Abstract
Staphylococcus aureus antimicrobial resistance, especially to beta-lactams, favors treatment failures and its persistence in herd environment. This work aimed to develop a more specific primer for mecA gene detection based on the comparison of the conserved regions from distinct host origins and also investigated the presence of homologue mecALGA251 in bovine strains. A total of 43 Staphylococcus spp. were included in this study, comprising 38 bovine S. aureus, two human and three equine coagulase-negative staphylococci (CNS). Phenotypical methicillin-resistance detection was performed through oxacillin agar-screening and cefoxitin disk-diffusion test. None isolate tested positive for mecALGA251 gene. For mecA gene PCR, new primers were designed based on the sequences of human S. aureus (HE681097) and bovine S. sciuri (AY820253) mecA. The new primers based on the S. aureus mecA sequence amplified fragments of human and equine CNS and the ones based on S. sciuri mecA sequence only yielded fragments for S. aureus bovine strains. Multiples alignments of mecA gene sequences from bovine, human and equine revealed punctual but significant differences in bovine strains that can lead to the mecA gene detection impairment. The observed divergences of mecA gene sequences are not a matter of animal or human origin, it is a specificity of bovine samples.
Keywords: antimicrobial resistance, bovine mastitis, Staphylococccus, mecA gene
Introduction
Staphylococcus spp. is the most commonly isolated agent in bovine mastitis, especially considering Staphylococcus aureus (Coelho et al., 2009). Its importance is not only because of its distribution and pathogenicity but also due to its ability to overcome antimicrobial effects. Its high antimicrobial resistance level, especially to beta-lactams, favors treatment failures and its persistence in herd environment. Bacterial resistance mechanisms to this antimicrobial class include production of β-lactamases and low-affinity penicillin-binding protein 2a (PBP2a) determined by the presence of the chromosomal genes bla and mecA, respectively. The latter, involved in oxacillin-resistance, precludes human therapy with any of the currently available β-lactam antibiotics, and is considered to predict resistance to several classes of antibiotics (Moon et al., 2007). Once phenotypic expression of beta-lactam resistance in Staphylococcus isolates is usually heterogeneous, the amplification of mecA gene is prescribed as a gold standard in the detection of this resistance (CLSI, 2012).
Otherwise, our research results in animal science field concerning to bovine staphylococci beta-lactam resistance does not support that mecA gene plays the same significant role in the detection of this resistance. As a matter of fact, our previous studies reported several phenotypically oxacillin-resistant isolates that tested negative for mecA gene (Coelho et al., 2009; Mendonça et al., 2012; Soares et al., 2012).
Theories have been proposed to explain the lack of correlation between the observed phenotypical methicillin-resistance and the detection of mecA gene. The occurrence of deletions, insertions or point mutations could have altered the original nucleotidic sequence in mecA gene primer alignment region in bovine isolates, as reported (Sakoulas et al., 2001). Also, a recently published article (García-Álvarez et al., 2011) reported the presence of MRSA strains with unusual features in bovine milk samples from the UK. These strains carried a novel mecA gene that was only 70% identical at the nucleotide level to the classical mecA gene, escaping detection by routine PCR assays. The novel mecA was named mecALGA251. The gene mecALGA251 is part of a novel SCCmec element, identified as type XI. Human clinical isolates with mecALGA251 were reported in the UK and Denmark. The ability of these strains to cause infections in humans was confirmed by a concurrent report describing two patients infected with similar strains in Ireland (Shore et al., 2011).
Considering these hypothesis and that the true scope of MRSA in animals and its impact on human health are still only superficially understood we decided to investigate the presence of mecALGA251 in the bovine isolates tested negative for mecA. Furthermore, mecA sequences from human and animal origins stored in NCBI databases were analyzed in order to develop a more specific primer considering distinct conserved regions in the gene.
