass cultivars were tested for contamination with S. homoeocarpa using culture-based and molecular detection methods. A viable, pathogenic isolate of S. homoeocarpa was isolated from one commercial seed lot and contamination of this lot was confirmed with nested PCR using S. homoeocarpa specific primers. A sensitive nested PCR assay detected S. homoeocarpa contamination in eight of twelve (75%) commercial seed lots. Seed source, but not cultivar or resistance to dollar spot, influenced contamination by S. homoeocarpa. Overall, this research suggests that seeds are a potential source of initial inoculum for dollar spot epidemics and presents the need for further research in this area.
Introduction
Dollar spot, caused by the fungus Sclerotinia homoeocarpa, is the most important disease of turfgrass in North America. It is responsible for the largest disease management expenditures in turfgrass settings throughout the world [1], [2], [3]. Under intense disease pressure, fungicide application costs for dollar spot management may exceed $170/ha per year on high-value golf course greens and fairways [4]. These high costs, coupled with concerns over the environmental impacts of fungicide use and the prevalence of fungicide resistance in S. homoeocarpa populations have motivated the development of alternative methods for dollar spot management [3], [5].
The initial descriptions of S. homoeocarpa by Bennett [6] reported three strain types for this fungus: Perfect, ascigerous, and nonsporiferous. Since that time, researchers have shown that only S. homoeocarpa isolates collected from fescues (Festuca sp.) in the United Kingdom are capable of producing conidia or fertile apothecia [6], [7], [8]. Original strains of S. homoeocarpa from Australia and the United States examined by Bennett [6] were categorized as nonsporiferous and subsequent studies found that S. homoeocarpa isolates from outside the United Kingdom produce apothecial initials but fail to mature and produce sexual ascospores [9], [10]. Consequently, vegetative mycelia and substratal stroma, which is formed by S. homoeocarpa under adverse conditions, are believed to be the primary means of survival for this pathogen [1], [11].
The stroma formed by S. homoeocarpa consists of a thin, plate-like mat of melanized hyphal cells and lacks the structural features of true sclerotia [10], [12]. With the exception of S. homoeocarpa, this type of indeterminate stroma is formed by saprophytic fungi and has been termed a survival structure [12]. Only a single report associates the presence of stroma on leaf blades with overwintering of S. homoeocarpa [9]. In that study, the pathogen was isolated from less than 10% of symptomatic leaf blades after a three-month winter incubation period and a considerable number of these isolations were made from leaf blades with visible stroma [9]. While this research provided support for the ability of S. homoeocarpa to survive the winter months in dormant turf, it was conducted in artificially constructed environments that may not reflect the true overwintering capacity of S. homoeocarpa and was restricted to a single year in a single location [9]. Since in planta overwintering is considered an important source of initial inoculum for this pathogen, additional years of data and a more in-depth investigation of S. homoeocarpa overwintering are warranted [3].
Dollar spot is ubiquitous in golf course settings and appears within a year of seeding, even following methyl bromide fumigation [14], [15]. The presence of dollar spot after methyl bromide application coupled with evidence that S. homoeocarpa grows poorly in soil without plant debris, such as a newly constructed turf green or fairway, suggests that this is not a soil-borne pathogen [16]. Further, lack of spore production by S. homoeocarpa isolates originating outside of the United Kingdom indicates that long-distance dispersal via wind or rain is unlikely [9], [10]. Geostatistical analysis of dollar spot epidemics demonstrated limited spread of this pathogen within a season, indicating that equipment does not contribute to spread of inoculum. Consequently, the authors concluded that epidemics initiated from individual clusters of infection centers [13].
Introduction of the pathogen with turfgrass seed is a possible source of long-distance dissemination for S. homoeocarpa that corresponds with initiation of epidemics from aggregated areas in a turfgrass sward. In the United States, the vast majority of cool-season turfgrass seed is produced in the Pacific Northwest [17], [18]. There is also limited genetic diversity in S. homoeocarpa isolates collected from cool-season grasses in the United States and Canada [19], [20], [21]. Conversely, S. homoeocarpa isolates collected from warm-season grasses, which are vegetatively propagated and cultivated in many regions, have much greater genetic diversity than their C3 counterparts [17], [22], [23]. We hypothesize that one explanation for this difference in genetic diversity of C3 and C4 isolates could be dissemination of the pathogen with seed or vegetative propagation material. The finding that seeding rate of both Kentucky bluegrass [24] and fine fescue [14] is positively correlated with dollar spot severity supports the hypothesis that S. homoeocarpa may be dispersed with seed. Although this correlative evidence suggests that seed may be a means spreading S. homoeocarpa, commercial seed has not previously been evaluated as a possible source of inoculum for S. homoeocarpa.
In the present study, we evaluated both in planta overwintering of S. homoeocarpa and the prevalence of S. homoeocarpa contamination in commercial creeping bentgrass seed. The goal of the study was to identify sources of primary inoculum for the dollar spot pathogen in order to advance understanding of dollar spot epidemiology. The specific objectives of this research were to determine: (1) the rate of pathogen overwintering in roots and shoots of symptomatic and asymptomatic creeping bentgrass turf; (2) the importance of stroma in S. homoeocarpa overwintering; and (3) the viability, pathogenicity, and prevalence of S. homoeocarpa on commercial creeping bentgrass seed.
Materials and Methods
Field studies were conducted at the O.J. Noer Turfgrass Research Facility. This site is owned by the University of Wisconsin-Madison and is available to all University of Wisconsin-Madison turfgrass research faculty and students. The O.J. Noer Turfgrass Research Facility is located in Verona, WI (43.024181, −89.534294). The turfgrass species sampled, creeping bentgrass, is not considered an endangered or protected species.
Study site and sample collection
The overwintering study was conducted on a United States Golf Association sand-based creeping bentgrass (Agrostis stolonifera L.; cv. ‘Penncross’) putting green maintained at a height of 0.40 cm and located at the University of Wisconsin-Madison's O.J. Noer Turfgrass Research and Education Facility in Verona, Wisconsin (43.024181, −89.534294). The same site was resampled in all three years of the study. In each year, late season dollar spot development permitted sampling from symptomatic turf at the margin of dollar spot infection centers and asymptomatic turf that was not within dollar spot infection centers. Fall sampling was conducted in December of all three years, after the ground had frozen but prior to lasting snow cover. Spring sampling was conducted in late-March to mid-April of each year, after the ground had thawed and snow cover had melted (Table 1). Samples were collected using a standard 2 cm diameter soil probe to a depth of approximately 8 cm to allow for collection of root tissue. In the first winter of the study, 20 samples were collected from both symptomatic and asymptomatic turf. Twenty-five samples were collected in the second and third years of the study. Symptomatic samples were collected from the perimeter of the arbitrarily selected dollar spot infection center. Asymptomatic samples were collected from symptomless turf approximately 15 cm from each infection center; thus, a symptomatic and an asymptomatic sample collected for each infection center. Infection centers were marked with golf tees and were resampled in the spring to allow direct comparisons of fall and spring isolation rates. All samples were collected in individual zip-lock plastic bags and stored at 4°C until plating (<72 hpi).
Table 1. Sampling dates for S. homoeocarpa overwintering sample collections.
| Year | Season | Sampling Date | No. of Samples |
| 2010–2011 | Fall | 12/1/2010 | 20 |
| 2010–2011 | Spring | 3/24/2011 | 20 |
| 2011–2012 | Fall | 12/7/2011 | 25 |
| 2011–2012 | Spring | 3/29/2012 | 25 |
| 2012–2013 | Fall | 12/17/2012 | 25 |
| 2012–2013 | Spring | 4/16/2013 | 25 |
Sample plating and S. homoeocarpa isolation
Intact soil cores were surface disinfested prior to plating by submerging in 10% Clorox (0.06% sodium hypochlorite) for 15 s and gently massaging the root area to remove attached sand. Disinfestation was immediately followed by two 30 s rinses in deionized water and samples were then dried on fresh paper towels. Sterile forceps were used to select and plate individual leaf blades and roots onto antibiotic-amended PDA (BD Difco, Sparks, MD; amended with 100 mg/L each tetracycline, chloramphenicol, and streptomycin sulfate). For each sample, four leaf blades and four root pieces were placed on two replicate 50 mm diameter petri dishes (Fisher Scientific, Hanover Park, IL), resulting in a total of eight organ subsamples per sample. All petri dishes were inverted and incubated at ambient temperature (22±2°C) for 48 h.
