Abstract
The interactions occurring between a virus and a host cell during a viral infection are complex. The purpose of this paper was to analyze altered cellular protein levels in porcine transmissible gastroenteritis coronavirus (TGEV)-infected swine testicular (ST) cells in order to determine potential virus-host interactions. A proteomic approach using isobaric tags for relative and absolute quantitation (iTRAQ)-coupled two-dimensional liquid chromatography-tandem mass spectrometry identification was conducted on the TGEV-infected ST cells. The results showed that the 4-plex iTRAQ-based quantitative approach identified 4,112 proteins, 146 of which showed significant changes in expression 48 h after infection. At 64 h post infection, 219 of these proteins showed significant change, further indicating that a larger number of proteomic changes appear to occur during the later stages of infection. Gene ontology analysis of the altered proteins showed enrichment in multiple biological processes, including cell adhesion, response to stress, generation of precursor metabolites and energy, cell motility, protein complex assembly, growth, developmental maturation, immune system process, extracellular matrix organization, locomotion, cell-cell signaling, neurological system process, and cell junction organization. Changes in the expression levels of transforming growth factor beta 1 (TGF-β1), caspase-8, and heat shock protein 90 alpha (HSP90α) were also verified by western blot analysis. To our knowledge, this study is the first time the response profile of ST host cells following TGEV infection has been analyzed using iTRAQ technology, and our description of the late proteomic changes that are occurring after the time of vigorous viral production are novel. Therefore, this study provides a solid foundation for further investigation, and will likely help us to better understand the mechanisms of TGEV infection and pathogenesis.
Introduction
Porcine transmissible gastroenteritis coronavirus (TGEV) is an animal coronavirus that causes severe gastroenteritis in young TGEV-seronegative pigs. Various breeds of pigs, regardless of age, are susceptible to TGEV; however, the mortality rate for piglets under 2 weeks of age is the highest, reaching almost 100%. Diseased pigs often present with vomiting, dehydration, and severe diarrhea. Further, the disease is known to affect pigs in many countries throughout the world and an outbreak can cause enormous losses in the pig industry [1], [2]. The pathogen, TGEV, which belongs to the Alphacoronavirus genus of the Coronavirinae subfamily within the family Coronaviridae, is an enveloped, non-segmented, single-stranded positive-sense RNA virus [3], [4]. The envelop, core, and nucleocapsid of the TGEV virion contain four major structural proteins: the nucleocapsid (N) protein, the membrane (M) glycoprotein, the small envelope (E) protein, and the spike (S) protein [5]. The tropism and pathogenicity of the virus are influenced by the S protein, which has four major antigenic sites, A, B, C, and D, with site A being the major inducer of antibody neutralization [3], [5]. The M protein, which plays a central role in virus assembly by interacting with viral ribonucleoprotein (RNP) and S glycoproteins [6], is embedded within the virus membrane and interacts with the nucleocapsid, forming the core of TGEV virion. In addition, the N-terminal domain of the M protein is essential for interferon alpha (IFN-α) induction [7], which is involved in the host’s innate immune response. The E protein, a transmembrane protein that acts as a minor structural component in TGEV and affects virus morphogenesis, is essential for virion assembly and release [8].
TGEV RNA, along with the N protein, is infectious and invades the organism through the digestive and respiratory tracts, resulting in infection of the small intestinal enterocytes, villous atrophy, and severe watery diarrhea. These changes in intestinal health are known to be important during the pathogenesis of TGEV infection [9]. Furthermore, corresponding to these pathologic changes observed in vivo, TGEV can also propagate and cause cytopathic effects (CPEs) in multiple types of cultured cells, such as swine testicular (ST) cells, PK-15 cells, and villous enterocytes. Notably, ST cells are more susceptible to TGEV, and higher levels of virus replication have been observed in this cell line [10], [11].
The full RNA genome of TGEV is approximately 28.5 kb in length and has a 5′-cap structure and a poly(A) tail at the 3′ end. The 9 open reading frame (ORF) genes included in the TGEV genome are arranged in the following order 5′-la- lb-S-3a-3b-E-M-N-7-3′. The first gene at the 5′ end consists of two large ORFs, ORF la and ORF lb, which constitute the replicase gene, known for its RNA-dependent RNA-polymerase and helicase activities, as well as other enzymes, such as endoribonuclease, 3′–5′exoribonuclease, 2′-O-ribose methyltransferase, ribose ADP 1” phosphatase, etc. [12]. ORF2, ORF4, ORF5, and ORF6 encode the S, E, M, and N proteins, respectively, while ORF3a, ORF3b, and ORF7 encode non-structural proteins [13]. Some investigators have suggested that ORF3 may be related to viral virulence and pathogenesis [12], while ORF7 may interact with host cell proteins and play a role in TGEV replication [14]. In fact, a recent study indicates that plasmid-transcribed small hairpin (sh) RNAs targeting the ORF7 gene of TGEV is capable of inhibiting virus replication and expression of the viral target gene in ST cells in vitro [15]. Although we have some knowledge concerning the translation and function of these viral proteins, the interactions that occur between these proteins and host cell proteins are not fully understood.
Importantly, recent advances in proteomic technology have allowed for more in depth investigation of virus-host interactions, and different techniques have been successfully applied to identify altered proteins in infected host cells and tissues. For example, Sun et al. [16] have identified 35 differentially expressed proteins in PK-15 cells infected with classical swine fever virus (CSFV) using two-dimensional polyacrylamide gel electrophoresis (2D PAGE) followed by matrix-assisted laser desorption-ionization time-of-flight tandem mass spectrometry (MALDI-TOF-MS/MS). In addition, two-dimensional fluorescence difference gel electrophoresis (2D-DIGE) and MS/MS proteomic approaches have been applied to characterize protein changes occurring in host cells in response to porcine circovirus type 2 (PCV2) infection [17]. The same methods have also been studied for many other pathogenic animal viruses, including porcine reproductive and respiratory syndrome virus (PRRSV) [18], coronavirus infectious bronchitis virus (IBV) [19], severe acute respiratory syndrome-associated coronavirus (SARS-CoV) [20], and TGEV [21]. However, these conventional approaches based on 2D gel electrophoresis are not suitable for detecting low abundance, hydrophobic, or very acidic/basic proteins. On the other hand, the isobaric tags for relative and absolute quantitation (iTRAQ) technique, in association with liquid chromatograph (LC), is a more advanced method for proteomic research, and is capable of detecting a much larger number of proteins, even those with low abundance, in addition to identifying and quantifying the proteins simultaneously [22]. To this end, Lu et al. [23] previously used the iTRAQ method to identify 160 significantly altered proteins in pulmonary alveolar macrophages (PAMs) infected with PRRSV. Similarly, this method has been used to investigate influenza virus infection in primary human macrophages [24], human immunodeficiency virus 1 (HIV-1) infection in CD4+ T cells [25], and Epstein–Barr virus (EBV) infection in nasopharyngeal carcinoma cell line [26].
Here, we report the first differential proteomic analysis of TGEV-infected and uninfected ST cells using iTRAQ labeling followed by 2D-LC-MS and bioinformatic analyses. The proteomic data obtained in this study will help to enhance our understanding of the host response to TGEV infection, but also provide new insights on the mechanisms of disease onset.
Materials and Methods
Cell culture and viral replication
ST cells were obtained from the American Type Culture Collection (ATCC). The cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; GIBCO, UK) containing 1% L-glutamine and 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) at 37°C in 5% CO2. Culture medium was replaced two to three times per week. The TGEV TH-98 strain was isolated from a suburb of Harbin, Heilongjiang province, China. The virus was propagated in ST cells and preserved at −70°C in our laboratory.
TGEV infection
The monolayer of confluent ST cells was dispersed with 0.25% trypsin and 0.02% ethylenediaminetetraacetic acid (EDTA) and seeded in 6-cm cell culture flasks. After a 24 h incubation period, the culture medium was removed and the ST cells were washed with phosphate buffered saline (PBS, pH 7.4). The cells were then infected with the TGEV TH-98 strain at a 50% tissue culture infectious dose (TCID50) of 1×103.53 viruses per well, with absorption for 2 h at 37°C. Maintenance medium (DMEM medium supplemented with 2% FBS) was then added to the cells. A mock group of ST cells that were not infected with TGEV was used as a negative control for each of the following experiments. Three replicates of virus-infected and mock-infected cultures with different passage numbers were prepared at each time point. The morphological changes were observed under the light microscope at 24, 40, 48, and 64 hours post infection (hpi).
Reverse transcription polymerase chain reaction (RT-PCR) and real time quantitative PCR (qRT-PCR)
To determine the extent of TGEV infection, conventional RT-PCR and qRT-PCR assays were performed to detect the viral N gene. Monolayers of ST cells were infected with TGEV as described above. Cells were collected from 24 to 80 hpi at 8 h intervals, and the total RNA of the infected cells was extracted using Trizol (Invitrogen). RNA samples were reverse-transcribed using PrimeScript RT reagent Kit (Takara Bio, Dalian, China), according to the manufacturer’s instructions. The RT reaction was incubated at 37°C for 15 min followed by 85°C for 5 s. A mixture of oligo dT primers and random 6 mers was used in the RT step. The cDNA was stored at −20°C until further use.
PCR was performed for the TGEV N gene in a 25 µl reaction mixture containing 1 µl of the cDNA, 0.5 µl of each forward (F) and reverse (R) primer, 12.5 µl of Premix Taq (Takara Bio, Dalian, China), and 10.5 µl DEPC water, starting with a 5 min denaturation at 95 C followed by 32 cycles of 30 s denaturation at 95 C, 30 s annealing at 56 C, and 40 s extension at 72 C. A final extension step was carried out at 72 C for 10 min. RT-PCR products were resolved on a 15 g/L agarose gel. The following PCR primers were used in this study: TGEV N (F, 5′-GAGCAGTGCCAAGCATTACCC-3′ and R, 5′-GACTTCTAT CTGGTCGCCATCTTC-3′) and β-actin (F, 5′-GCAAGGACCTCTACGCCAA-3′ and R, 5′-CTGGAAGGTGGACAGCGAG-3′).
The mRNA expression level of the TGEV N gene was quantified using a SYBR Green assay on a Bio-Rad iQ5 real time PCR detection system as described previously [27]. We used the same primers listed above for qRT-PCR. Reactions were carried out in 50 µl volumes containing 0.5 µl of 20 × SYBR Green I, 2 µl of cDNA template, 1 µl of each F and R primer, 25 µl of 2 × PCR buffer, and 20.5 µl DEPC water. The cycling conditions were 94°C for 4 min, followed by 35 cycles of 94°C for 20 s, 60°C for 30 s, 72°C for 30 s, and then a final extension of 10 min at 72°C. The relative gene expression was determined with the 2(−ΔΔCt) method [28], and the tests were performed in triplicate.