Materials and Methods
Bacterial strains
A total of 43 previously identified isolates were included in this study. Thirty eight Staphylococcus aureus from milk and two Coagulase-Negative Staphylococcus (CNS) from milker’s hands and noses from dairy cattle farm properties at the state of Rio de Janeiro, Brazil. Also, three CNS strains, two S. sciuri and one S. lentus, collected from horses in the Army Biology Institute, Rio de Janeiro provided by Silva et al. (2013, unpublished results). A human S. aureus standard strain (ATCC43300) was used as control.
Phenotypic detection of methicillin-resistance
Methicillin-resistance detection in Staphylococcus strains was performed according to the recommendations of the Clinical Laboratory Standard Institute (CLSI, 2008; CLSI, 2012). The agar-screening test was performed on Mueller-Hinton agar (Himedia) supplemented with 4% (w/v) NaCl containing oxacillin at 6 μg/mL where bacterial growth 24 h at 35 °C indicates oxacillin- resistance. Also, strains were screened by cefoxitin disk-diffusion test considering an inhibition zone diameter ≤ 21 mm as resistant. S. aureus standard strains ATCC43300 and ATCC29213 were used as control.
Detection of mecA and mecALGA251 gene by PCR
A 1.5 mL overnight culture of a single Staphylococcus colony was centrifuged (three times) and the cell pellet was suspended in 600 μL of lysis solution (200 mM TrisHCl, 25 mM EDTA, 25 mM NaCl, 1% SDS, pH8.0) at 65 °C for 30 min. The DNA was extracted with Chloroform:Isoamyl Alcohol 25:24:1 twice and precipitated by ice-cold ethanol two volumes. DNA pellet was washed with 70% ethanol and resuspended in 30 μL of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH8.0) and stored at −20 °C until used. PCR for mecA gene was initially carried out using the primers and methodology outlined by Murakami et al. (1991): 5′-AAA ATC GAT GGT AAA GGT TGG C-3′ and 5′-AGT TCT GCA GTA CCG GAT TTG C-3′. PCR screening for mecALGA251 gene was carried out using the primers and methodology outlined by Cuny et al. (2011): 5′-GCT CCT AAT GCT AAT GCA-3′ and 5′-TAG CAA TAA TGA CTA CC-3′. Amplicons were detected by 1.5% agarose gel, stained with SYBR Green (Invitrogen) and examined under UV transilluminator (UvTrans).
Design of primers based on GenBank database sequences of mecA gene from Staphylococcus genus
Firstly, a range of nucleotide sequences of Staphylococcus spp. mecA gene was obtained from the GenBank database (http://www.ncbi.nlm.nih.gov/) and aligned using ClustalW (Higgins et al., 1994) within the MEGA version 4.0 program (Tamura et al., 2007). From the resultant alignment, a representative sequence of human S. aureus mecA (HE681097) was used to design the primers using the Primer3 website (http://frodo.wi.mit.edu/) (Rozen and Skaletsky 2000). Four distinct primer pairs were designed to amplify different but overlapping segments of the whole mecA gene. Table 1 displays the primer sequences considering the four covered regions: anterior (mecAant), internal 2 (mecAint2), internal 1 (mecAint1) and posterior (mecApos). The unique primer that tested positive for both human and bovine isolates was the internal 1 (mecAint1). The alignment of its sequenced amplicon yielded a maximal level of identity with the bovine S. sciuri mecA (AY820253) on BLASTn. In a further step, a new set of three distinct primers was designed considering the following regions: anterior (mecSsciuriAnt), internal (mecSsciuriInt), and posterior (mecSsciuriPos). PCR assays were carried out using the newly designed primers plus the previous designed mecAint1 in order to cover the whole mecA gene (Table 1). Figure 1 displays the schemes of the highly conserved regions used to design mecA primers for S. aureus (1A) and S. sciuri (1B).
Table 1.