From 48–96 h after plating, fungal colonies resembling S. homoeocarpa were marked and sub-cultured onto fresh antibiotic-amended PDA to obtain pure cultures. In the first year of the study, colony morphology and sequencing of the internal transcribed spacer (ITS) 1 region with the primer pair ITS1/ITS2 were used to positively confirm S. homoeocarpa isolations. Fungal DNA was extracted from freshly harvested mycelia of S. homoeocarpa using a modified CTAB extraction protocol [25] and fungal ITS region-specific primers ITS1 and ITS2 [26] were used for amplification and sequencing of fungal DNA. DNA sequencing was performed at the University of Wisconsin-Madison DNA Sequencing Facility (Madison, WI). In subsequent years, colony morphology alone was used to confirm S. homoeocarpa isolation.
Stroma plating experiments used a procedure similar to that described above. A dissecting microscope was used to visually identify leaf blades with and without stroma-like material from samples previously identified as positive for S. homoeocarpa isolation in fall 2012 and spring 2013. Because dollar spot lesions on closely mown creeping bentgrass leaf blades rarely have the distinct reddish brown borders associated with stroma formation in other grasses, dark material embedded in tissue with dollar spot lesions was considered stroma-like. Leaf blades with and without stroma-like material were plated individually in 50 mm diameter petri dishes and S. homoeocarpa isolation and identification were repeated as above. Colony diameters were measured at 48 h after plating and were calculated by averaging the lengths of two perpendicular transects across the colony. In total, 20 leaf blades with and 20 leaf blades without stroma-like material were used in these studies. Sample size was constrained by the limited number of leaf blades with stroma-like material.
Seed sources
Samples from twelve commercial CRB seed lots were obtained directly from seed production companies. CRB cultivars for seed assays were selected based on their resistance to dollar spot in National Turfgrass Evaluation Program trials from 2009–2011 (http://www.ntep.org; Table 2). A minimum of two seed lots per cultivar were tested for S. homoeocarpa contamination by both culture-based and molecular detection methods.
Table 2. Seed sources, cultivars, and National Turf Evaluation Program performance results for creeping bentgrass commercial seed lots used in S. homoeocarpa culture-based and molecular seed detection studies.
| Source | Cultivar | NTEP Rating2 | Resistance Designation |
| Seed Research of Oregon | 96-2 | 4.63 | VS |
| Mountain View Seed | Shark | 5.3 | MS |
| Tee-2-Green | Penncross | 5.5 | MS |
| Seed Research of Oregon | SR1150 | 6.3 | R |
| Lebanon Turf | Declaration | 7.4 | VR |
| Seed Research of Oregon | Focus1 | 7.9 | VR |
The experimental name for cv. ‘Focus’ in NTEP trials was ‘GMC comp’.
NTEP ratings provided are the average from dollar spot trials in 2009, 2010, and 2010 that included all six cultivars.
Ratings are based on turfgrass quality and are given on a 0–9 scale; 9 = highest turf quality/resistance to dollar spot and 0 = lowest turf quality/resistance to dollar spot.
Culture-based detection
Individual seed plating
In an experimental replicate, a total of 1000 seeds from each seed lot were plated on two fungicide-amended media semi-selective for S. homoeocarpa [27], one with 5 ppm azoxystrobin and the other with 0.1 ppm triticonazole, in 100 mm diameter petri plates (Fisher Scientific, Hanover Park, IL). Twenty-five seeds were placed on each petri dish, resulting in a total of 20 plates of each medium and 40 plates total per seed lot in each experimental replicate. A dissecting needle was used to select and plate individual seeds. The entire experiment was repeated three times; thus, an overall total of 3,000 seeds per lot were plated. For each experimental replicate, plates were sealed, inverted, and incubated at 22±2°C for 72–120 h. Fungal colonies resembling S. homoeocarpa were marked and positively identified as described above for overwintering samples. To allow for identification of other fungal species commonly present in commercial CRB seed, the ten most common fungal colony morphotypes were noted and isolated in pure culture. DNA was extracted, using methods described above [25], from fresh mycelia of these cultures and the ITS 1 region was sequenced with the fungal ITS-region primers ITS1/ITS2 [25].
Enrichment and dilution plating
Potato dextrose broth (PDB) was prepared according to the manufacturer's instructions and amended with 100 mg/L of the antibiotics streptomycin sulfate, chloramphenicol, and tetracycline and 25 ppm azoxystrobin (Heritage TL; Syngenta, Greensboro, NC). HCl was added to the medium to decrease the pH to four. Twenty mL of antibiotic and fungicide-amended PDB at pH 4 and 0.25 g of seed from each commercial seed lot were added to 12 sterile flasks. Flasks were sealed with a double layer of parafilm and incubated with constant shaking for 72 h at ambient temperature (22±2°C). After 72 h, the suspension was homogenized in a commercial blender and four 10-fold serial dilutions were made for each sample. Three 0.5 mL aliquots of each dilution were plated onto separate azoxystrobin-amended PDA plates prepared as described above, except that PDA was substituted for PDB. All plates were incubated for 24–48 h. Potential S. homoeocarpa colonies were then marked and sub-cultured to obtain pure cultures, as previously described.
Pathogenicity assays with S. homoeocarpa isolates collected from seed were conducted as follows. Four-week-old CRB (cv. ‘Penncross’) plants were inoculated by placing two agar plugs, collected from the edge of four day-old fungal cultures, deep within the center of the turfgrass canopy. Mock-inoculated controls were included for comparison. All pots in the experiment were randomized in a plastic flat and covered with a humidity dome to promote infection. Four-week-old barley (cv. ‘Parkland’) plants were inoculated using the parafilm sachet method [28] and control plants were mock-inoculated with PDA plugs. Disease symptoms on both CRB and barley were photographed at 120 hpi. Both CRB and barley plants were cultivated in a growth room at 24±2°C with a 14 h day-length and inoculations were performed under these conditions. Barley was used in these assays because characteristic S. homoeocarpa lesions form on this host in controlled environment conditions and are more easily visualized than in creeping bentgrass.
Molecular detection
For nested and quantitative PCR, primer sets were designed to the outside and inside, respectively, of a previously described primer set specific to the S. homoeocarpa EF1α gene [29] (Table 3). The NCBI Primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to design primers specific to the S. homoeocarpa EF1α DNA sequence (GenBank Accession DQ448301).
Table 3. Primers used for molecular detection of S. homoeocarpa in creeping bentgrass commercial seed lots.
| Primer Name | Sequence | Prod. Length (bp) | Use | Reference |
| EF1α_F | CGGTATGACTTCTCCACCTTTC | 219 | Nested PCR | Al-Elmagid et al. 2013 |
| EF1α_R | GAACCCTTTCCCATCTCCTT | Nested PCR | Al-Elmagid et al. 2013 | |
| EF1α_Outer_F | CGGTAAGCAGAACCCTCGAC | 554 | Primary PCR | This paper1 |
| EF1α_Outer_R | CAGCTTGGGAGGTACCAGTG | Primary PCR | This paper | |
| EF1α_Nest_F | TTATCGGGTTGCGTTTTCTC | 102 | Q-PCR | This paper |
| EF1α_Nest_R | AACGGGTTAGCAAAGGGATT | Q-PCR | This paper |
Primers developed in the present research were designed using the NCBI Primer-BLAST tool with the DNA sequence for the S. homoeocarpa EF1α input as the sole target sequence. Primer specificity was checked by sequencing of primary and nested PCR products and qPCR melt curve analysis.