Protein isolation, digestion, and labeling with iTRAQ reagents
Following ST cell infection, cells were collected at 48 and 64 hpi by centrifugation at 3,000 rpm for 5 min at 4°C, washed twice with PBS, and 1 mL of iTRAQ lysis solution (8 M urea, 1% (w/v) dithiothreitol (DTT)) containing protease inhibitor was added. Then, the cells were put in an ice bath and broken up by sonication. The solution was then mixed for 30 min at 4°C. The soluble protein fraction was harvested by centrifugation at 40,000 × g for 30 min at 4°C and the debris was discarded. The protein concentration was determined with the Bradford protein assay (2-D Quant Kit, Bestbio, China). A 100 µg aliquot of protein from each sample was reduced, alkylated, and trypsin-digested as described in the iTRAQ protocol (AB Sciex, American), followed by labeling with the 4-plex iTRAQ Reagents Multiplex Kit according to the manufacturer’s instructions (AB Sciex, American). Two virus-free samples at 48 h and 64 h were labeled with iTRAQ tags 114 and 115, while two TGEV-infected samples at 48 h and 64 h were labeled with tags 116 and 117. The labeled digests were then pooled, dried using a vacuum freeze drier (Christ RVC 2−25, Germany), and preserved at −20°C for later use.
2D LC-MS/MS analysis
The combined peptide mixtures were separated by reversed phase high-performance liquid chromatography (HPLC) (Ekspert ultraLC 100, AB Sciex, USA) on a Durashell-C18 reverse phase column (4.6 mm × 250 mm, 5 µm 100 Å, Agela). The mobile phases used were composed of 20 mM ammonium formate (pH 10) in water (labeled mobile phase A) and 20 mM ammonium formate (pH 10) in acetonitrile(ACN) (mobile phase B). The flow rate was 0.8 mL/min, and the elutant was collected into 48 centrifuge tubes at each minute after the first 5 min. Each aliquot was then dried by vacuum freezing.
The peptides were then analyzed with a nanoflow reversed-phase liquid chromatography-tandem mass spectrometry (nano-RPLC-MS/MS) system (TripleTOF 5600, AB Sciex, USA). The above 48 tubes were merged into 10 components dissolved in 2% ACN and 0.1% formic acid (FA), then centrifuged at 12,000 × g for 10 min. The supernatant (8 µl) was used for loading at a rate of 2 µl/min, with a separation rate of 0.3 µl/min. The mobile phase A used in this analysis was composed of 2% ACN and 0.2% FA, while mobile phase B was composed of 98% ACN and 0.1% FA. The following MS parameters were utilized: source gas parameters (ion spray voltage: 2.3 kV, GS1∶4, curtain gas: 30 or 35, DP: 100 or 80); TOF MS (m/z: 350–1250, accumulation time: 0.25 s); and product ion scan (IDA number: 30, m/z: 100–1500, accumulation time: 0.1 s, dynamic exclusion time: 25 s, rolling CE: enabled, adjust CE when using iTRAQ reagent: enabled, CES: 5).
Data analysis and bioinformatics
Protein identification and quantification were performed with the ProteinPilot software (version 4.0, AB Sciex) using the Paragon algorithm. Each MS/MS spectrum was searched against a database of Sus scrofa protein sequences (NCBI nr, released in March 2011, downloaded from ftp://ftp.ncbi.nih.gov/genomes/Sus_scrofa/protein/). The following search parameters were used: iTRAQ 4-plex (peptide labeled), cysteine alkylation with methyl methanethiosulfonate(MMTS), trypsin digestion, biological modifications allowed, a thorough search, a detected protein threshold of 95% confidence (unused Protscore ≥1.3), and a critical false discovery rate (FDR) of 1%. The peptide and protein selection criteria for relative quantitation were performed as described previously, whereby only peptides unique for a given protein were considered [29]. In addition, proteins with an iTRAQ ratio higher than 20 or lower than 0.05 as well as proteins in reverse database were removed [30].
To assign enriched Gene Ontology (GO) terms to the identified proteins, the differentially expressed proteins identified from iTRAQ experiments and all of the 4,112 measured proteins were classified based on their GO annotations using QuickGO (http://www.ebi.ac.uk/QuickGO/), with UniProt ID (http://www.uniprot.org/?tab=mapping) as the data source. GO enrichment analysis of the differentially regulated proteins was evaluated using all of the 4,112 quantified proteins as background with hypergeometric distribution [31]. Categories belonging to biological processes, molecular functions, and cellular components that were identified at a confidence level of 95% were included in the analysis. The protein-protein interaction network for a select group of proteins was analyzed using the STRING 9.1 database (http://string-db.org/). Network analysis was set at medium confidence (STRING score >0.4).
Western blot analysis
Following ST cell infection with TGEV, the culture medium was removed after incubating for 48 h and 64 h; then, the cells were washed with cold PBS and collected after centrifugation at 3,000 rpm for 10 min. Cells were then lysed in RIPA lysis buffer with protease inhibitors (Applygen Technologies Inc., China). Cellular debris was removed by centrifugation at 12,000 × g for 5 min at 4°C, and the protein concentration was measured by Coomassie blue G250 staining. An equal amount (20 µg) of cell lysate from each sample was separated using 10% SDS-PAGE and then transferred to polyvinyl difluoride (PVDF) membranes (Millipore, Bedford, USA). The PVDF membranes were then blocked with 5% (w/v) de-fatted milk powder dissolved in tris buffered saline and tween 20 (TBST) buffer (150 mM NaCl, 50 mM Tris, 0.05% Tween 20) for 1 h at 37°C. After blocking, membranes were incubated with anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mouse monoclonal antibody (1∶3000; Western Biotechnology, China), anti-heat shock protein 90 alpha (Hsp90α/HSP90AA1) antibody (1∶300; Abcam, Cambridge, UK), anti-caspase 8 antibody (1∶300; Abcam, Cambridge, UK), or anti-transforming growth factor β 1 (TGF-β1/TGFB1) antibody (1∶300; Abcam, Cambridge, UK) overnight at 4°C, followed by HRP-conjugated secondary antibody (1∶5000; Western Biotechnology, China) for 1.5 h at 37°C. The membranes were then washed four times in TBST buffer for 5 min each time. Protein band detection was performed using ECL reagents (Applygen Technologies Inc., China), and the band intensities were analyzed using Labworks 4.6 software.
Results
Confirmation of TGEV infection in ST Cells
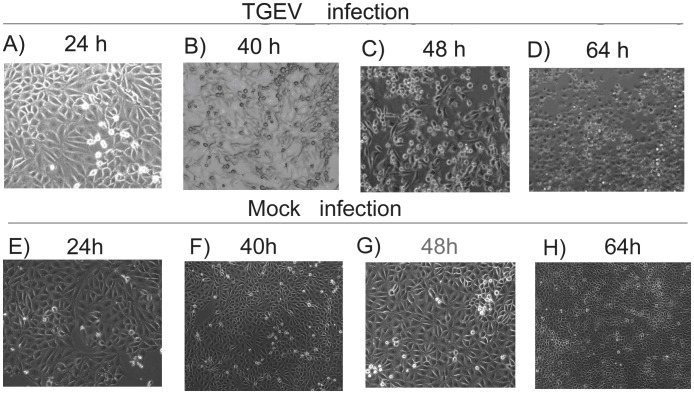
After introducing TGEV into the ST cells, we observed the induction of typical CPEs, including cell rounding, swelling, granular degeneration of the cytoplasm, cell detachment, and severely diseased cell morphology, from 40 to 64 h after inoculation (Figure 1 A–D) compared to the non-infected control cells (Figure 1 E–H). Virus infection at 48 and 64 h was also confirmed by RT-PCR detection of the viral N gene in the sample (Figure 2A).
Figure 1. Morphological changes in TGEV-infected cells.
ST cells were seeded into 6-cm culture plates, infected with TGEV, and the cytopathic effects (CPEs) were imaged at 24 (A), 40 (B), 48 (C), and 64 (D) hours following infection. Images of non-infected cells (mock infection) are shown for comparison at each time point (E, F, G, H).
Figure 2. Validation of TGEV virus infection of ST cells.
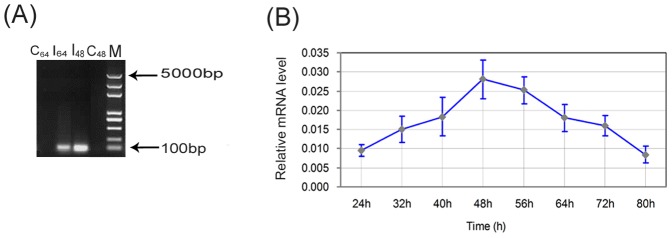
(A) RT-PCR validation of TGEV infection in ST cells at 48 hpi (I48) and 64 hpi (I64) compared to the control at 48 h (C48) and 64 h (C64). A marker (M) was used to identify fragment size. (B) qRT-PCR analysis of changes in TGEV mRNA expression levels in the ST cells over time. The changes in mRNA expression level at the various time points is indicated, and show that the expression level of TGEV increased gradually, reaching a peak at 48 h, then decreased dramatically. Values are the means of three repeated experiments. The error bars in the graphs represent the standard deviation.
Dynamic changes in viral gene expression in infected cells
To further identify the extent of TGEV infection, the mRNA expression levels of viral genes in infected cells were determined using qRT-PCR. Comparative threshold (Ct) cycle values in three independent experiments were calculated and the results indicated that the average Ct value for the TGEV N gene ranged from 25.2 to 27.5. Correspondingly, the average Ct value observed for the β-actin control gene ranged from 19.6 to 21.0. The relative expression of TGEV N mRNA was calculated using the 2(–ΔΔCT) method [28], and the change in expression at each time point is indicated in Figure 2B. These data show that, following infection, the viral mRNA levels increased gradually over time, and reached a peak at 48 hpi. Following this time point, the viral mRNA levels appear to decrease.