Sequences of the primer sets designed in this study for the amplification of mecA gene.
| Primer | Sequence (5′ - 3′) | PCR product | Program* |
|---|---|---|---|
| mecAantF mecAantR | CAT ATC GTG AGC AAT GAA CTG A GGC CAA TTC CAC ATT GTT TC 641 bp 1 | ||
| mecAint2F mecAint2R | TCC AGG AAT GCA GAA AGA CC TCA CCT GTT TGA GGG TGG A 669 bp | 1 | |
| mecAint1F mecAint1R | GGC TAT CGT GTC ACA ATC GTT TCA CCT TGT CCG TAA CCT GA | 689 bp | 1 |
| mecAposF mecAposR | GCA CTC GAA TTA GGC AGT AAG AA AGC AAC CAT CGT TAC GGA TT | 631 bp | 1 |
| mecSciuriAntF mecSsciuriAntR | AGC CAT CGT GGT TGT AAT CAT CAA TGC CAA CTT CAT GTG CT 500 bp 2 | ||
| mecSsciuriIntF mecSsciuriIntR | CAG GCA TGC AGA AAA ATC AA TTG AGT CGA ACC AGG TGA TG 809 bp 2 | ||
| mecSsciuriPosF mecSsciuriPosR | AGG TTA TGG ACA AGG CGA AA AAA TCG TCA TAC ACT TTT CCA GA 417 bp 2 |
1. 5 min at 94 °C, 30 x (1 min at 94 °C, 1 min at 55 °C, 1 min at 72 °C), and 10 min at 72 °C, 2. 5 min at 94 °C, 30 x (1 min at 94 °C, 1 min at 50 °C, 1 min at 72 °C) and 10 min at 72 °C.
Figure 1.
Schematic diagram of mecA gene of Staphylococcus aureus (HE681097) (A) and Staphylococcus sciuri (AY820253) (B) displaying the location of designed primers.
Sequencing and sequence analysis
PCR products were purified using Exo-Sap (USB Corporation) according to the manufacturer’s recommended protocol. To assure fidelity, sequencing of both strands was performed at Helixxa Company, Brazil.
Nucleotide sequences were edited using the software Bioedit version 7.0.9.0 (Pichon et al., 2011) and compared to sequences deposited in GenBank using the Blastn algorithm (Altschul et al., 1997). The sequences were imported into the program MEGA version 4.0, and aligned in Clustal W program. The resulting multiple alignments were optimized visually. The final alignment of this work is available on request. The sequence contigs of mecA gene were assembled using the Mobyle@Pasteur program - (http://mobyle.pasteur.fr/cgi-bin/portal.py?#forms::merger) (Rice et al., 2000). The sequences were deposited in a GenBank database and compared with other sequences (Table 2). Dendogram was performed by a neighbor joining (NJ) algorithm method using p-distance model with MEGA (Tamura et al., 2007). The robustness of each branch was determined using the non-parametric bootstrap test (Felsenstein 1985) with 1000 replicates.
Table 2.
GenBank accession numbers for mecA sequences of Staphylococcus spp. isolates and respective hosts used for dendogram assembly.
| Species | Host | GenBank accession number |
|---|---|---|
| S. pseudintermedius | Dog | AM904731 |
| S. sciuri | Rodent | Y13096 |
| S. pseudintermedius | Dog | AM904732 |
| S. sciuri | Rodent | Y13095 |
| S. kloosii | Equine | AM048803 |
| S. vitulinus | Equine | AM048802 |
| S. pseudintermedius | Dog and Cat | EU929082 |
| S. pseudintermedius | Dog and Cat | EU929081 |
| S. capitis | Equine | AM048805 |
| S. kloosii | Equine | AM048804 |
| S. sciuri | Bovine | AY820253 |
| S. aureus | Human | HE681097 |
| S. aureus* | Human | KF058908 |
| CNS* | Human | KF058904 |
| CNS* | Human | KF058903 |
| S. aureus* | Bovine | KF058901 |
| S. aureus* | Bovine | KF058902 |
| S. aureus* | Bovine | KF058900 |
| S. sciuri* | Equine | KF058905 |
| S. sciuri* | Equine | KF058906 |
| S. lentus* | Equine | KF058907 |
Obtained from this study.