Seed DNA was extracted from ten 50 mg subsamples of each seed lot using the Qiagen DNeasy Plant Mini-Kit (Qiagen, Carlsbad, CA) according to the manufacturer's instructions except with slight modifications in sample preparation. Specifically, seeds were ground in Lysing Matrix A tubes (MP Biomedicals, Irvine, CA) containing two 0.64 cm diameter ceramic beads with a FastPrep-24 (MP Biomedicals, Irvine, CA). Samples were homogenized twice for 40 s at 6 m s−1 in Qiagen DNeasy kit buffer AP1. RNAse A was added to samples after homogenization but otherwise the manufacturer's protocol was followed. DNA quantity and quality were assessed with an ND-1000 Spectrophotomer (NanoDrop Technologies, Wilmington, DE).
Both primary and nested PCR reactions were performed in 25 µL reaction volumes with GoTaq Colorless PCR Master Mix (Promega, Madison, WI), following the manufacturers protocol. Primary reactions contained: 12.5 µL GoTaq Colorless PCR Master Mix, 2 µL seed DNA, 0.2 µM of primers EF1α_outerF and EF1α_outerR, and nuclease free water (NFW). Nested reaction mixtures were identical, except that 2 µL of primary PCR products were used as template and the EF1α primer set was used. Positive control primary PCR products were diluted 1∶30 with NFW. Cycling conditions for primary and nested PCR included an initial two minute denaturation period at 95°C; 30 cycles of 30 s denaturation at 95°C, 30 s annealing at 56°C and 52°C for primary and nested PCR, respectively, and 30 s elongation at 72°C; and a final five minute elongation period. A positive control of seed DNA spiked with S. homoeocarpa genomic DNA and a no-template negative control were included in all runs and PCR was performed in a MasterCycle Pro S (Eppendorf, Hamburg, Germany). Five µL of all primary and nested PCR products were mixed with 1 µL of 6× DNA loading dye and subject to electrophoresis in a 1% agarose gel stained with SYBR Safe DNA Gel Stain (Invitrogen, Carlsbad, CA). Gels were visualized with UV light in a Universal Hood II Gel Doc System (BioRad, Hercules, CA) and analyzed with Quantity One imaging software (BioRad, Hercules, CA). Positive bands were excised from the gel with a sterile razor blade and cleaned up with the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI) then sequenced at the University of Wisconsin-Madison DNA Sequencing facility (Madison, WI). NCBI BLASTn was used to confirm sequences were specific to the S. homoeocarpa EF1- α gene sequence.
Quantitative PCR (qPCR) reactions were performed in hard-shell 96-well skirted PCR plates and sealed with Microseal ‘B’ adhesive seals (BioRad, Hercules, CA). qPCR reactions contained 1× SsoFast EvaGreen Supermix, 8 µL template (seed DNA or EF1α plasmid DNA standard), 0.2 µM EF1α_nest_F and EF1α_nest_R, and NFW for a final reaction volume of 20 µL. A standard curve with concentrations of EF1α plasmid DNA ranging from 106 to one copy/µL was included on each plate and all DNA samples were assayed in triplicate. Cycling conditions included an initial 2 minute denaturation at 98°C, followed by 40 cycles of 98°C for 2 s and annealing at 55°C for 2 s. A melt curve was created after the final cycle by increasing the temperature from 70–90°C in 0.2°C intervals and holding at each temperature for 10 s. qPCR reactions were performed in a CFX-96 Real-Time System (Bio-Rad, Hercules, CA). The plasmid used for generation of standard curves and copy number estimation was constructed by cloning S. homoeocarpa EF1α sequence amplified by our primers into a pGEM-T Easy plasmid (Promega, Madison, WI) according to the manufacturer's instructions. Escherichia coli DH5α was transformed with plasmid containing the target sequence and plasmid DNA was purified from overnight cultures of E. coli containing the cloned target using the Wizard Plus SV Miniprep DNA purification system (Promega, Madison, WI), as instructed by the manufacturer. A NanoDrop was used to quantify purified plasmid and copy number was calculated according to the method of Frost and colleagues [30].
Statistical analysis
Statistical analyses were performed using the generalized linear mixed model procedure (PROC GLIMMIX) in SAS version 9.3 [31]. Studentized residuals and tests in PROC UNIVARIATE were used to assess normality and determine the best fitting distribution for each data set. Full models with all random effects and their interactions with fixed effects were then fit to the appropriate distribution and model fitting criteria (AIC of -2RLL, depending on distribution) were used to select the best fitting model.
Overwintering data was coded as a binary variable (0 = no S. homoeocarpa detected; 1 = S. homoeocarpa detected) and subject to analysis of variance using the binary distribution. Sample (block) was considered a random effect and all other factors (treatment, organ, and season) were fixed. The full data set was analyzed to test for a season effect. Individual years were then analyzed independently and factors (season or treatment) were sliced to compare specific levels of each factor. Stroma isolation data were also subject to analysis of variance under the binary distribution. Stroma colony diameter data were analyzed using the normal distribution. In both cases, a simple one-way ANOVA was used to test for the main effect of the presence or absence of stroma.
Nested PCR seed detection data were subject to analysis of variance under a normal distribution. Because lots were exclusive to the cultivar from which they came and cultivars were exclusive to the source from which they came, both of these factors were treated as nested in statistical analyses. Source and cultivar (source) were considered fixed effects while lot (cultivar) was treated as a random effect. Fixed treatment effects were assessed with a simple two-way ANOVA and a single degree of freedom orthogonal contrast was used to test for a difference between resistant and susceptible cultivars.
Results
Pathogen overwintering
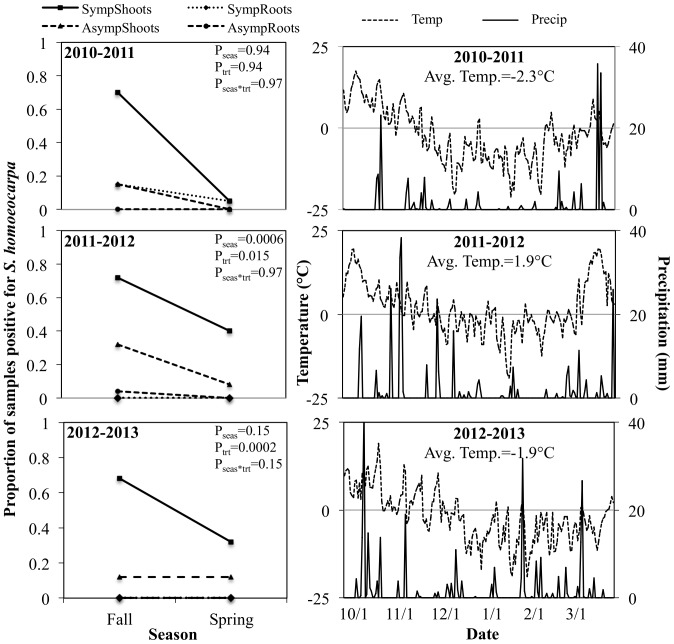
In all three years of this study, the pathogen was isolated from 70% of symptomatic shoots (Fig. 1). S. homoeocarpa was isolated from asymptomatic shoots in the fall of all years, but the percentage of positive samples varied, with 15%, 32%, and 12% of samples positive for S. homoeocarpa in 2010, 2011, and 2012, respectively (Fig. 1). The percentage of root samples positive for S. homoeocarpa was 15% in fall 2010 but negligible in all other seasons. Consequently, roots were not included in statistical analyses of S. homoeocarpa overwintering. In the first year of the study, identification by colony morphology was confirmed by sequencing results (File S1); consequently, colony morphology alone was used to identify S. homoeocarpa isolates in future years of the study.