Protein identification by MS
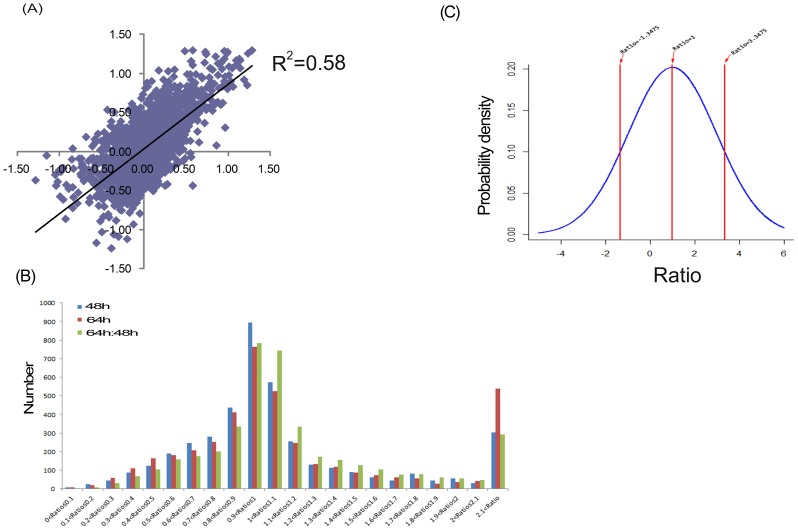
In the infected ST cells, a total of 29,214 peptides and 4,364 proteins were detected (Table S1); however, only 4,112 proteins were quantified reliably (Table S2). Notably, the abnormal proteins, such as the proteins with iTRAQ ratio higher than 20 or lower than 0.05, which are not quantifiable [30], were removed and only proteins with reasonable ratios across all channels were investigated further. Figure 3A depicts the scatter plots for the log10 116/114 and log10 117/115 ratios in the iTRAQ experiment. Linear regression analysis showed that correlation (R 2) was 0.58, with a p-value less than 0.05. These results suggest that the alterations in protein abundance due to virus infection were near-linear dependency between the two time points. In order to identify the proteins that were significantly different at each time point (infected/uninfected) or between the different time points, we analyzed the distribution of ratios for the identified proteins as shown in the Figure 3B. For the distribution range of the differentially expressed proteins identified at 48 hpi, shown in Figure 3C, a ratio higher than 3.35 or lower than −1.35 was defined as a statistically significant difference in protein expression. At 64 hpi, a ratio higher than 4.55 or lower than −2.15 was defined as a statistically significant difference in protein expression. According to analyses, the differentially expressed proteins identified were considered to show a significant upward or downward trend if their expression ratios were greater than 4.0 or less than 0.25 compared to the control group.
Figure 3. Results of the iTRAQ ratios analysis.
(A) A scatter plot showing the correlation between the log10 infection/mock ratios at 48 hpi and 64 hpi for the 4,112 reliably quantified proteins in the iTRAQ experiment. Linear regression analysis shows that correlation (R 2) was 0.58, with a p-value less than 0.05. (B) Histograms showing the distribution of protein ratios identified at 48 and 64 hpi. (C) The distribution range of differentially expressed proteins identified at 48 hpi. iTRAQ ratios higher than 3.3475 (p = 0.975) or lower than −1.3475 (p = 0.025) were defined as statistically significant.
Using the criterion listed above, the expression of 146 proteins was significantly changed at 48 hpi (95 upregulated and 51 downregulated), while 219 proteins were significantly changed at 64 hpi (172 upregulated and 47 downregulated). Further, 72 proteins were identified to be significantly different between the two time points (54 upregulated and 18 downregulated), resulting in a total of 316 unique proteins being significantly altered during TGEV infection, including 162 predicted proteins (Table S3 and Table 1 (excluding the predicted proteins)). Because the current pig genome database is poorly annotated compared to the human genome database, there were numerous proteins that were unassigned or uncharacterized, resulting in a large number of predicted proteins in our analysis. However, our ability to detect the unannotated proteins by MS demonstrates that they do existence in this species, and additional research concerning their function is warranted.
Table 1. Differentially expressed proteins identified by iTRAQ analysis of ST cells infected with TGEV.
| Accession number | Protein name | Gene symbol | Unused ProtScore | Infected/uninfected (48 h) | Infected/uninfected (64 h) | ||
| Ratio | P-value | Ratio | P-value | ||||
| Upregulated proteins | |||||||
| gi|359811347 | 60 kDa heat shock protein, mitochondrial | – | 139.52 | 3.16 | 0.00 | 5.86↑ | 0.00 |
| gi|227430407 | Keratin, type II cytoskeletal 8 | KRT8 | 110.35 | 4.02↑ | 0.00 | 6.49↑ | 0.00 |
| gi|347300243 | Glutamate dehydrogenase 1, mitochondrial | GLUD1 | 102.28 | 1.79 | 0.18 | 4.17↑ | 0.00 |
| gi|297591975 | ATP synthase subunit alpha, mitochondrial | ATP5A1 | 96.61 | 1.17 | 0.67 | 4.66↑ | 0.00 |
| gi|417515796 | Hypoxia up-regulated protein 1 precursor | – | 92.84 | 3.91 | 0.01 | 6.92↑ | 0.00 |
| gi|349732227 | Heterogeneous nuclear ribonucleoprotein M | – | 89.74 | 7.66↑ | 0.00 | 9.64↑ | 0.00 |
| gi|56748897 | Heat shock 70 kDa protein 1B | HSPA1B | 62.37 | 4.33↑ | 0.21 | 4.02↑ | 0.12 |
| gi|47522630 | Aspartate aminotransferase, mitochondrial precursor | GOT2 | 60.36 | 1.43 | 0.01 | 4.66↑ | 0.00 |
| gi|387912908 | Calreticulin | CALR | 55.58 | 2.40 | 0.11 | 4.61↑ | 0.00 |
| gi|346421378 | Serpin H1 precursor | – | 52.10 | 3.22 | 0.00 | 4.06↑ | 0.00 |
| gi|2506849 | Malate dehydrogenase, mitochondrial | MDH2 | 49.19 | 3.28 | 0.00 | 6.98↑ | 0.00 |
| gi|148230268 | Galectin-3 | LGALS3 | 48.39 | 3.08 | 0.15 | 5.06↑ | 0.01 |
| gi|417515899 | 2-oxoglutarate dehydrogenase, mitochondrial | – | 45.01 | 2.99 | 0.02 | 5.25↑ | 0.00 |
| gi|8745552 | Voltage-dependent anion channel 1 | VDAC1 | 43.46 | 6.19↑ | 0.01 | 11.59↑ | 0.00 |
| gi|330417958 | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial | PCK2 | 42.89 | 1.96 | 0.09 | 5.75↑ | 0.00 |
| gi|353468887 | Signal transducer and activator of transcription 1 | STAT1 | 42.79 | 1.80 | 0.33 | 6.98↑ | 0.00 |
| gi|21264506 | Succinyl-CoA ligase [GDP-forming] subunit beta, mitochondrial | SUCLG2 | 41.68 | 1.53 | 0.00 | 4.06↑ | 0.00 |
| gi|47716872 | Galectin-1 | – | 41.49 | 5.20↑ | 0.05 | 4.66↑ | 0.06 |
| gi|342349346 | Lon peptidase 1, mitochondrial | – | 41.02 | 2.49 | 0.03 | 6.03↑ | 0.00 |
| gi|210050415 | Mx2 protein | Mx2 | 40.44 | 3.08 | 0.79 | 18.88↑* | 0.00 |
| gi|342349319 | Calnexin precursor | – | 37.71 | 4.79↑ | 0.00 | 6.14↑ | 0.00 |
| gi|72535198 | Histone H1.3-like protein | – | 36.51 | 1.69 | 0.38 | 8.32↑* | 0.12 |
| gi|347300207 | Nucleobindin-1 precursor | NUCB1 | 35.30 | 3.40 | 0.00 | 5.01↑ | 0.00 |
| gi|347800693 | Ferredoxin reductase | FDXR | 33.41 | 1.56 | 0.05 | 4.57↑ | 0.00 |
| gi|417515788 | Prolow-density lipoprotein receptor-related protein 1 precursor | – | 32.44 | 1.60 | 0.13 | 5.65↑ | 0.00 |
| gi|297747350 | FAT tumor suppressor homolog 1 | – | 32.04 | 6.08↑ | 0.00 | 7.24↑ | 0.00 |
| gi|298104076 | Enoyl-CoA hydratase, mitochondrial | – | 30.50 | 2.21 | 0.23 | 6.55↑ | 0.00 |
| gi|7939586 | Dihydrolipoamide succinyltransferase | DLST | 30.42 | 1.71 | 0.17 | 4.06↑ | 0.00 |
| gi|7404364 | Hydroxyacyl-coenzyme A dehydrogenase, mitochondrialPrecursor | HADH | 29.29 | 1.92 | 0.00 | 5.50↑ | 0.00 |
| gi|346644866 | Coiled-coil-helix-coiled-coil-helix domain-containing protein 3, mitochondrial | CHCHD3 | 28.98 | 2.51 | 0.01 | 4.61↑ | 0.00 |
| gi|47522814 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial precursor | – | 28.30 | 1.98 | 0.32 | 4.92↑ | 0.00 |
| gi|6165556 | Long-chain 3-ketoacyl-CoA thiolase | LCTHIO | 26.88 | 3.98 | 0.00 | 7.59↑ | 0.00 |
| gi|156720190 | Mx1 protein | Mx1 | 26.26 | 3.80 | 0.98 | 19.41↑* | 0.00 |
| gi|47522770 | Clusterin precursor | CLU | 25.75 | 13.80↑ | 0.00 | 14.59↑ | 0.00 |
| gi|347300323 | Thioredoxin-dependent peroxide reductase, mitochondrial | PRDX3 | 24.24 | 2.72 | 0.00 | 6.92↑ | 0.00 |
| gi|47522610 | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial precursor | OXCT1 | 23.94 | 1.72 | 0.22 | 6.37↑ | 0.00 |
| gi|346986361 | Electron-transfer-flavoprotein, alpha polypeptide | ETFA | 22.41 | 2.42 | 0.34 | 5.55↑ | 0.00 |
| gi|172072653 | Lactadherin precursor | MFGE8 | 22.33 | 10.19↑ | 0.00 | 9.64↑ | 0.00 |