Results
From thirty-eight bovine S. aureus isolates, twenty were found oxacillin-resistant at the phenotypical assays, but none tested positive for mecA gene using primers set specified by Murakami et al. (1991). Also, bovine mecA-negative isolates also tested negative for mecALGA251 gene described by Cuny et al. (2011). Otherwise, the oxacillin-resistant CNS isolates of human origin tested positive for mecA gene (Table 3).
Table 3.
Distribution of phenotypical beta-lactam resistance pattern with the corresponding detection of mecA gene and homologue mecALGA251.
| Pattern/ isolates (n) | OAS | CDD | mecA gene* | mecALGA251† |
|---|---|---|---|---|
| 1(7) | R | R | − | − |
| 2(2) | R | S | − | − |
| 3(11) | S | R | − | − |
| 4(18) | S | S | − | − |
| 5 (1 CNS) | R | R | + | − |
| 6 (1 CNS) | S | R | + | − |
R - resistant; S - sensible; OAS - oxacillin agar-screening test; CDD - cefoxitin disk-diffusion test; (−) negative; (+) positive.
All 43 Staphylococcus isolates were tested for mecA gene by PCR using the primers set (mecAant, mecAint2, mecAint1, and mecApos) based on the human S.aurues mecA sequence (HE681097). These primers successfully amplified mecA gene fragments from the two CNS and the standard strain from human and for the two S. sciuri and one S. lentus from equine whereas only three bovine Staphylococcus strains were positive with only one (mecAint1) of the four primers set (Table 4).
Table 4.
Staphylococcal mecA gene detected for human, equine and bovine isolates.
| Origin | Isolates | mecA** | mecA ant | mecA int2 | mecA int1 | mecA pos | mec Ssciuri Ant | mec Ssciuri Int | mec Ssciuri Pos |
|---|---|---|---|---|---|---|---|---|---|
| Milk (Bovine) | S. aureus (1) | − | − | − | + | − | + | + | + |
| Milk (Bovine) | S. aureus (2) | − | − | − | + | − | + | + | + |
| Milk (Bovine) | S. aureus (3) | − | − | − | + | − | + | + | + |
| Hands (Human) | ECN (1) | + | + | + | + | + | − | − | − |
| Nasal (Human) | ECN (2) | + | + | + | + | + | − | − | − |
| Standard Strain (Human) | S. aureus | + | + | + | + | + | − | − | − |
| Nasal (Equine) | S. sciuri (1) | + | + | + | + | + | − | − | − |
| Nasal (Equine) | S. sciuri (2) | + | + | + | + | + | − | − | − |
| Nasal (Equine) | S. lentus | + | + | + | + | + | − | − | − |
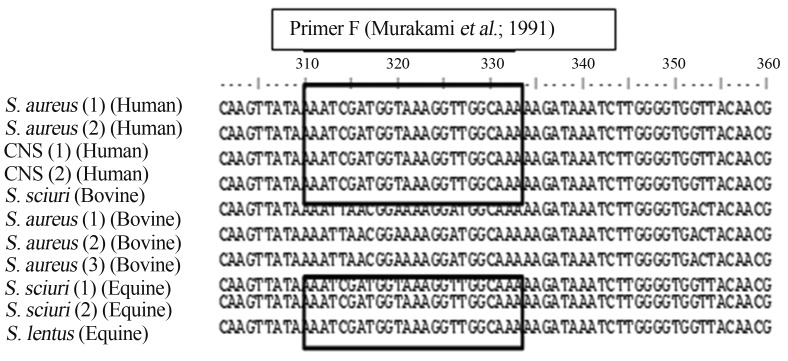
Fragments originated from the mecAInt1 primer set amplification were sequenced and aligned in order to evaluate the differences in the nucleotide sequences. It was observed punctual differences in the alignment when comparing the bovine to the human and equine strains. Also it was observed differences in the primer annealing site of the established forward primer specified by Murakami et al. (1991) (Figure 2). The comparison of the sequences from our bovine strains with others sequences available in NCBI databases provided a fully identity (100%) to the mecA gene from bovine S. sciuri (AY820253).