Figure 1. Sclerotinia homoeocarpa isolation and weather data for pathogen overwintering studies.
Roots and shoots were collected from symptomatic and asymptomatic turf in the fall of each year to determine starting S. homoeocarpa populations. The same areas were resampled in the spring to assess pathogen survival. ANOVA values for season, treatment, and season by treatment effects are reported for each year. Weather data includes daily air temperature (dashed line) and precipitation (solid line) averages. The average temperature in 2011–2012 was c. 4°C higher than in the other two years and a major snow events occurred in early November 2011–2012 and early February of 2012–2013 (arrows).
The majority of spring isolations of S. homoeocarpa were made from samples taken from areas with dollar spot symptoms the previous fall (Fig. 1). The rate of S. homoeocarpa spring isolation from symptomatic tissue ranged from 0.05% in spring of 2010 to 40% in spring of 2011. In two of three years of this study, spring isolations of S. homoeocarpa were also made from tissue collected from asymptomatic areas in the previous fall (Fig. 1). S. homoeocarpa was isolated from 8 and 12% of asymptomatic samples in the spring of 2012 and 2013, respectively. In the 2012–2013 season, there was no difference in the percentage of samples positive for S. homoeocarpa in the fall and spring. A single spring isolation of S. homoeocarpa was made from roots in spring 2011. S. homoeocarpa was not recovered from roots at any other spring sampling date.
Across all years, season had an effect on S. homoeocarpa isolation from both symptomatic (P<0.001) and asymptomatic (P = 0.03) samples. In individual years, a significant effect of season across sample types (symptomatic or asymptomatic) was detected only in Year 2 (Fig. 1). By separating sample types, an effect of season was detected in symptomatic samples for all three years (P<0.05) and for asymptomatic samples in Year 2 (P = 0.03). Sample type had an effect on S. homoeocarpa isolation in the fall of each year (P<0.05), but had no effect on spring isolations (P>0.1) in any year. No season by sample type interaction was detected in any year of the study (P>0.1).
Stroma
Leaf blades with black to brown stroma-like material on or within epidermal tissue were readily identified with a dissecting microscope and were selected as potentially harboring S. homoeocarpa stroma (Fig. 2A). Leaf blades serving as non-stroma controls were green to yellow in color and did not appear to contain fungal material. S. homoeocarpa was isolated from leaf blades with and without stroma, but the presence of stroma-like material significantly increased both the rate of isolation (P = 0.03; Fig. 2B) and growth rate (P = 0.0002; Fig. 2C) of S. homoeocarpa.
Figure 2. Stroma on creeping bentgrass leaf blades and its effect on isolation on S. homoeocarpa.
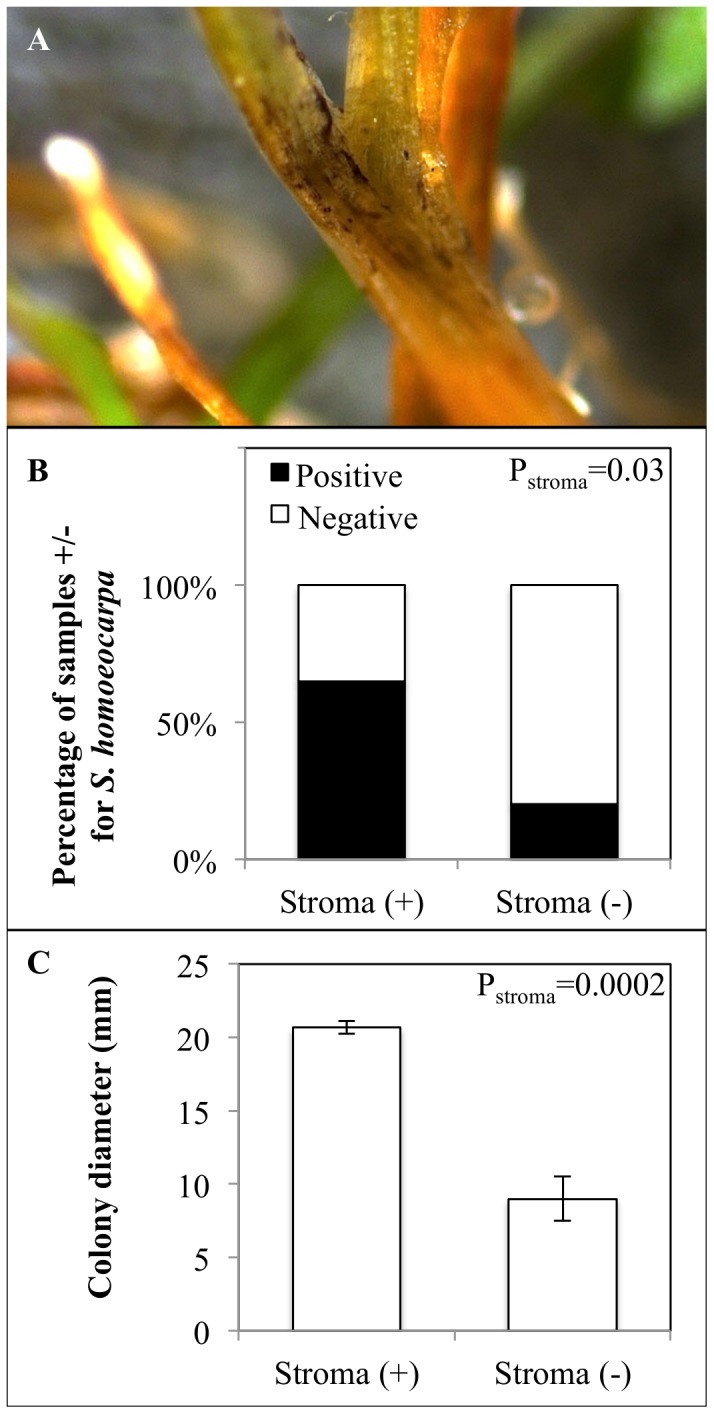
A, typical stroma-like material observed on a turfgrass leaf blade was brown in color and appeared as flaky, plate-like material. B, The percentage of CRB leaf blades with and without visible stroma from which S. homoeocarpa was isolated. P-value from ANOVA using the binary distribution for presence/absence of S. homoeocarpa and α = 0.05. C, Average diameter of S. homoeocarpa colonies 48 h after plating leaf blades with or without visible stroma. P-value is from ANOVA with the normal distribution and α = 0.05. Error bars represent ± one standard error of the mean (n = 5).
Culture-based detection
Individual seed plating
A single isolate of S. homoeocarpa was collected from commercial seed with the culture-based detection method (Table 4). The isolate was collected from Lot 1 of cv. ‘Shark’ on the triticonazole–amended semi-selective medium [27]. Colony morphology (Fig. S1A) and amplification of the ITS1 region by universal primers ITS1/ITS2 [26] (File S1) confirmed the identification of this isolate. Tests on CRB and barley demonstrated pathogenicity and resulting symptoms were similar to those observed with a virulent isolate of S. homoeocarpa (Fig. S1B–C). Specifically, symptoms on CRB included water-soaking around the site of inoculation, accompanied with reddish brown lesions on individual leaf blades. Light-tan lesions with reddish brown borders, characteristic of S. homoeocarpa infection, developed on barley.