| gi|56417363 | Cathepsin D protein | – | 21.95 | 0.54 | 0.0 | 2.38* | 0.02 |
| gi|87047636 | ATP synthase H+-transporting mitochondrial F1 complex O subunit | ATP5O | 21.83 | 1.11 | 0.66 | 7.66↑* | 0.00 |
| gi|89573851 | Succinate dehydrogenase complex subunit B | SDHB | 21.18 | 2.00 | 0.04 | 5.97↑ | 0.00 |
| gi|5921142 | Amyloid precursor protein | APP | 20.29 | 13.43↑ | 0.00 | 15.14↑ | 0.00 |
| gi|347658971 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d | – | 20.26 | 3.34 | 0.01 | 9.82↑ | 0.00 |
| gi|75052621 | Transcription factor A, mitochondrial | TFAM | 19.14 | 2.01 | 0.00 | 5.50↑ | 0.00 |
| gi|312283580 | Superoxide dismutase [Mn], mitochondrial | – | 18.40 | 1.79 | 0.18 | 5.35↑ | 0.00 |
| gi|6093657 | Propionyl-CoA carboxylase beta chain, mitochondria | PCCB | 17.92 | 2.33 | 0.13 | 6.85↑ | 0.00 |
| gi|346716275 | DnaJ homolog subfamily B member 11 precursor | DNAJB11 | 17.48 | 2.63 | 0.01 | 4.74↑ | 0.00 |
| gi|118403762 | Extracellular superoxide dismutase precursor | - | 16.78 | 11.27↑ | 0.01 | 11.48↑ | 0.01 |
| gi|150251019 | Adenylate kinase 3-like 1 | AK3L1 | 15.85 | 2.05 | 0.12 | 4.06↑ | 0.00 |
| gi|158517860 | Thymosin beta-10 | TMSB10 | 13.45 | 6.25↑ | 0.30 | 7.52↑ | 0.30 |
| gi|47522698 | Cathepsin L1 precursor | CTSL | 12.72 | 4.45↑ | 0.01 | 5.11↑ | 0.01 |
| gi|329744622 | Low-density lipoprotein receptor precursor | LDLR | 12.69 | 3.50 | 0.04 | 4.06↑ | 0.01 |
| gi|346644882 | Reticulocalbin 2, EF-hand calcium binding domain precursor | RCN2 | 12.05 | 2.00 | 0.04 | 4.09↑ | 0.00 |
| gi|284519712 | Caspase-8 | – | 11.38 | 7.11↑ | 0.13 | 16.14↑ | 0.00 |
| gi|211578396 | Nitrogen fixation 1-like protein | LOC100156145 | 11.29 | 3.60 | 0.02 | 5.45↑ | 0.00 |
| gi|346644830 | Sulfide:quinone oxidoreductase, mitochondrial | SQRDL | 10.92 | 1.87 | 0.38 | 4.53↑ | 0.02 |
| gi|417515419 | Semaphorin-3C precursor | – | 10.76 | 4.92↑ | 0.08 | 3.94 | 0.24 |
| gi|75064988 | Syndecan-4 | SDC4 | 10.29 | 18.88↑ | 0.00 | 19.59↑ | 0.00 |
| gi|346716228 | Histidine triad nucleotide-binding protein 2, mitochondrial isoform 2 precursor | HINT2 | 10.06 | 3.77 | 0.20 | 12.71↑ | 0.03 |
| gi|85720739 | Beta-enolase 3 | ENO3 | 9.83 | 15.42↑ | 0.20 | 8.32↑ | 0.25 |
| gi|223634702 | Succinyl-CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | SUCLG1 | 9.76 | 4.74 ↑ | 0.00 | 9.04↑ | 0.00 |
| gi|4579751 | 130 kDa regulatory subunit of myosin phosphatase, partial | – | 9.64 | 8.39↑ | 0.00 | 3.94 | 0.27 |
| gi|76781337 | ADAMTS1 | ADAMTS1 | 9.61 | 6.79 ↑ | 0.00 | 7.38↑ | 0.00 |
| gi|417515625 | Interferon-induced protein with tetratricopeptide repeats 2 | – | 9.52 | 1.53 | 0.68 | 10.76↑* | 0.00 |
| gi|47522640 | CD97 antigen | – | 8.47 | 5.40↑ | 0.01 | 5.20↑ | 0.02 |
| gi|55247591 | Granulin precursor | GRN | 8.43 | 12.71↑ | 0.00 | 14.59↑ | 0.01 |
| gi|8347147 | Inflammatory response protein 6 | RSAD2 | 8.22 | 0.72 | 0.88 | 4.06↑* | 0.00 |
| gi|148234138 | Cytochrome c oxidase subunit 6B1 | COX6B | 8.11 | 2.65 | 0.11 | 4.97↑ | 0.01 |
| gi|343790890 | Acyl-CoA dehydrogenase family, member 8 | – | 8.06 | 1.56 | 0.30 | 5.55↑ | 0.02 |
| gi|9957597 | Probable ATP-dependent RNA helicase DDX58 | DDX58 | 7.83 | 1.22 | 0.52 | 8.02↑* | 0.00 |
| gi|347300255 | DAZ-associated protein 1 | DAZAP1 | 7.32 | 7.52↑ | 0.01 | 4.29↑ | 0.05 |
| gi|148887343 | ATP synthase subunit e, mitochondrial | ATP5I | 7.00 | 1.11 | 0.97 | 4.21↑ | 0.01 |
| gi|297632426 | Signal sequence receptor, alpha | – | 6.36 | 4.49 ↑ | 0.03 | 5.20↑ | 0.03 |
| gi|6919844 | Transforming growth factor-beta-induced protein ig-h3 | TGFBI | 6.12 | 4.92 ↑ | 0.01 | 3.56 | 0.20 |
| gi|47523704 | Double stranded RNA-dependent protein kinase | PKR | 6.07 | 5.65↑ | 0.19 | 6.67↑ | 0.15 |
| gi|339895859 | Lipase, endothelial precursor | LIPG | 5.14 | 4.06↑ | 0.04 | 3.25 | 0.05 |
| gi|6226834 | 2'-5'-oligoadenylate synthase 1 | OAS1 | 5.03 | 1.96 | 0.09 | 10.47↑* | 0.01 |
| gi|21636588 | ATP synthase gamma subunit 1 | – | 4.61 | 2.78 | 0.16 | 4.49↑ | 0.05 |
| gi|56392985 | Asparagine-linked glycosylation 2 | ALG2 | 4.31 | 2.65 | 0.30 | 4.57↑ | 0.23 |
| gi|52346216 | Fibroleukin precursor | FGL2 | 4.22 | 3.13 | 0.11 | 4.33↑ | 0.07 |
| gi|154147577 | Interferon-induced helicase C domain-containing protein 1 | MDA5 | 4.20 | 2.09 | 0.78 | 6.67↑ | 0.06 |
| gi|343098453 | Chromatin target of PRMT1 protein | CHTOP | 4.10 | 8.47↑ | 0.05 | 6.79↑ | 0.24 |
| gi|343478189 | Tubulin beta-2B chain | TUBB2B | 4.04 | 5.25↑ | 0.30 | 5.40↑ | 0.24 |
| gi|47523638 | Nexin-1 precursor | PN-1 | 4.01 | 5.97↑ | 0.16 | 8.95↑ | 0.14 |
| gi|346716354 | Protein lunapark | – | 4.00 | 10.76↑ | 0.17 | 7.94↑ | 0.23 |
| gi|87047624 | C-C motif chemokine 5 | CCL5 | 3.80 | 5.35↑ | 0.31 | 18.71↑ | 0.12 |
| gi|75056555 | Integral membrane protein 2B | ITM2B | 3.70 | 12.82↑ | 0.20 | 12.94↑ | 0.18 |
| gi|264681460 | Acyl carrier protein, mitochondrial | NDUFAB1 | 3.13 | 2.21 | 0.25 | 4.33↑ | 0.09 |
| gi|456752927 | Lectin, galactoside-binding, soluble, 3 binding protein | – | 2.94 | 1.04 | 0.13 | 6.43↑* | 0.06 |
| gi|116175255 | Regulator of differentiation 1 | ROD1 | 2.79 | 2.68 | 0.23 | 4.29↑ | 0.14 |
| gi|164664468 | ATP synthase subunit epsilon, mitochondrial | ATP5E | 2.74 | 3.66 | 0.14 | 14.06 ↑ | 0.02 |
| gi|47522704 | Vascular cell adhesion protein 1 precursor | – | 2.72 | 3.56 | 0.11 | 6.79 ↑ | 0.02 |
| gi|417515517 | Solute carrier family 2,facilitated glucose transporter member 1 | – | 2.52 | 4.06↑ | 0.17 | 2.83 | 0.23 |
| gi|346644790 | Eukaryotic translation initiation factor 4E-binding protein 1 | – | 2.15 | 11.48↑ | 0.05 | 6.73↑ | 0.17 |
| gi|346644828 | Nuclear ubiquitous casein and cyclin-dependent kinases substrate | NUCKS1 | 2.01 | 5.70↑ | 0.24 | 3.60 | 0.37 |
| gi|35208827 | Macrophage colony-stimulating factor 1 precursor | MCSF alpha | 2.01 | 6.37↑ | 0.24 | 7.73↑ | 0.21 |
| gi|158726687 | IGFBP-6 | – | 2.00 | 9.29 ↑ | 0.11 | 9.20↑ | 0.11 |
| gi|146345485 | Plasminogen | PLG | 2.00 | 7.94 ↑ | 0.12 | 13.30↑ | 0.10 |
| gi|63809 | Transforming growth factor beta-1 | TGFB1 | 2.00 | 8.32↑ | 0.31 | 13.43↑ | 0.21 |
| gi|239504564 | Claudin-4 | CLDN4 | 1.97 | 4.92 ↑ | 0.27 | 8.63↑ | 0.16 |
| gi|75049861 | C-X-C motif chemokine 16 | CXCL16 | 1.96 | 3.02 | 0.22 | 4.92↑ | 0.15 |
| gi|158514029 | ATP synthase lipid-binding protein, mitochondrial | ATP5G1 | 1.45 | 1.38 | 0.49 | 5.81↑* | 0.34 |
| gi|872313 | Monocyte chemoattractant protein 1 | CCL2 | 1.32 | 3.44 | 0.25 | 4.79↑ | 0.19 |
| gi|81295909 | Mitochondrial aldehyde dehydrogenase 2 | ALDH2 | 34.88 | 0.72 | 0.14 | 3.16* | 0.00 |
| gi|224593280 | Tyrosine-protein phosphatase non-receptor type 1 | PTPN1 | 12.65 | 0.33 | 0.01 | 1,37* | 0.11 |
| gi|83415439 | MHC class I antigen | PD1 | 7.05 | 0.45 | 0.43 | 3.13* | 0.04 |
| gi|148747492 | Keratin, type II cytoskeletal 2 epidermal | KRT2A | 6.68 | 0.67 | 0.98 | 3.40* | 0.10 |
| gi|75054309 | N-acetylgalactosamine-6-sulfatase | GALNS | 6.61 | 0.34 | 0.05 | 1.72* | 0.11 |
| gi|343791025 | Lysosomal protective protein precursor | – | 5.84 | 0.81 | 0.80 | 3.25* | 0.05 |
| gi|262204920 | Peroxisomal trans-2-enoyl-CoA reductase | PECR | 5.77 | 0.26 | 0.13 | 1.25* | 0.32 |
| gi|75063982 | Alpha-crystallin B chain | CRYAB | 4.92 | 0.37 | 0.19 | 3.40* | 0.07 |
| gi|456753359 | Mevalonate (diphospho) decarboxylase, partial | – | 4.01 | 0.26 | 0.44 | 1.41* | 0.77 |
| gi|343478257 | Peptidase M20 domain containing 1 | – | 3.19 | 0.31 | 0.36 | 1.34* | 0.69 |