Figure 2.
Multiple alignments of sequences of mecA gene fragments generated by amplification using the first-step primers set mecAint1. Analyzes of the sequences segments of mecA genes from different hosts (bovine, human and equine) revealed that the ones from bovine origin present punctual differences. The highlighted box displays the primer F’s annealing site based on human S. aureus mecA gene specified by Murakami et al. (1991) detaching the punctual nucleotide differences that possibly impaired the annealing and amplification of mecA gene from our bovine strains. S. aureus (1) (Human): (HE681097); S. aureus (2) (Human): standard strain (ATCC43300) (KF058908); CNS (1) and (2): CNS isolates 1 and 2 (human origin) (KF058904, KF058903, respectivaly); S. aureus (1, 2 and 3): S. aureus isolates 1, 2 and 3 (bovine origin) (KF058901, KF058902, KF058900, respectivaly); S. sciuri (1and 2): S.sciuri isolates 1 and 2 (equine origin) (KF058905, KF058906, respectivaly) and S. lentus: S. lentus isolate (equine origin) (KF058907, respectivaly).
All eight isolates comprising two CNS and one standard strain from human, three CNS from equine, and three S. aureus bovine strain that tested positive for the mecA gene amplification carried out with at least one of the first-step designed primers set (mecAant, mecAint2, mecAint1, and mecApos) were submitted to a PCR using the primers designed at the second-step (mecSsciuriAnt, mecSsciuriInt and mecSsciuriPos) based on S. sciuri sequence of mecA gene (AY820253) (Table 1, Figure 1B). In such a different way, the result of this PCR assay showed that only the three bovine strains amplified mecA gene segments (Table 4). The resulting bovine, human and equine mecA gene fragments were sequenced and the overlapping reads were assembled in contigs.
The alignment of the mecA gene showed that the nucleotide sequences were sorted into 2 different groups, one comprising the bovine strains and the other containing human and equine strains (Figure 3). Punctual mutations in the primer annealing sites to detect mecA described by Murakami et al. (1991) explain why only the human and equine mecA gene were amplified and the fail to detect this gene from bovine isolates (Table 4, Figure 3). Moreover, all primer annealing sites for mecA gene available in the literature were evaluated and do not aligned in none of the mecA sequences obtained from the bovine isolates used in this study (Oliveira and Lencastre, 2002; Tan, 2002; Zhang et al., 2004; Baddour and Abuelkeir, 2007). Also the in silico analysis of the primer annealing sites of the newly designed primers set used in this study confirm PCR results shown in Table 4.
Figure 3.
Multiple alignments of contigs of mecA gene sequences generated by amplification using both the first (mecAant, mecAint1, mecAint2, mecApos) and second-step (mecSsciuriAnt, mecSsciuriInt, mecSsciuriPos) primers set. Analyzes of the mecA genes sequences of different hosts (bovine, human and equine) revealed that the ones from bovine origin present punctual differences. The highlighted boxes display the primer annealing sites(Primer mecAint2 F, Primer mecSsciuriInt F and Primer R from Murakami et al. (1991)). The in silico analysis of these regions detaches punctual nucleotide differences that can lead to the mecA gene detection impairment in bovine strains. S. aureus (1) (Human): (HE681097); S. aureus (2) (Human): standard strain (ATCC43300) (KF058908); CNS (1) and (2): CNS isolates 1 and 2 (human origin) (KF058904, KF058903, respectivaly); S. aureus (1, 2 and 3): S. aureus isolates 1, 2 and 3 (bovine origin) (KF058901, KF058902, KF058900, respectivaly); S. sciuri (1and 2): S.sciuri isolates 1 and 2 (equine origin) (KF058905, KF058906, respectivaly) and S. lentus: S. lentus isolate (equine origin) (KF058907).