Table 4. Results for S. homoeocarpa contamination of creeping bentgrass commercial seed lots by culture-based and molecular detection methods.
| Cultivar | Lot1 | Culture-based Detection2 | Nested PCR3 (Subsamples +) | Q-PCR4 (Subsamples +) |
| 96-2 | 1 | − | + (1) | n/a |
| 2 | − | − (0) | n/a | |
| Shark | 1 | + | + (5) | − (0) |
| 2 | − | + (3) | − (0) | |
| Penncross | 1 | − | + (1) | − (0) |
| 2 | − | + (2) | n/a | |
| SR1150 | 1 | − | − (0) | − (0) |
| 2 | − | − (0) | n/a | |
| Declaration | 1 | − | + (2) | n/a |
| 2 | − | − (0) | n/a | |
| Focus | 1 | − | + (1) | − (0) |
| 2 | − | + (1) | n/a |
Two representative lots of each cultivar were selected by for detection of S. homoeocarpa contamination by each method; additional lots of ‘Shark’ and ‘Penncross’ seed were tested by the nested PCR detection method only.
Three 1,000 seed replicates for all lots were performed by plating 500 seeds from each lot onto two different media semi-selective for S. homoecarpa (1,000 seeds total) and repeating three times (3,000 seeds/lot total).
Nested PCR was repeated twice for each seed lot; numbers shown in parentheses are the number of subsamples testing positive for S. homoeocarpa contamination in each run.
Q-PCR detection was ceased after it was determined that PCR inhibitors in seed DNA samples rendered the assay ineffective.
Overall, 16 different fungal colony morphotypes were commonly isolated during seed plating. The most commonly isolated seed contaminant was a pink yeast in the genus Rhodotorula. The fungus Epicoccum nigrum was the second most commonly isolated seed contaminant. Other frequently isolated fungi included Trichoderma sp., Penicillium polonicum, Mucor fragilis, and Aureobasidium proteae. Lack of morphological characteristics needed for identification, inability to extract quality DNA, or poor sequencing results made it difficult to identify many of the fungi isolated.
Enrichment and dilution plating
After 24 h of incubation, plates from the enrichment experiment were overrun with Zygomycete fungi; consequently, no S. homoeocarpa colonies were identified by this method.
Molecular detection
Nested PCR
Nested PCR using the primary primer set EF1α_Outer and nested primer set EF1α was sensitive enough to detect a single S. homoeocarpa-infested seed in 50 mg of seed (Fig. S2). A minimum of two seed lots per CRB cultivar were tested for S. homoeocarpa contamination with nested PCR. Eight of the twelve seed lots tested were positive for S. homoeocarpa DNA (Table 4), including both seed lots from cultivars ‘Penncross’ and ‘Shark’. The highest contamination was present in ‘Shark’ Lot 1, from which S. homoeocarpa was also isolated using the culture-based detection method. Three additional ‘Shark’ seed lots were subsequently tested for S. homoeocarpa contamination and two were positive (Table 4). A subset of positive nested PCR products were sequenced (File S1) and their identity confirmed through sequence similarity to the S. homoeocarpa EF1α sequence (GenBank Accession DQ448301).
Seed source affected detection of S. homoeocarpa (P = 0.01) based on ANOVA of nested PCR data. CRB cultivar did not affect detection (P>0.05). Cultivars were nested within seed source; thus, source by cultivar interaction was not tested. An orthogonal contrast between cultivars with high and low dollar spot resistance, according to NTEP creeping bentgrass variety trials, indicated that dollar spot resistance did not influence detection levels for S. homoeocarpa (P>0.05; Fig. 3).
Figure 3. Results of nested PCR detection of Sclerotinia homoeocarpa DNA in creeping bentgrass commercial seed lots.
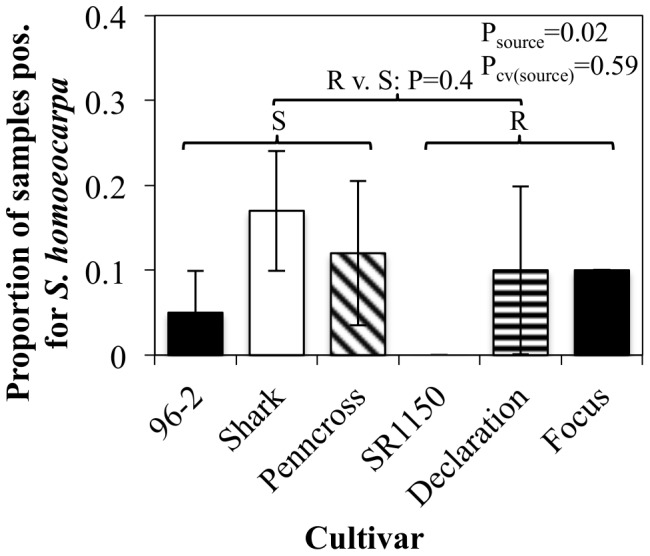
Columns reflect the average proportion of samples positive for S. homoeocarpa contamination across the two lots of each cultivar. Shading indicates cultivar source: ‘96-2’, ‘Focus,’ and ‘SR1150’ were from a single source while ‘Shark,’ ‘Penncross,’ and ‘Declaration’ were from different seed distributors. Error bars represent ± one standard error of the mean. Source and cv(source) P-values are from ANOVA of the data with the normal distribution and α = 0.05. The P-value for R versus S cultivars was obtained from a pre-planned orthogonal contrast.
Quantitative PCR
The primer set EF1α_Nest_F/EF1α_Nest_R had >99% efficiency in standard curves constructed from plasmid DNA and a detection limit as low as 10 target copies/µL template (Fig. S3A). Six seed lots positive for S. homoeocarpa by nested PCR were also tested by qPCR. Five of the six lots, including Shark Lot 1, tested negative for S. homoeocarpa contamination by qPCR (Table 4). The low success rate of qPCR suggested the presence of PCR inhibitors in seed DNA samples. Decreased detection threshold values following sample dilution and increased threshold values in spiked seed DNA samples over standards with the same target concentration confirmed the presence of PCR inhibitors in seed DNA (Fig. S3B).
Discussion
This is the first multi-year study on in planta overwintering of S. homoeocarpa. Overall, our results indicate that S. homoeocarpa overwinters moderately well in infected plant material. Previously, the only study on S. homoeocarpa overwintering was conducted on Kentucky bluegrass (Poa pratensis) at the University of Rhode Island in the winter of 1973–1974 [9]. In that study, S. homoeocarpa was recovered from less than 10% of symptomatic leaf blades and never from asymptomatic leaf blades [9]. This research differs significantly from this previous experiment because a different turf species (creeping bentgrass as opposed to Kentucky bluegrass) and a much harsher winter climate were evaluated. Most importantly, this research evaluated S. homoeocarpa overwintering under entirely natural conditions, without any manipulation of the host or pathogen until the time of sample collection. In the present research, spring recovery of S. homoeocarpa ranged from 5–40% and 0–12% in symptomatic and asymptomatic samples, respectively. The authors of the previous study [9] noted the presence of numerous saprophytic fungi in their samples and degradation of leaf blades by fungal contaminants. While saprophytes were frequently recovered in our study, they did not appear to degrade host tissue in the course of our study. This may be one reason for the higher rates of S. homoeocarpa isolation in the present research. It is also plausible that the higher rates of overwintering in our study are a reflection of the methods used—undisturbed dormant infection centers in our experiment, symptomatic leaf blades placed inside plastic straws in the previous experiment.
The rate of fall isolation from symptomatic shoots was similar in all three years of our study, but spring isolation rates varied, indicating that environmental factors may influence winter survival of S. homoeocarpa. Consequently, average daily temperature and precipitation data were collected and compared for the three years of our study (Fig. 1B,D,F). S. homoeocarpa survival rates were highest in the second year of the study, during which average winter temperatures were approximately 4°C higher than in years one and three. The warmer winter temperatures in 2011–2012 may have promoted overwintering of S. homoeocarpa relative to the other two years of this study. Warmer winter temperatures are also associated with increased survival of the turfgrass stem rust pathogen, Puccinia graminis subsp. graminis on tall fescue and perennial ryegrass [32]. Spring isolations of S. homoeocarpa were higher in the third year of our study than in the first year, despite similar average winter temperatures in both years. A heavy snowfall event in late January of 2013 may have insulated S. homoeocarpa from fluctuating air temperatures, resulting in a greater number of spring isolations in year three. Direct comparisons of these studies to prior research are difficult because most research on fungal pathogen overwintering focuses on survival of spores and often involves artificial manipulations of the pathogen itself or of infected host material [33], [34], [35]. However, a few studies have shown that fungal pathogens survive harsh winter conditions in host debris that then serves as primary inoculum in the following spring [36], [37], [38].