| gi|90024980 | Peroxisomal enoyl coenzyme A hydratase 1 | ECH1 | 17.09 | 0.79 | 0.88 | 3.37* | 0.00 |
| Downregulated proteins | |||||||
| gi|346986428 | Heat shock 90kD protein 1, beta | HSPCB | 130.10 | 0.70 | 0.52 | 0.21↓ | 0.00 |
| gi|48675927 | Tropomyosin alpha-3 chain | TPM3 | 91.83 | 0.53 | 0.01 | 0.20↓ | 0.00 |
| gi|28948618 | Chain A, structure of full-length annexin A1 in the presence of calcium | ANXA1 | 72.35 | 0.42 | 0.00 | 0.06↓* | 0.00 |
| gi|6016267 | Heat shock protein HSP 90-alpha | HSP90AA1 | 53.06 | 0.74 | 0.10 | 0.19↓* | 0.00 |
| gi|47523720 | Glucose-6-phosphate isomerase | GPI | 50.00 | 0.54 | 0.00 | 0.18↓ | 0.00 |
| gi|57527982 | Radixin | RDX | 44.08 | 0.53 | 0.00 | 0.22↓ | 0.00 |
| gi|51702768 | Peptidyl-prolyl cis-trans isomerase A | PPIA | 41.51 | 0.75 | 0.35 | 0.24↓ | 0.00 |
| gi|7650140 | Gag-pol precursor | – | 40.78 | 0.23↓ | 0.00 | 0.82 | 0.04 |
| gi|262263205 | Triosephosphate isomerase 1 | TPI1 | 37.70 | 0.47 | 0.02 | 0.13↓ | 0.00 |
| gi|1927 | Cardiac alpha tropomyosin | TPM1 | 36.76 | 0.50 | 0.01 | 0.08↓* | 0.00 |
| gi|75074817 | Peroxiredoxin-6 | PRDX6 | 35.65 | 0.90 | 0.03 | 0.16↓* | 0.00 |
| gi|94962086 | Aldo-keto reductase family 1 member C4 | AKR1C4 | 34.49 | 0.12↓ | 0.00 | 0.37 | 0.00 |
| gi|473575 | Lactate dehydrogenase-B | LDHB | 24.97 | 0.63 | 0.01 | 0.08↓* | 0.00 |
| gi|164414678 | Alternative pig liver esterase | APLE | 23.65 | 0.19↓ | 0.06 | 0.64 | 0.43 |
| gi|343780946 | D-dopachrome decarboxylase | DDT | 19.05 | 0.15↓ | 0.26 | 0.60* | 0.61 |
| gi|347300176 | Peroxiredoxin-2 | PRDX2 | 24.03 | 0.74 | 0.30 | 0.24↓ | 0.01 |
| gi|302372516 | Heart fatty acid-binding protein | FABP3 | 23.79 | 0.60 | 0.00 | 0.23↓ | 0.00 |
| gi|343887360 | Proteasome (prosome, macropain) subunit, alpha type | – | 21.65 | 0.42 | 0.00 | 0.25↓ | 0.00 |
| gi|47522644 | Acylamino-acid-releasing enzyme | APEH | 20.10 | 0.38 | 0.01 | 0.15↓ | 0.00 |
| gi|346716148 | Importin-5 | - | 18.84 | 0.83 | 0.27 | 0.22↓ | 0.00 |
| gi|47523046 | Acyl-CoA-binding protein | DBI | 18.36 | 0.44 | 0.05 | 0.23↓ | 0.00 |
| gi|47523158 | Glutathione S-transferase A2 | – | 15.78 | 0.31 | 0.00 | 0.09↓ | 0.00 |
| gi|297591959 | Farnesyl pyrophosphate synthase precursor | FDPS | 15.29 | 0.67 | 0.32 | 0.16↓* | 0.00 |
| gi|56384247 | Ribosomal protein L7 | – | 15.34 | 0.09↓ | 0.01 | 0.37 | 0.05 |
| gi|347300398 | Core histone macro-H2A.1 isoform 1 | H2AFY | 14.24 | 0.25↓ | 0.24 | 1.16* | 0.58 |
| gi|417515487 | Collectin sub-family member 12 | – | 14.04 | 0.19↓ | 0.00 | 0.48 | 0.14 |
| gi|94471896 | signal transducer and activator of transcription 3 | STAT3 | 13.42 | 0.21↓ | 0.00 | 0.58 | 0.14 |
| gi|417515866 | KIAA0196 | – | 12.91 | 0.39 | 0.00 | 0.14↓ | 0.00 |
| gi|584724 | Aminoacylase-1 | ACY1 | 12.26 | 0.30 | 0.00 | 0.13↓ | 0.00 |
| gi|158514030 | 60S ribosomal protein L14 | RPL14 | 10.79 | 0.14↓ | 0.00 | 0.79* | 0.85 |
| gi|187606917 | 40S ribosomal protein S26 | RPS26 | 6.00 | 0.19↓ | 0.07 | 0.45 | 0.15 |
| gi|89257972 | Protein phosphatase 1 catalytic subunit beta isoform | PPP1CB | 5.27 | 0.11↓ | 0.25 | 0.61* | 0.52 |
| gi|417515889 | FK506-binding protein 15 | – | 4.06 | 0.05↓ | 0.12 | 0.43* | 0.25 |
| gi|48474224 | Scavenger receptor class B member 1 | SCARB1 | 2.98 | 0.24↓ | 0.08 | 0.41 | 0.13 |
| gi|83778524 | Beta-tropomyosin | TPM2 | 2.55 | 0.28 | 0.21 | 0.07↓* | 0.04 |
| gi|298104074 | Protein FAM54A | – | 2.08 | 0.46 | 0.30 | 0.21↓ | 0.16 |
| gi|342349308 | Calmegin precursor | – | 2.00 | 0.16↓ | 0.14 | 0.21↓ | 0.16 |
| gi|1016311 | Cytochrome P450 2C33v3, partial | – | 1.96 | 0.10 ↓ | 0.11 | 0.41* | 0.26 |
| gi|346716298 | Heterogeneous nuclear ribonucleoprotein G | RBMX | 32.46 | 3.63 | 0.00 | 0.86* | 0.41 |
| gi|262263201 | Squalene epoxidase | SQLE | 2.02 | 1.37 | 0.67 | 0.34* | 0.32 |
Note: *means the proteins significantly differed in expression level between 48 and 64 hpi. The three proteins verified by Western blot analysis are highlighted in bold. The corresponding ratios at each time point (infected/uninfected) are given. Gene symbols were retrieved from UniProt.
GO enrichment analysis
Biological process-based enrichment analysis of the differentially expressed proteins revealed that six common GO terms were significantly enriched in this set of proteins (p<0.05). Thus, it appears that in TGEV-infected ST cells at 48 and 64 hpi there are expression changes in proteins that are related to cell adhesion, neurological system processes, extracellular matrix organization, locomotion, cell junction organization, and cell-cell signaling. Moreover, at the later time point, 64 hpi, our GO term analysis also indicated that a significant number of the differentially expressed proteins were related to cellular stress (p = 8.18E-4), generation of precursor metabolites and energy (p = 2.74E-3), cell motility (p = 6.71E-3), protein complex assembly (p = 4.69E-2), growth (p = 3.87E-2), developmental maturation (p = 1.53E-2), and immune system processes (p = 4.67E-2) (Table 2).
Table 2. Biological process-based GO term enrichment analysis.
| GO term | Gene symbol or protein name (48 hpi) | P-value(48 hpi) | Gene symbol or protein name (64 hpi) | P-value(64 hpi) |
| Cell adhesion | MFGE8, CYR61, ITGA5, FN1, TGFBI, TGFB1, PN-1, CCL5, APP, PPP1CB, SCARB1 | 2.57E-3 | MFGE8, CYR61, ITGA5, FN1, TGFB1, CALR, APP, TACSTD2, PN-1, Vascular cell adhesion molecule, CCL5 | 2.57E-3 |
| Response to stress | NUDT9, CYR61, ITGA5, FN1, PLG, TGFB1, Extracellular superoxide dismutase precursor, CLU, CCL5, HSPA1B, PN-1, VDAC1 | 7.98E-1 | CCL2, NUDT9, CYR61, ITGA5, FN1, LOC100516779, Mitochondrial heat shock 60 kDa protein 1, CCDC47, PLG, TGFB1, CALR, Extracellular superoxide dismutase precursor, OAS1, HSPA1B, PN-1, DDX58, VDAC1, RSAD2, HSP90AA1, HSPCB, DBI, PRDX2, PRDX3, CLU, CCL5, CXCL16, PRDX6 | 8.18E-4 |
| Generation of precursor metabolites and energy | ENO3, SUCLG1, PPP1CB | 6.51E-1 | ENO3, TPI1, IDH3A, SUCLG1, LDHB, MDH2, GPI, SUCLG2, SDHB, DLST | 2.74E-3 |
| Extracellular matrix organization | CYR61, TGFB1, APP | 1.22E-2 | LGALS3, CYR61, TGFB1, APP | 1.51E-3 |
| Locomotion | TGFB1, APP, CCL5 | 1.22E-2 | TGFB1, APP, CCL5, CXCL16 | 1.51E-3 |
| Cell motility | STAT3, CYR61, ITGA5, TUBB2B, TGFB1, CCL5 | 2.07E-1 | CCL2, TACSTD2, CYR61, ITGA5, TUBB2B, TGFB1, CALR, CCL5, CXCL16, DDX58 | 6.71E-3 |
| Cell-cell signaling | ITPR3, APP, PN-1, VDAC1, CCL5 | 2.58E-2 | GLUD1, APP, PN-1, VDAC1, CCL5 | 2.58E-2 |
| Neurological system process | ITPR3, ITGA5, APP, VDAC1, PN-1 | 7.91E-3 | ITGA5, APP, PN-1, VDAC1 | 3.34E-2 |
| Protein complex assembly | H2AFY, SLAIN2, HIST1H2BF, TUBB2B, HIST1H2BJ, TMSB10, TGFB1, CLU, CCL5 | 2.66E-1 | SLAIN2,HIST1H2BF,TUBB2B,HIST1H2BJ, TGFB1, TMSB10, RDX, CALR,CLU, CCL5, Histone H1.3-like protein, TFAM | 4.69E-2 |
| Cell junction organization | ITGA5, FN1, TGFB1 | 1.22E-2 | ITGA5, FN1, TGFB1 | 1.22E-2 |
| Growth | STAT3, CYR61, TGFB1, APP, PN-1 | 1.01E-1 | COL9A1, CYR61, TGFB1, PN-1, APP, CXCL16 | 3.87E-2 |
| Developmental maturation | APP | 1.13E-1 | ARCN1, APP | 1.53E-2 |
| Immune system process | TGFB1, CCL5 | 9.81E-1 | CCL2, HSPCB, PRDX3, LOC100516779, TGFB1, CALR, OAS1, CCL5, CXCL16, DDX58, RSAD2 | 4.67E-2 |
Note: P-values were calculated in the hypergeometric test. Gene symbols were retrieved from UniProt. The significantly common processes affected are highlighted in bold.