Sequences of mecA gene of Staphylococcus spp. from different hosts provided by our study and available at NCBI GenBank were used to generate a dendogram (Table 2, Figure 4). Genomic divergences between mecA genes originated two different clusters of Staphylococcus spp., one comprising dog, cat, rodent, equine and human isolates and the other just bovine isolates.
Figure 4.
Dendogram showing the genetic divergence of nucleotide sequences of bovine (dotted line) and others species (continuous line) Staphylococcal mecA gene. *Obtained from this study.
Discussion
Livestock associated Staphylococcus spp. seems to be an additional challenge for veterinary research. After a decade of dairy science research concerning antimicrobial resistance we were really puzzled with the low correlation observed between phenotypic assays for detection of beta-lactam resistance and mecA gene detection.
Once literature reported the existence of a homologue gene, mecALGA251, with unusual features in bovine milk samples that was not possibly detected by routine PCR assays (Cuny et al., 2011; García-Álvarez et al., 2011), all phenotypic oxacillin-resistant strains of this study were tested for this gene, no one was positive.
Assuming that all steps of PCR techniques were well-established and its efficiency and reliability were not in question, a critical point was left for us to investigate: the selection of the primers for mecA gene detection.
Recently, literature reports that the majority of Staphylococcus aureus isolates that are recovered from humans or cattle represent genetically distinct sets of clonal groups with evidence of host specialization among them (Herron-Olson et al., 2007; Sakwinsk et al., 2011).
This statement supports the idea that the use of human parameters in the analysis of beta-lactam resistance considering detection of mecA gene could lead to the misidentification of this resistance. This idea was supported by our results that the primer F’s annealing site based on human S. aureus mecA gene specified by Murakami et al. (1991) presented punctual nucleotide differences that possibly impaired the annealing and amplification of mecA gene from our bovine strains.
The two-step procedure for primers design took in account sequences of distinct conserved regions in the mecA gene. In the first step, the newly synthesized primers were based on the nucleotide sequences of mecA gene of S. aureus (HE681097). Those primers were successful in amplifying mecA gene segment of human and equine Staphylococcus but mostly failed in bovine strains that tested positive only for the first internal segment primers set (mecAint1). Otherwise, the second-step primers set based on sequence of S. sciuri mecA gene (AY820253) only yielded mecA gene segments for bovine strains. Analyzes of the multiple alignments of the contigs of mecA gene sequences from bovine, human and equine origins revealed that the ones from bovine origin presented punctual differences. As far as we can see, these differences in the nucleotide sequences are significant, detaching that the primer annealing sites described in literature (Murakami et al., 1991; Oliveira and Lencastre, 2002; Tan, 2002, Zhang et al., 2004, Baddour and Abuelkeir, 2007) were not able in the in silico analysis to anneal to the bovine sequence what can lead to the mecA gene detection impairment in bovine strains.
The impairment of the mecA gene detection in bovine isolates is related to the punctual but significant differences in primer annealing sites once mostly primers set are usually based on human mecA gene sequences. This impairment was not noticed for equine isolates since that the divergence of mecA gene sequences are not a matter of animal or human origin, it is a specificity of bovine samples.
Acknowledgments
This study was supported by the National Council for Scientific and Technological Development (CNPq, Rio de Janeiro, Brazil - process 472119/2011-7) and Foundation for Research Support in the State of Rio de Janeiro (FAPERJ; process E-26/110.526/2011).