Increased isolation of S. homoeocarpa from leaf blades with stroma in our study agrees with the findings of Fenstermacher [9] on Kentucky bluegrass and supports the general conclusion that stroma is a survival structure for this pathogen [3], [6], [11]. Stroma was not observed on creeping bentgrass roots and isolations of S. homoeocarpa from roots were limited, indicating that overwintering in roots does not contribute to primary inoculum for dollar spot epidemics. To our knowledge, this is the first report that stroma aids overwintering of S. homoeocarpa in creeping bentgrass. The atypical nature of S. homoeocarpa stroma in comparison with other pathogenic fungi in the Sclerotiniaceae family, including the fact that this survival structure is generally restricted to saprophytic fungi, warrant further investigation of the role of this structure in S. homoeocarpa biology and epidemiology [12]. Potentially, control methods aimed at decreasing the formation of stroma in the fall could decrease overwintering of S. homoeocarpa and reduce initial inoculum for dollar spot epidemics.
The isolation of S. homoeocarpa from asymptomatic samples in the spring of 2011–2012 and 2012–2013 was unexpected. Since diseased tissue from the previous year's epidemic was believed to be the source of inoculum for subsequent years [3], [11], [39], detection of this pathogen in asymptomatic material indicates that latent infections may be an important source of overwintering and inoculum. Isolation of S. homoeocarpa from asymptomatic turfgrass also may help explain the detection of S. homoeocarpa in commercial creeping bentgrass seed. In the Pacific Northwest, where the majority of cool-season turfgrass seed is produced, environmental conditions are generally not suitable for dollar spot epidemics [40]. Asymptomatic infection with S. homoeocarpa may allow contamination of seed fields to go unnoticed; thus, leading to harvesting and distribution of seed that appears to be pathogen-free yet is latently contaminated with S. homoeocarpa. Symptomless infection arising from seeds has previously been described for Botrytis cinerea in lettuce [41] and for many bacterial pathogens. The frequency of asymptomatic colonization of turfgrasses by S. homoeocarpa should be investigated further.
To our knowledge, this is the first report of a viable, pathogenic isolate of S. homoeocarpa obtained from commercial seed of any turfgrass species. This finding provides evidence that commercial seed may be an important source of inoculum for dollar spot epidemics. A single isolation from the 36,000 total seeds plated in this study may seem trivial, but a kilogram of CRB seed contains two to four million individual seeds [42]. This translates to approximately 70 S. homoeocarpa-infected seeds per kilogram of creeping bentgrass seed or a minimum of thousands of infected seeds on a freshly seeded golf course putting green at a standard seeding rates [42]. In the present study, we only detected the presence of S. homoeocarpa in commercial seed and did not directly relate this to establishment of dollar spot epidemics in the field. Studies investigating the influence of seed contamination with S. homoeocarpa on the development of dollar spot epidemics are a logical and necessary next step.
Eight of twelve (75%) commercial seed lots in our study tested positive for S. homoeocarpa with nested PCR. In these lots, the number of positive subsamples ranged from 1–5. The discrepancy between S. homoeocarpa detection by the culture-based and molecular methods could be a result of amplification of non-viable pathogen DNA with nested PCR or of contaminant fungi outcompeting S. homoeocarpa with the culture-based method. With the nested PCR method, a single S. homoeocarpa infested seed in a 50 mg sample, approximately 1 in 450 creeping seeds (0.002% infestation), was detected. This detection rate is higher than the 0.01–0.1% detection rates reported for other molecular detection methods [43], [44], [45] though not as high as that of some qPCR detection methods [46], [47]. We found that qPCR was less sensitive than nested PCR for our samples because turfgrass seed has many associated PCR inhibitors. Though qPCR detection methods are currently popular, regular and nested PCR-based detection methods are still highly effective and provide an efficient means of qualitative pathogen detection [44], [48], [49], [50]. In the future, the nested PCR method could lead to development of a loop-mediated isothermal amplification (LAMP) assay for detection of S. homoeocarpa in seed. LAMP assays are less sensitive to PCR inhibitors than conventional and real-time PCR methods and can be performed in the field with minimal equipment or training [51], [52], [53], [54].
Analysis of nested PCR data indicated that seed source but not cultivar resistance to dollar spot had an effect on contamination with S. homoeocarpa. This was somewhat surprising as an influence of host cultivar resistance on seed contamination has previously been demonstrated for other cereal pathogens [55], [56]. However, the sample size of two lots from each of six cultivars limited our power to detect significant differences between cultivars. In the future, it will be necessary to screen more seed lots for S. homoeocarpa contamination. A number of factors could explain the effect of source on seed lot contamination. Previous field history or environmental conditions may differ between production fields used by individual seed companies. Climatic conditions in the Pacific northwest, where the majority of creeping bentgrass seed is grown, are fairly stable and unlikely to vary drastically from one company's fields to the next [17]. It is possible, however, that certain fields or areas within a field have microclimates conducive to plant infection by S. homoeocarpa. Alternatively, management or harvest practices may vary between companies and contribute to the differences observed. It is unclear whether S. homoeocarpa is harbored on or in the seed itself or associated with debris mixed in with the seed. If the latter is the case, seed with minimal debris may also have lower rates of S. homoeocarpa contamination. Future experiments to determine the source of S. homoeocarpa in commercial seed will help to identify practices that promote or limit contamination of seed with this pathogen.
Supporting Information
Sclerotinia homoeocarpa isolate ‘Shark’ obtained from Shark Lot 1 using culture-based detection of semi-selective medium. A, Colony morphology of the isolate ‘Shark’ obtained from CRB cv. ‘Shark’ commercial seed lot 1. B, Symptoms produced by mock-inoculated control, virulent S. homoeocarpa, and seed S. homoeocarpa isolate ‘Shark’ on creeping bentgrass (cv. ‘Penncross’) at 5 dpi. Similar symptoms were produced for each treatment in six biological replicates. C, Symptoms produced by mock-inoculated control, virulent S. homoeocarpa, and seed S. homoeocarpa isolate ‘Shark’ on barley (cv. ‘Parkland’) at 5 dpi. Similar symptoms were produced for each treatment in six biological replicates.
(TIF)
Sensitivity of Sclerotinia homoeocarpa -specific primers in primary and nested PCR. Primary PCR was run with S. homoeocarpa specific primers EF1α_OuterF/EF1α_OuterR, with an expected product size of 554 bp, and contained 2 µL of seed DNA from seed samples spiked with the indicated number of artificially-infested CRB seeds prior to DNA extraction. Nested PCR was performed using S. homoeocarpa specific primers EF1α_F/EF1α_R, with a product size of 219 bp internal to EF1α_OuterF/EF1α_OuterR, and contained 2 µL of primary PCR products diluted 1∶30 in nuclease free water. The negative control was treated exactly as samples except that 2 µL of NFW replaced template DNA in the primary PCR. Five µL of sample were mixed with 1 µL of DNA loading dye and run in a 1% agarose gel in TBE buffer along with 5 µL of TrackIt 100 bp DNA ladder (Invitrogen, Carlsbad, CA).