To further investigate the localization pattern of these differentially expressed genes, a cellular component-based enrichment analysis was performed. At 48 hpi, we observed the significant enrichments in extracellular region (p = 1.29E-4), proteinaceous extracellular matrix (p = 1.62E-4), and extracellular space (p = 1.52E-2) (Table S4). In addition, 37 differentially expressed proteins were also significantly enriched (p = 8.65E-3) in mitochondrion at 64 hpi (Table S5).
The final step of our GO enrichment analysis consisted of investigating the mechanistic role these genes play in the cell. To do so, we performed a molecular function-based enrichment analysis. This analysis showed that two GO terms, unfolded protein binding (p = 2.67E-2) and transmembrane transporter activity (p = 3.55E-2), were significantly enriched at 64 hpi (Table S5). Further GO analysis of the differentially expressed proteins between the two time points indicated that there were no significant enriched terms.
Protein–protein interaction analysis
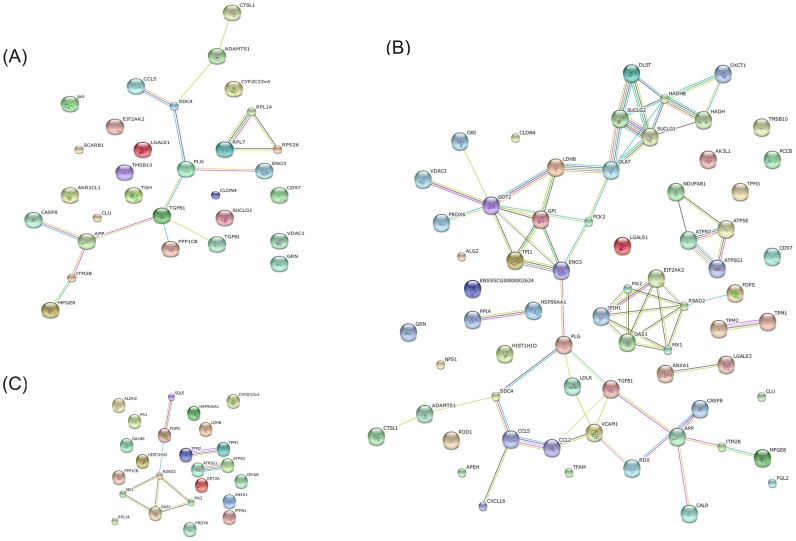
In order to understand the interactions between TGEV and host cell proteins, we further analyzed the differentially expressed proteins by searching the STRING 9.1 database (http://string-db.org/) for protein-protein interactions (Figure 4). In this STRING analysis, the interactions (edges) of the submitted proteins (nodes) were scored according to known and predicted protein-protein interactions. We created three protein network maps: one for proteins changed significantly at 48 hpi (30 nodes and 15 edges; Figure 4A), one for proteins changed significantly at 64 hpi (66 nodes and 70 edges; Figure 4B), and one for the proteins that were significantly changed when the viral infection was prolonged from 48 to 64 h (24 nodes and 9 edges; Figure 4C). Notably, the protein network constructed for the 64 hpi time point is clearly much more extensive than the two other networks, and these protein-protein interactions suggest the existence of reported functional linkages. GO enrichment analysis for the STRING protein network at 64 hpi showed that several biological processes were significantly affected (p<0.05 based on the FDR correction) in this network, including the regulation of viral genome replication, the innate immune response, negative regulation of viral genome replication, positive and negative regulation of viral processes, and ATP biosynthetic processes (Table 3). However, at 48 hpi, the most enriched biological process was related to cell recognition during phagocytosis(p = 8.02E-1). In Figure 4C, we have shown that the majority proteins in these protein networks, such as radical S-adenosyl methionine domain containing protein 2 (RSAD2), Mx dynamin-like GTPase 1 (Mx1), 2′-5′-oligoadenylate synthetase 1 (OAS1), Mx dynamin-like GTPase 2 (Mx2), are involved in the innate immune response. These data suggest that some entirely different host proteins, interactions, or processes, including the immune response, were perturbed at these times during TGEV infection.
Figure 4. Protein-protein interaction network created using the STRING database.
(A) Network of the differentially expressed proteins at 48 hpi. The network includes 30 nodes (proteins) and 15 edges (interactions). (B) Network of differentially expressed proteins at 64 hpi. The network includes 66 nodes and 70 edges. (C) Network of differentially expressed proteins between the two time points. The network includes 24 nodes and 9 edges. Network analysis was set at medium confidence (STRING score = 0.4). Seven different colored lines were used to represent the types of evidence for the association: green, neighborhood evidence; red, gene fusion; blue, co-occurrence; black, co-expression; purple, experimental; light blue, database; yellow, text mining.
Table 3. List of the GO biological processes enriched for the proteins present in the STRING protein network.
| GO biological process | P-value |
| Regulation of viral genome replication | 1.33E-2 |
| Innate immune response | 1.35E-2 |
| Negative regulation of viral genome replication | 2.36E-2 |
| Regulation of viral process | 2.70E-2 |
| Negative regulation of viral process | 2.83E-2 |
| ATP biosynthetic process | 2.89E-2 |
Note: The significance of the GO biological process is derived from the network in Figure 4B and was determined using the FDR correction (p<0.05).
Western blot confirmation of altered expression for three of the differentially expressed proteins
To further confirm the proteomic data for three of the proteins, western blot analysis was performed to investigate the changes in the expression of HSP90α, caspase 8, and TGF-β1. The proteins were selected based on three criteria: 1) the expression of the protein was increased or decreased during TGEV infection according to our proteomics data; 2) the protein is known to be relevant during viral infection; and 3) each protein analyzed needs to be involved in a special biological process as determined by our GO enrichment analysis [32]. HSP90α, caspase 8, and TGF-β1 all filled these criteria and their protein expression was analyzed via western blot analysis of the cell lysate. As shown in Figure 5, the expression of HSP90α was significantly downregulated in TGEV-infected cells at 64 hpi, while the expression of caspase-8 was upregulated from 48 to 64 hpi in these cells. The expression of TGF-β1 was also significantly induced in TGEV-infected cells following infection. Thus, these results confirm the altered expression observed in the proteomic data for these three representative proteins during TGEV infection.
Figure 5. Western blot confirmation for three differentially expressed proteins (caspase-8, HSP90α, and TGF-β1).
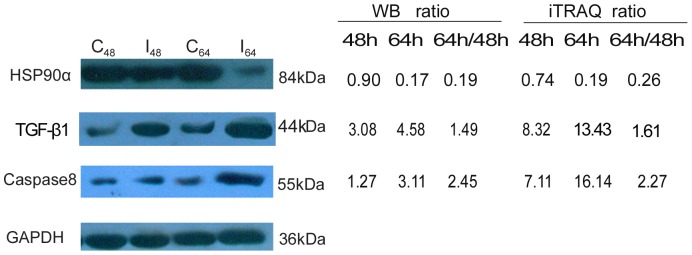
Following TGEV and mock infection of the ST cells, equal amounts of protein were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were then probed with the specified antibody, and the identified bands were visualized. GAPDH was used as an internal control to normalize the quantitative data. The representative images shown are typical of two independent experiments. At 48 hpi (I48), integrated optical density (IOD) analysis showed an upregulation of caspase-8 (1.27 fold) and TGF-β1 (3.08 fold), but HSP90α was almost unchanged (0.90 fold). At 64 hpi (I64), we observed an upregulation in both caspase-8 (3.11 fold) and TGF-β1 (4.58 fold), but a 5.82 fold downregulation of HSP90α. The IOD was normalized against GAPDH.
Discussion
The interactions between a virus and a host cell during a viral infection are complex, involving numerous genes and signaling pathways. ST cells are known to be sensitive to TGEV, resulting in increased viral multiplication and CPEs [15]. In order to better understand the interactions between the host proteome and TGEV, we adopted an iTRAQ quantitative proteomic approach to investigate the altered cellular proteins of the ST cells during TGEV infection in vitro. Compared with the 2-DE and 2D-DIGE methods often used, the 2D-LC-MS/MS method utilized here provides more quantitative and qualitative information about the proteins, and can also detect membrane proteins, hydrophobic proteins, higher molecular weight proteins, and low-abundance proteins, which are often missed by other methods. iTRAQ also has more advantages compared to isotope-coded affinity tags (ICAT) and stable isotope labeling by amino acids in cell culture (SILAC) methods, which both allow multiple labeling and quantitation of four to eight samples simultaneously with high sensitivity [22], [33], [34]. Further, the iTRAQ technique has been widely used for quantitative proteomics, including protein expression analysis and biomarker identification [23]–[26], [35].
Prior to proteomic analysis, we determined which time points to investigate following infection by observing the morphological changes and analyzing viral gene expression dynamics in the TGEV infected cells. The results indicated that TGEV induced significant CPEs from 40 to 64 hpi in infected cells compared to the mock infected cells. At 40 hpi, less than 50% of the infected cells were morphologically altered, while at 48 hpi more than 80% infected cells showed rounding and granular degeneration. Further, the mRNA level of the viral N gene in ST cells continuously increased in the infected cells until 48 h, at which time we observed the highest viral replication level. At 64 hpi, the morphological effects observed were much more pronounced, characterized by even more cellular rounding and detachment. However, the mRNA levels of the viral N gene decreased rapidly from 48 to 64 h, a phenomenon we believe may be attributed to the host's immune response or a decrease in infected cell viability as the TGEV infection progressed. Based on our qRT-PCR and CPE analyses, we choose to more deeply investigate the proteomic changes occurring in the TGEV-infected ST cells at 48 hpi and 64 hpi using a 4-plex iTRAQ analysis.