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddour MM, Abuelkeir MM. Comparison of mecA Polymerase Chain Reaction with phenotypic methods for the detection of methicillin-resistant Staphylococcus aureus. Curr Microbiol. 2007;55:473–479. doi: 10.1007/s00284-007-9015-6. [DOI] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:26–31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI. Informational Supplement, M45-P. CLSI; Wayne, PA Clinical and Laboratory Standards Institute: 2008. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. [Google Scholar]
- CLSI. 20th Informational Supplement, M100-S20. CLSI; Wayne, PA Clinical and Laboratory Standards Institute: 2012. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- Coelho SMO, Reinoso E, Pereira IA, Soares LC, Demo M, Bogni C, Souza MMS. Virulence factors and antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis in Rio de Janeiro. Pesq Vet Bras. 2009;29:369–374. [Google Scholar]
- Cuny C, Layer F, Strommenger B, Witte W. Rare Occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS ONE. 2011;6(9):e24360. doi: 10.1371/journal.pone.0024360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Mol Biol and Evol. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS ONE. 2007;2(10):e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins ET, Roney C, Crowe E, Hymes C. Ideal vs. ought predilections for approach and avoidance: Distinct self-regulatory systems. J Pers Soc Psychol. 1994;66:276–286. doi: 10.1037//0022-3514.66.2.276. [DOI] [PubMed] [Google Scholar]
- Mendonça ECL, Marques VF, Melo DA, Alencar TA, Coelho IS, Coelho SMO, Souza MMS. Caracterização fenogenotípica da resistência antimicrobiana em Staphylococcus spp. isolados de mastite bovina. Pesq Vet Bras. 2012;32(9):859–864. [Google Scholar]
- Moon JS, Lee AR, Kang HM, Lee ES, Kim MN, Paik YH, Park YH, Joo YS, Koo HS. Phenotypic and genetic antibiogram of methicillin-resistant Staphylococci isolated from bovine mastitis in Korea. J Dairy Sci. 2007;90:1176–1185. doi: 10.3168/jds.S0022-0302(07)71604-1. [DOI] [PubMed] [Google Scholar]
- Murakami KW, Minamide K, Wada W, Nakamura E, Teraoka H, Watanbe S. Identification of methicillin resistant strains of staphylococci by Polymerase Chain Reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Néron B, Ménager H, Maufrais C, Joly N, Letort MS, Carrere S, Tuffery P, Letondal C. Mobyle a new full web bioinformatics framework. Bioinformatics Advance. 2009;25:3005–3011. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira DC, Lencastre H. Multiplex PCR Strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. TIG. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the www for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sakoulas G, Gold HS, Venkataraman L, Degirolami PC, Eliopoulos GM, Qian Q. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J Clin Microbiol. 2001;39:3946–3951. doi: 10.1128/JCM.39.11.3946-3951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakwinsk O, Morisset D, Madec JY, Waldvogel A, Moreillon P, Haenni M. Link between genotype and antimicrobial resistance in bovine mastitis-related Staphylococcus aureus strains, determined by comparing swiss and french isolates from the Rhône Valley. Appl Environ Microbiol. 2011;77:3428–3432. doi: 10.1128/AEM.02468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Mizrachi I, Ostell J, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Yaschenko E, Ye J. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37(Database issue):D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore AC, Deasy EC, Slickers P, Brennan GO, Connell B, Monecke S, Ehricht R, Coleman DC. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:3765–3773. doi: 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva ACC, Melo DA, Medeiros PTC, Carvalho BO, Coelho IS, Coelho SMO, Souza MMS. Evaluation of mec A gene sequences and detection of homologue pbp D correlated to phenotypic oxacilin resistance in Staphylococcus sciuri group. Braz J Microbiol. 2013 Submitted. [Google Scholar]
- Soares LC, Pereira IA, Pribul BR, Oliva MS, Coelho SMO, Souza MMS. Antimicrobial resistance and detection of mecA and blaZ genes in coagulase-negative Staphylococcus isolated from bovine mastitis. Pesq Vet Bras. 2012;32:692–696. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tan TYA. Comparison of PCR detection of mecA with two standard methods of oxacillin disk susceptibility testing for coagulase-negative staphylococci. J Med Microbiol. 2002;51:83–85. doi: 10.1099/0022-1317-51-1-83. [DOI] [PubMed] [Google Scholar]
- Zhang K, Sparling J, Chow BL, Elsayed S, Hussain Z, Church DL, Gregson DB, Louie T, Conly JM. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from Coagulase-Negative Staphylococci. J Clin Microbiol. 2004;42:4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]