(TIF)
Q-PCR for molecular detection of Sclerotinia homoeocarpa DNA in creeping bentgrass commercial seed lots. A, Standard curve of EF1α plasmid DNA with the primer set EF1α_NestF/EF1α_NestR indicating near 100% primer efficiency. B, Q-PCR run with various controls to reveal the presence of PCR inhibitors: Spike, Spike−1, Spike−2—Ten-fold dilution series of seed DNA mixed with 1/10 volume EF1α plasmid DNA at a starting concentration 1×106.; Check, Check−1, Check−2—Ten-fold dilution series of a seed DNA sample positive by nested PCR but negative by Q-PCR. Decreasing Cq value with dilution indicates presence of inhibitors in the original DNA sample.
(TIF)
DNA sequences from overwintering Sclerotinia homoeocarpa isolates, isolate ‘Shark’ obtained from commercial seed, and S. homoeocarpa EF1α sequences from seed lots positive by nested PCR.
(DOCX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
These authors have no support or funding to report.
References
- 1. Allen TW, Martinez A, Burpee LL (2005) Dollar spot of turfgrass. Plant Health Instr doi:101094/PHI-I-2005-0217-02 [Google Scholar]
- 2.Vargas JMJ (1994) Management of turfgrass diseases. 2nd edition. Lewis Publishing.
- 3. Walsh B, Ikeda SS, Boland GJ (1999) Biology and management of dollar spot (Sclerotinia homoeocarpa); an important disease of turfgrass. HortScience 34: 13–21. [Google Scholar]
- 4. Goodman DM, Burpee LL (1991) Biological control of dollar spot. Phytopathology 81: 1438–1446. [Google Scholar]
- 5. Watkins JE, Shearman RC, Gaussoin RE, Cecil WK, Vaitkus M, et al. (2001) An integrated approach to dollar spot management on a bentgrass fairway. Int Turfgrass Soc Res J 9: 729–735. [Google Scholar]
- 6. Bennett FT (1937) Dollar spot disease of turfgrass and its causal organism Sclerotinia homoeocarpa n sp. Ann Appl Biol 24: 236–257. [Google Scholar]
- 7. Baldwin NA, Newell AJ (1992) Field production of fertile apothecia by Sclerotinia homoeocarpa in Festuca turf. J Sport Turf Res Inst 68: 73–76. [Google Scholar]
- 8. Jackson N (1973) Apothecial production in Sclerotinia homoeocarpa FT Bennett. Int Sport Turf Res Inst 49: 58–63. [Google Scholar]
- 9.Fenstermacher JM (1980) Certain features of dollar spot disease and its causal organism Sclerotinia homoeocarpa. In: Joyner BJ, Larsen PO, editors. Advances in Turfgrass Pathology. Harcourt Brace Jovanovich. pp 49–53.
- 10. Orshinsky AM, Boland GJ (2011) Ophiostoma mitovirus 3a, ascorbic acid, glutathione and photoperiod affect the development of stromata and apothecia by Sclerotinia homoeocarpa . Can J Microbiol 407: 398–407. [DOI] [PubMed] [Google Scholar]
- 11.Smiley RW, Dernoeden PH, Clark BB (2005) Compendium of Turfgrass Diseases. 3rd edition. APS Press.
- 12. Novak LA, Kohn LM (1991) Electrophoretic and immunological comparisons of developmentally regulated proteins in members of the Sclerotiniaceae and other sclerotial fungi. Appl Envir Microbiol 57: 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horvath BJ, Kravchenko AN, Robertson GP, Vargas JM (2007) Geostatistical analysis of dollar spot epidemics occurring on a mixed sward of creeping bentgrass and annual bluegrass. Crop Sci 47: 1206–1216. [Google Scholar]
- 14. Brede AD (1991) Interaction of management factors on dollar spot disease severity in tall fescue turf. HortScience 26: 1391–1392. [Google Scholar]
- 15. Golembiewski RC, Danneberger TK (1998) Dollar spot severity as influenced by trinexapac-ethyl creeping bentgrass cultivar and nitrogen fertility. Agron J 90: 466–470. [Google Scholar]
- 16.Wilson CM (2011) Changing the paradigm of dollar spot management and biology. MSc Thesis. University of Wisconsin-Madison.
- 17.Bonos SA, Huff DR (2013) Cool season grasses: Biology and breeding. In: Stier JC, Horgan BP, Bonos SA, editors. Turfgrass: Biology Use and Management American Society of Agronomy, Crop Science Society of America, Soil Science Society of America. pp. 591–660.
- 18.Breuninger JM, Welterlen MS, Augustin BJ, Cline V, Morris K (2013) The turfgrass industry. In: Stier JC, Horgan BP, Bonos SA, editors. Turfgrass: Biology Use and Management American Society of Agronomy, Crop Science Society of America, Soil Science Society of America. pp. 37–103.
- 19. Hsiang T, Mahuku JS (1999) Genetic variation within and between southern Ontario populations of Sclerotinia homoeocarpa . Plant Pathol 48: 83–94. [Google Scholar]
- 20. Powell JF, Vargas JM (2001) Vegetative compatibility and seasonal variation among isolates of Sclerotinia homoeocarpa . Plant Dis 85: 377–381. [DOI] [PubMed] [Google Scholar]
- 21. Putman A, Tredway L, Carbone I (2013) Signatures of global dispersal and population structure in Sclerotinia homoeocarpa . Phytopathology 103: 116. [Google Scholar]
- 22. Liberti D, Rollins JA, Harmon PF (2012) Evidence for morphological vegetative genetic and mating-type diversity in Sclerotinia homoeocarpa . Phytopathology 102: 506–518. [DOI] [PubMed] [Google Scholar]
- 23. Raina K, Jackson N, Chandlee JM (1997) Detection of genetic variation in Sclerotinia homoeocarpa isolates using RAPD analysis. Mycol Res 101: 585–590. [Google Scholar]
- 24. Brede AD, Dunfield TJ (1988) Seeding rate: its effect on disease and weed encroachment. Proc 42nd Northwest Turfgrass Conf 42: 90–97. [Google Scholar]
- 25. Rioux R, Manmathan H, Singh P, de los Reyes B, Jia Y, et al. (2011) Comparative analysis of putative pathogenesis-related gene expression in two Rhizoctonia solani pathosystems. Curr Genet 57: 391–408. [DOI] [PubMed] [Google Scholar]
- 26. White TJ, Bruns T, Lee SJWT, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR-Protocols: A Guide to methods and applications 18: 315–322. [Google Scholar]
- 27. Rioux R, Van Ryzin B, Kerns J (2014) Development of a semi-selective medium for improved isolation of the turfgrass dollar spot pathogen Sclerotinia homoeocarpa from host tissues. Can J Plant Pathol In press. [Google Scholar]
- 28. Jia Y, Liu G, Park D, Yang Y (2013) Inoculation and scoring methods for rice sheath blight diease. Methods Mol Biol 956: 257–68. [DOI] [PubMed] [Google Scholar]
- 29. Abd-Elmagid A, Garrido PA, Hunger R, Lyles JL, Mansfield MA, et al. (2013) Discriminatory simplex and multiplex PCR for four species of the genus Sclerotinia . J Microbiol Methods 92: 293–300. [DOI] [PubMed] [Google Scholar]
- 30. Frost KE, Willis DK, Groves RL (2011) Detection and variability of aster yellows phytoplasma in its insect vector, Macrosteles quadrilineatus (Hemiptera: Cicadellidea). J Econ Entomol 104: 1800–1815. [DOI] [PubMed] [Google Scholar]
- 31.SAS Institute Inc (2011) Base SAS 9.3 Procedures Guide. SAS Institute Inc.