In our analysis, we observed a statistically significant change in the expression of 316 proteins during TGEV infection in vitro. This number includes protein changes that were unique for a specific time point as well as those shared at these different time conditions. For example, the expression level of HSP90α expression was unchanged at 48 hpi, but decreased at 64 hpi, making this change unique for the latter time point. On the other hand, TGF-β1 was observed to increase at both of the time points, and was thus labeled a shared protein change. Moreover, the 316 altered proteins also includes proteins that changed from 48 hpi to 64 hpi, rather than one of these time points compared to non-infected cells. For example, mitochondrial aldehyde dehydrogenase 2 (ALDH2) and MHC class I antigen (PD1) were not changed at 48 or 64 hpi compared to the control group, but increased at 64 hpi compared with 48 hpi. We also observed a larger proteomic shift at 64 hpi compared to the 48 hpi time point in the infected ST cells.
Further, some proteins previously reported to play a role in virus-induced host cell death, such as caspase-8, caspase-3, caspase-9, and porcine aminopeptidase-N (pAPN) [36]–[38], were also identified using this iTRAQ technique. These caspase proteins are known to be involved in TGEV-induced cell apoptosis processes, while pAPN is the cell receptor for TGEV. Our results indicate that TGEV infection caused significant upregulation of caspase-8 expression at two time points (approximately 7-fold at 48 hpi and 16-fold at 64 hpi) in the virus-infected ST cells, and this change was verified by western blotting analysis. However, the expression of caspase-3, caspase-9, and pAPN was not significantly altered, indicating that the pathways involving these genes are not altered or that other proteins are compensating for their lack of change. In this regard, we identified an additional 15 proteins involved in cell death pathways that had significantly altered expression levels (p = 4.46E-2) (Table S6), including melanoma differentiation associated protein-5 (MDA5), monocyte chemoattractant protein 1 (CCL2), thioredoxin- dependent peroxide reductase, mitochondrial (PRDX3), peroxiredoxin-2 (PRDX2), predicted protein CYR61 (CYR61), keratin, type II cytoskeletal 8 (KRT8), predicted bcl-2-like protein 13 (BCL2L13), predicted integrin alpha-5 isoform 1 (ITGA5), TGF-β1, amyloid beta A4 protein (APP), clusterin (CLU), C–C motif chemokine 5 (CCL5), heat shock 70 kDa protein 1B (HSPA1B), alpha-crystallin B chain (CRYAB), voltage-dependent anion-selective channel protein 1 (VDAC1), all of which, with the exception of PRDX2 and BCL2L13 were upregulated at one or two time points. Regulation of cell death is known to be important for replication and pathogenesis in various coronaviruses [39], and we believe that further research on these proteins will lead to a better understanding of cell death regulation during TGEV infection.
In order to determine what other processes, in addition to cell death, were affected by TGEV infection, we performed a GO enrichment analysis for the different temporal conditions. This analysis indicated that six biological processes were significantly affected at 48 and 64 hpi, and the differentially expressed proteins involved in these processes were almost the same. The large overlap between the two time points suggests that some of the same sets of host proteins or processes were disturbed at these times. However, it is also likely that some processes were affected solely at one time point or the other. At 48 hpi, serine/threonine-protein phosphatase PP1-beta-catalytic subunit (PPP1CB), scavenger receptor class B member 1 (SCARB1), transforming growth factor-beta-induced protein ig-h3 (TGFBI), and predicted inositol 1,4,5-trisphosphate receptor type 3 (ITPR3) were uniquely altered, likely indicating changes in cell adhesion and/or cell-cell signaling processes. At 64 hpi, on the other hand, calreticulin (CALR), predicted tumor- associated calcium signal transducer 2-like (TACSTD2), vascular cell adhesion molecule, galectin-3 (LGALS3), glutamate dehydrogenase 1 (GLUD1), and C–X-C motif chemokine 16 (CXCL16) were uniquely changed, also indicating changes in cell adhesion and/or cell-cell signaling as well as extracellular matrix organization and locomotion. We believe that these uniquely altered proteins reflect changes in specific/specialized processes at each time point that are tightly linked to the temporal changes observed in the host cell morphology and gene/protein expression after TGEV infection.
The most significantly enriched GO category related to the differentially expressed proteins was stress, which included 12 differentially expressed proteins at 48 hpi and 27 different proteins at 64 hpi. The increased number of proteins association with this GO term at 48 hpi likely highlights the initial upregulation of the cellular stress response, while the higher number at 64 hpi indicates that the stress response to TGEV infection is likely more fully induced at this later stage. HSPs, also known as stress proteins, are often involved in the cellular response to stress, influencing changes in the state or activity of the cell or organism. HSP90, which has two isoforms (HSP90α and HSP90β), is one of the most abundant molecular chaperones that is induced in response to cellular stress, and it functions to stabilize proteins involved in cell growth and anti-apoptotic signaling [40]. The expression of HSP90α has been reported to play an important role in the replication of some viruses, such as Ebola virus (EBOV) [41], hepatitis C virus (HCV) [42], influenza virus [43], and Japanese encephalitis virus [44]. On the other hand, the reduction of HSP90β has been reported to decrease the correct assembly of human enterovirus 71 viral particles [40]. In this study, HSP90α and heat shock 90kD protein 1, beta (HSPCB/HSP90β) were significantly downregulated at 64 hpi in the TGEV-infected ST cells, but were unchanged at 48 hpi, indicating that they may play a similar role in TGEV infection. Interestingly, a member of the HSP70 protein family, heat shock 70 kDa protein 1B (HSPA1B), as well as mitochondrial 60 kDa heat shock protein (HSP60) were both upregulated in infected ST cells at 48 and/or 64 hpi. HSP60 is a mitochondrial chaperonin protein involved in protein folding and a number of extracellular immunomodulatory activities. Elevated expression of HSP60 is associated with a number of inflammatory disorders [45]. HSP70 plays an important role in multiple processes within cells, including protein translation, folding, intracellular trafficking, and degradation. A previous study has revealed that HSP70 is involved in all steps of the viral life cycle, including replication, and is highly specific in regards to viral response, differing from one cell to another for any given virus type [46]. For example, silencing HSP70 expression has been associated with an increase in viral protein levels, while an increase in HSP70 has been suspected to be the initial cellular response to protect against viral infection in rotavirus-infected cells [47]. Further, a recent study showed that HSP70 is an essential host factor for the replication of PRRSV as the silence of HSP70 significantly reduced PRRSV replication [48]. Our results provide new experimental evidence relating the expression of HSP90, HSP70, and HSP60 to TGEV infection, and we speculate that these proteins play a potential role in TGEV replication. Additional work is required to investigate the detailed role of these proteins during TGEV infection.
Furthermore, another significantly enriched GO process we observed that 11 significantly altered proteins was immune system processes. Most of these proteins were significantly upregulated at 64 hpi in response to the viral infection, while some were first upregulated at 48 hpi, including CCL5 and TGF-β1. Chemokines, such as CCL2, CCL5, and CXCL16, whose main function is macrophage recruitment and activation, are potentially involved in host-mediated immunopathology. A recent study showed that coronavirus infection of transgenic mice expressing CCL2 led to a dysregulated immune response without effective virus clearance and enhanced death [49]. In additional, TGEV-infection can induce the expression of proinflammatory genes, including CCL2, CCL5, and probable ATP-dependent RNA helicase DDX58 (DDX58/RIG-1), in cell culture and in vivo in the absence of viral protein 7 [50]. In this study, we observed an upregulation of CCL2, CCL5, CXCL16, TGF-β1, and DDX58 expression. TGF-β1 is a multifunctional cytokine, secreted from various cells, and, in immunology, it regulates cellular proliferation, differentiation, and other cellular functions for a variety of cell types, especially regulatory T cells [51]. Some research has indicated that SARS-CoV papain-like protease (PLpro) increases TGF-β1 mRNA expression and protein production in human promonocytes [52]. Further, Gomez-Laguna et al. [53] inferred that the upregulation of the TGF-β may impair the host immune response during PRRSV infection by limiting the overproduction of proinflammatory cytokines necessary to decrease PRRSV replication. In response to viral infection, DDX58 plays important roles in the recognition of RNA viruses in various cells, and has been identified as a candidate for a cytoplasmic viral dsRNA receptor [54]. Further, upregulation of this gene activates cells to produce type I interferons, which may increase the antiviral status of cells to protect against viral infection. In this regard, we found that interferon-inducible antiviral proteins, RSAD2, OAS1, were also upregulated in the period of late infection, suggesting that many of the proteins identified in this study are associated with inflammation, IFN activation, and the innate immune response. Increased expression of these proteins may help the virus enter the cell as well as potentially enhance TGEV replication or the host response against the virus, during the late stages of infection.
In conclusion, we used the iTRAQ method to identify 316 significantly altered proteins in TGEV-infected ST cells. A larger number of protein expression changes occurred at 64 hpi compared to 48 hpi, indicating a larger shift in the proteome in the later stages of infection. GO analysis of these differentially expressed proteins indicated that a number of diverse biological processes are affected. In addition, many of the significant immune response related changes in protein expression we discovered are novel and, to our knowledge, have not been detected in previous proteome study. Results from this study complement the previous proteomics data obtained concerning the host response to a viral infection, and further facilitates a better understanding of the pathogenic mechanisms of TGEV infection and molecular responses of host cells to this virus.
Supporting Information
Total proteins (4,364) identified and quantified by iTRAQ.
(XLSX)
Differentially expressed proteins identified under different conditions.
(XLSX)
GO enrichment analysis of differentially expressed proteins identified at 48 hpi.
(XLSX)
GO enrichment analysis of differentially expressed proteins identified at 64 hpi.
(XLSX)
GO enrichment of all the differentially expressed proteins.
(XLSX)
Acknowledgments
We thank Qiangqiang Zhao, Chen Lou, Wulong Liang, and Helin Li for their technical support, and Shuo Chen for his valuable advice.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 31172339). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones T, Pritchard G, Paton D (1997) Transmissible gastroenteritis of pigs. Vet Rec 141: 427–428. [PubMed] [Google Scholar]
- 2. Wesley RD, Lager KM (2003) Increased litter survival rates, reduced clinical illness and better lactogenic immunity against TGEV in gilts that were primed as neonates with porcine respiratory coronavirus (PRCV). Vet Microbiol 95: 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim L, Hayes J, Lewis P, Parwani AV, Chang KO, et al. (2000) Molecular characterization and pathogenesis of transmissible gastroenteritis coronavirus (TGEV) and porcine respiratory coronavirus (PRCV) field isolates co-circulating in a swine herd. Arch Virol 145: 1133–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vlasova AN, Halpin R, Wang S, Ghedin E, Spiro DJ, et al. (2011) Molecular characterization of a new species in the genus Alphacoronavirus associated with mink epizootic catarrhal gastroenteritis. J Gen Virol 92: 1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spaan W, Cavanagh D, Horzinek MC (1988) Coronaviruses: structure and genome expression. J Gen Virol 69: 2939–2952. [DOI] [PubMed] [Google Scholar]
- 6. Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, et al. (2011) A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol 174: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H (1998) Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J Virol 72: 8636–8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Curtis KM, Yount B, Baric RS (2002) Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J Virol 76: 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weingartl HM, Derbyshire JB (1993) Binding of porcine transmissible gastroenteritis virus by enterocytes from newborn and weaned piglets. Vet Microbiol 35: 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weingartl HM, Derbyshire JB (1994) Evidence for a putative second receptor for porcine transmissible gastroenteritis virus on the villous enterocytes of newborn pigs. J Virol 68: 7253–7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sirinarumitr T, Paul PS, Kluge JP, Halbur PG (1996) In situ hybridization technique for the detection of swine enteric and respiratory coronaviruses, transmissible gastroenteritis virus (TGEV) and porcine respiratory coronavirus (PRCV), in formalin-fixed paraffin-embedded tissues. J Virol Methods 56: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Galán C, Sola I, Nogales A, Thomas B, Akoulitchev A, et al. (2009) Host cell proteins interacting with the 3′end of TGEV coronavirus genome influence virus replication. Virology 391: 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Penzes Z, Gonzalez JM, Calvo E, Izeta A, Smerdou C, et al. (2001) Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the purdue virus cluster. Virus Genes 23: 105–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ortego J, Sola I, Almazán F, Ceriani JE, Riquelme C, et al. (2003) Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology 308: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He L, Zhang YM, Dong LJ, Cheng M, Wang J, et al. (2012) In vitro inhibition of transmissible gastroenteritis coronavirus replication in swine testicular cells by short hairpin RNAs targeting the ORF 7 gene. Virol J 9: 176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun J, Jiang Y, Shi Z, Yan Y, Guo H, et al. (2008) Proteomic alteration of PK-15 cells after infection by classical swine fever virus. J Proteome Res 7: 5263–5269. [DOI] [PubMed] [Google Scholar]
- 17. Zhang X, Zhou J, Wu Y, Zheng X, Ma G, et al. (2009) Differential proteome analysis of host cells infected with porcine circovirus type 2. J Proteome Res 8: 5111–5119. [DOI] [PubMed] [Google Scholar]
- 18. Yang Y, An T, Gong D, Li D, Peng J, et al. (2012) Identification of porcine serum proteins modified in response to HP-PRRSV HuN4 infection by two-dimensional differential gel electrophoresis. Vet Microbiol 158: 237–246. [DOI] [PubMed] [Google Scholar]
- 19. Cao Z, Han Z, Shao Y, Liu X, Sun J, et al. (2012) Proteomics analysis of differentially expressed proteins in chicken trachea and kidney after infection with the highly virulent and attenuated coronavirus infectious bronchitis virus in vivo. Proteome Sci 10: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang XS, Tang LY, Dai J, Zhou H, Li SJ, et al. (2005) Quantitative analysis of severe acute respiratory syndrome (SARS)-associated coronavirus-infected cells using proteomic approaches implications for cellular responses to virus infection. Mol Cell Proteomics 4: 902–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X, Shi HY, Chen JF, Shi D, Lang HW, et al. (2013) Identification of cellular proteome using two-dimensional difference gel electrophoresis in ST cells infected with transmissible gastroenteritis coronavirus. Proteome Sci 11: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu WW, Wang G, Baek SJ, Shen RF (2006) Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome Res 5: 651–658. [DOI] [PubMed] [Google Scholar]
- 23. Lu Q, Bai J, Zhang L, Liu J, Jiang Z, et al. (2012) Two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. J Proteome Res 11: 2890–2903. [DOI] [PubMed] [Google Scholar]
- 24. Lietzen N, Ohman T, Rintahaka J, Julkunen I, Aittokallio T, et al. (2011) Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog 7: e1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng X, Zhang J, Chen WN, Ching CB (2011) Proteome profiling of Epstein-Barr virus infected nasopharyngeal carcinoma cell line: identification of potential biomarkers by comparative iTRAQ-coupled 2D LC/MS-MS analysis. J proteomics 74: 567–576. [DOI] [PubMed] [Google Scholar]
- 27. Liu W, Saint DA (2002) Validation of a quantitative method for real time PCR kinetics. Biochem Biophys Res Commun 294: 347–353. [DOI] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 29. Ruppen I, Grau L, Orenes-Pinero E, Ashman K, Gil M, et al. (2010) Differential protein expression profiling by iTRAQ-two-dimensional LC-MS/MS of human bladder cancer EJ138 cells transfected with the metastasis suppressor KiSS-1 gene. Mol Cell Proteomics 9: 2276–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun L, Bertke MM, Champion MM, Zhu G, Huber PW, et al. (2014) Quantitative proteomics of Xenopus laevis embryos: expression kinetics of nearly 4000 proteins during early development. Sci Rep 4: 4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rivals I, Personnaz L, Taing L, Potier MC (2007) Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23: 401–407. [DOI] [PubMed] [Google Scholar]
- 32. Chiu HC, Hannemann H, Heesom KJ, Matthews DA, Davidson AD (2014) High-Throughput Quantitative Proteomic Analysis of Dengue Virus Type 2 Infected A549 Cells. PloS One 9: e93305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, et al. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169. [DOI] [PubMed] [Google Scholar]
- 34. Munday DC, Surtees R, Emmott E, Dove BK, Digard P, et al. (2012) Using SILAC and quantitative proteomics to investigate the interactions between viral and host proteomes. Proteomics 12: 666–672. [DOI] [PubMed] [Google Scholar]
- 35. Li H, DeSouza LV, Ghanny S, Li W, Romaschin AD, et al. (2007) Identification of candidate biomarker proteins released by human endometrial and cervical cancer cells using two-dimensional liquid chromatography/tandem mass spectrometry. J Proteome Res 6: 2615–2622. [DOI] [PubMed] [Google Scholar]
- 36. Ding L, Xu X, Huang Y, Li Z, Zhang K, et al. (2012) Transmissible gastroenteritis virus infection induces apoptosis through FasL- and mitochondria-mediated pathways. Vet Microbiol 158: 12–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eleouet JF, Chilmonczyk S, Besnardeau L, Laude H (1998) Transmissible gastroenteritis coronavirus induces programmed cell death in infected cells through a caspase-dependent pathway. J Virol 72: 4918–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Delmas B, Gelfi J, L'Haridon R, Vogel LK, Sjostrom H, et al. (1992) Aminopeptidase N is a major receptor for the enteropathogenic coronavirus TGEV. Nature 357: 417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan YJ, Lim SG, Hong W (2007) Regulation of cell death during infection by the severe acute respiratory syndrome coronavirus and other coronaviruses. Cell Microbiol 9: 2552–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang RYL, Kuo RL, Ma WC, Huang HI, Yu JS, et al. (2013) Heat shock protein-90-beta facilitates enterovirus 71 viral particles assembly. Virology 443: 236–247. [DOI] [PubMed] [Google Scholar]
- 41. Smith DR, McCarthy S, Chrovian A, Olinger G, Stosselet A, et al. (2010) Inhibition of heat-shock protein 90 reduces Ebola virus replication. Antivir Res 87: 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, et al. (2006) Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J 25: 5015–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, et al. (2002) Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J Biol Chem 277: 45306–45314. [DOI] [PubMed] [Google Scholar]
- 44. Hung CY, Tsai MC, Wu YP, Wang RY (2011) Identification of heat-shock protein 90 beta in Japanese encephalitis virus-induced secretion proteins. J Gen Virol 92: 2803–2809. [DOI] [PubMed] [Google Scholar]
- 45. Johnson BJ, Le TTT, Dobbin CA, Banovic T, Howard CB, et al. (2005) Heat shock protein 10 inhibits lipopolysaccharide-induced inflammatory mediator production. J Biol Chem 280: 4037–4047. [DOI] [PubMed] [Google Scholar]
- 46. Lahaye X, Vidy A, Fouquet B, Blondel D (2012) Hsp70 protein positively regulates rabies virus infection. J virol 86: 4743–4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Broquet AH, Lenoir C, Gardet A, Sapin C, Chwetzoff S, et al. (2007) Hsp70 negatively controls rotavirus protein bioavailability in caco-2 cells infected by the rotavirus RF strain. J virol 81: 1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gao J, Xiao S, Liu X, Wang L, Ji Q, et al. (2014) Inhibition of HSP70 reduces porcine reproductive and respiratory syndrome virus replication in vitro. BMC microbiol 14: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trujillo JA, Fleming EL, Perlman S (2013) Transgenic CCL2 expression in the central nervous system results in a dysregulated immune response and enhanced lethality after coronavirus infection. J virol 87: 2376–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cruz JLG, Becares M, Sola I, Oliveros JC, Enjuanes L, et al. (2013) Alphacoronavirus protein 7 modulates host innate immune response. J virol 87: 9754–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yang Y, Zhang N, Lan F, Crombruggen K, Fang L, et al. (2014) Transforming growth factor-beta 1 pathways in inflammatory airway diseases. Allergy 69: 699–707. [DOI] [PubMed] [Google Scholar]
- 52. Li SW, Yang TC, Wan L, Lin YJ, Tsai FJ, et al. (2012) Correlation between TGF-β1 expression and proteomic profiling induced by severe acute respiratory syndrome coronavirus papain-like protease. Proteomics 12: 3193–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gomez-Laguna J, Rodriguez-Gomez IM, Barranco I, Pallares FJ, Salguero FJ, et al. (2012) Enhanced expression of TGFβ protein in lymphoid organs and lung, but not in serum, of pigs infected with a European field isolate of porcine reproductive and respiratory syndrome virus. Vet Microbiol 158: 187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takeuchi O, Akira S (2008) MDA5/RIG-I and virus recognition. Curr Opin Immunol 20: 17–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total proteins (4,364) identified and quantified by iTRAQ.
(XLSX)
Differentially expressed proteins identified under different conditions.
(XLSX)
GO enrichment analysis of differentially expressed proteins identified at 48 hpi.
(XLSX)
GO enrichment analysis of differentially expressed proteins identified at 64 hpi.
(XLSX)
GO enrichment of all the differentially expressed proteins.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.