- 32. Pfender WF, Vollmer SS (1999) Freezing temperature effect on survival of Puccinia graminis subsp. graminicola in Festuca arundinacea and Lolium perenne . Plant Dis 83: 1058–1062. [DOI] [PubMed] [Google Scholar]
- 33. Gross PL, Venette JR (2001) Overwinter survival of bean rust urediniospores in North Dakota. Plant Dis 85: 226–227. [DOI] [PubMed] [Google Scholar]
- 34. Holb IJ, Heijne B, Jeger MJ (2007) Overwintering of conidia of Venturia inaequalis and the contribution to early epidemics of apple scab. Plant Dis 88: 751–757. [DOI] [PubMed] [Google Scholar]
- 35. Xu Z, Gleason ML, Mueller DS, Esker PD, Bradley CA, et al. (2008) Overwintering of Sclerotium rolfsii and S rolfsii var delphinii in different latitudes of the United States. Plant Dis 92: 719–724. [DOI] [PubMed] [Google Scholar]
- 36. Foster WR, Henry AW (1937) Overwintering of certain cereal pathogens in Alberta. Can J Res 15c: 547–559. [Google Scholar]
- 37. Gossen BD, Miller PR (2004) Survival of Ascochyta rabiei in chickpea residue on the Canadian prairies. Can J Plant Pathol 26: 142–147. [Google Scholar]
- 38. Yoshida S, Shirata A (1999) Survival of Colletotrichum dematium in soil and infected mulberry leaves. Plant Dis 83: 465–468. [DOI] [PubMed] [Google Scholar]
- 39.Couch HB (1995) Disease of turfgrasses. 3rd ed Malabar FL: Krieger Publishing.
- 40.Smith JD, Jackson N, Woolhouse AP (1989) Fungal diseases of amenity turfgrasses. 3rd edition. E & FN Spon Ltd.
- 41. Sowley ENK, Dewey FM, Shaw MW (2009) Persistent symptomless systemic and seed-borne infection of lettuce by Botrytis cinerea . Eur J Plant Pathol 126: 61–71. [Google Scholar]
- 42.Emmons R (2005) Turfgrass science and management 4th ed Independence KY: Delmar Cengage Learning.
- 43. Duressa D, Rauscher G, Koike ST, Mou B, Hayes RJ, et al. (2012) A real-time PCR assay for detection and quantification of Verticillium dahliae in spinach seed. Phytopathology 104: 443–451. [DOI] [PubMed] [Google Scholar]
- 44. Mbofung GCY, Pryor BM (2010) A PCR-based assay for detection of Fusarium oxysporum f sp lactucae in lettuce seed. Plant Dis 94: 860–866. [DOI] [PubMed] [Google Scholar]
- 45. Pryor BM, Gilbertson RL (2001) A PCR-based assay for detection of Alternaria radicina on carrot seed. Plant Dis 85: 18–23. [DOI] [PubMed] [Google Scholar]
- 46. Ioos R, Fourrier C, Wilson V, Webb K, Schereffer J-L, et al. (2012) An optimized duplex real-time PCR tool for sensitive detection of the quarantine oomycete Plasmopara halstedii in sunflower seeds. Phytopathology 102: 908–917. [DOI] [PubMed] [Google Scholar]
- 47. Montes-borrego M, Muñoz-Ledesma FJ, Jiménez-Díaz RM, Landa BB (2011) Real-time PCR quantification of Peronospora arborescens the opium poppy downy mildew pathogen in seed stocks and symptomless infected plants. Plant Dis 95: 143–152. [DOI] [PubMed] [Google Scholar]
- 48. Flowers J, Hartman J, Vaillancourt L (2003) Detection of latent Sphaeropsis sapinea infections in Austrian pine tissues using nested-polymerase chain reaction. Phytopathology 93: 1471–1477. [DOI] [PubMed] [Google Scholar]
- 49. Glen M, Smith AH, Langrell SRH, Mohammed CL (2007) Development of nested polymerase chain reaction detection of Mycosphaerella spp and its application to the study of leaf disease in Eucalyptus plantations. Phytopathology 97: 132–44. [DOI] [PubMed] [Google Scholar]
- 50. Testen AL, Jimenez-Gasco MDM, Ochoa JB, Backman PA (2014) Molecular detection of Peronospora variabilis in quinoa seeds and phylogeny of the quinoa downy mildew pathogen in South America and the United States. Phytopathology 4: 379–386. [DOI] [PubMed] [Google Scholar]
- 51. Hamburger J, Abbasi I, Kariuki C, Wanjala A, Mzungu E, et al. (2013) Evaluation of loop-mediated isothermal amplification suitable for molecular monitoring of schistosome-infected snails in field laboratories. Am J Trop Med Hyg 88: 344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kaneko H, Kawana T, Fukushima E, Suzutani T (2007) Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods 70: 499–501. [DOI] [PubMed] [Google Scholar]
- 53. Mori Y, Notomi T (2009) Loop-mediated isothermal amplification (LAMP): a rapid accurate and cost-effective diagnostic method for infectious diseases. J Infect Chemother 15: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Notomi T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28: 63e–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee HK, Tewari JP, Turkington TK (2001) A PCR-based assay to detect Rhynchosporium secalis in barley seed. Plant Dis 85: 220–225. [DOI] [PubMed] [Google Scholar]
- 56. Lee HK, Tewari JP, Turkington TK (2001) Symptomless infection of barley seed by Rhynchosporium secalis . Can J Plant Pathol 23: 315–317. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sclerotinia homoeocarpa isolate ‘Shark’ obtained from Shark Lot 1 using culture-based detection of semi-selective medium. A, Colony morphology of the isolate ‘Shark’ obtained from CRB cv. ‘Shark’ commercial seed lot 1. B, Symptoms produced by mock-inoculated control, virulent S. homoeocarpa, and seed S. homoeocarpa isolate ‘Shark’ on creeping bentgrass (cv. ‘Penncross’) at 5 dpi. Similar symptoms were produced for each treatment in six biological replicates. C, Symptoms produced by mock-inoculated control, virulent S. homoeocarpa, and seed S. homoeocarpa isolate ‘Shark’ on barley (cv. ‘Parkland’) at 5 dpi. Similar symptoms were produced for each treatment in six biological replicates.
(TIF)
Sensitivity of Sclerotinia homoeocarpa -specific primers in primary and nested PCR. Primary PCR was run with S. homoeocarpa specific primers EF1α_OuterF/EF1α_OuterR, with an expected product size of 554 bp, and contained 2 µL of seed DNA from seed samples spiked with the indicated number of artificially-infested CRB seeds prior to DNA extraction. Nested PCR was performed using S. homoeocarpa specific primers EF1α_F/EF1α_R, with a product size of 219 bp internal to EF1α_OuterF/EF1α_OuterR, and contained 2 µL of primary PCR products diluted 1∶30 in nuclease free water. The negative control was treated exactly as samples except that 2 µL of NFW replaced template DNA in the primary PCR. Five µL of sample were mixed with 1 µL of DNA loading dye and run in a 1% agarose gel in TBE buffer along with 5 µL of TrackIt 100 bp DNA ladder (Invitrogen, Carlsbad, CA).
(TIF)
Q-PCR for molecular detection of Sclerotinia homoeocarpa DNA in creeping bentgrass commercial seed lots. A, Standard curve of EF1α plasmid DNA with the primer set EF1α_NestF/EF1α_NestR indicating near 100% primer efficiency. B, Q-PCR run with various controls to reveal the presence of PCR inhibitors: Spike, Spike−1, Spike−2—Ten-fold dilution series of seed DNA mixed with 1/10 volume EF1α plasmid DNA at a starting concentration 1×106.; Check, Check−1, Check−2—Ten-fold dilution series of a seed DNA sample positive by nested PCR but negative by Q-PCR. Decreasing Cq value with dilution indicates presence of inhibitors in the original DNA sample.
(TIF)
DNA sequences from overwintering Sclerotinia homoeocarpa isolates, isolate ‘Shark’ obtained from commercial seed, and S. homoeocarpa EF1α sequences from seed lots positive by nested PCR.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.