Abstract
Edwarsiella tarda is a zoonotic bacterium that can be isolated from humans, animals and the environment. Although E. tarda is primarily considered a fish pathogen, it is the only species of its genus considered to be pathogenic for humans as well. A survey of zoonotic intestinal bacteria in fresh feces from South American sea lions (SASL) Otaria flavescens, reported E. tarda as the most frequently isolated species. In this study, we used HEp-2 cells to establish in vitro the adherence and invasive ability of 17 E. tarda strains isolated from SASL fecal material. All the strains were able to adhere and invade HEp-2 cells with adhesion and invasion percentages ranging from 56 to 100% and 21 to 74%, respectively. Despite the expression of these pathogenic factors, further investigation is needed to determine whether this bacterium could play a role as primary pathogen for this and other species of pinnipeds.
Keywords: Edwarsiella tarda, adherence, invasion, Otaria flavescens
Introduction
The South American sea lion [SASL] Otaria flavescens is a marine mammal found along the coasts of South America and the Falkland Islands. On the Pacific side, breeding colonies are found south of Zorritos (03°40’ S), further down to Tierra del Fuego and, on the Atlantic side, they are found in Isla de Los Estados (54°45’ S) in Argentina, and further up to Recife dos Tôrres (29°21’ S) in Brazil. The type of sea habitat for SASL seems to be associated to the distribution and abundance of food. In general the species tends to feed in shallow waters in coastal areas, and around the continental slope, where it finds the greatest concentrations of pelagic fish and/or marine invertebrates. There are also records of the species entering river mouths (basin of the Uruguay River, basin of the Santa Lucía River, Arroyo Pando and Arroyo Solís in Uruguay) and Atlantic coastal lagunes. The sporadic presence of SASL individuals in the Valdivia River is a phenomenon observed through decades and scientifically documented since 1976 (González et al., 2011). In the last decade, a colony of about 30 to 40 animals was established in a coastal area of the urban course of the Valdivia River (39°47’Southern latitude, 73°15’ Western latitude), with the river being their feeding ground; whilst they use the sandy beach by the river, the lawn and the pavement of the street in the surrounding areas as resting ground. This prompted us to look for the presence of zoonotic intestinal bacteria in fresh sea lions feces placed on the pavement. During the course of this investigation, Campylobacter insulaenigrae (González et al., 2011) and several species of the Family Enterobacteriaceae were isolated (González et al., 2010), with Edwardsiella tarda being the most frequently found (73%) (González et al., 2010). This is a zoonotic bacterium that has been isolated from humans, animals, and the environment and it is particularly considered a fish pathogen, being the only species of its genus that has been reported to be pathogenic to humans (Leung et al., 2012).
In human beings it has been associated to infectious diarrhea, sepsis, and generalized infections predominantly in immunocompromised individuals (Marques et al., 1984; Janda & Abbott, 1993; Nucci et al., 2002; Leung et al., 2012).
Coles et al. (1978) recognize that marine animals are an important reservoir of this bacterium, with E. tarda being part of their microbiota. However, it could also be the etiological agent of several infectious diseases of freshwater and marine fish, including septicemia with extensive skin lesions, affecting internal organs such as liver, kidney, spleen and muscle, being pacu (Myleus micans) one of the affected fishes in their natural fresh water habitat. On the other hand, it could also be an experimental pathogen for tilapias (Oreochromis spp.) and common carp (Cyprinus carpio) (Lima et al., 2008). Recently, it was associated to an outbreak of bacterial septicemia with high mortality in turbots (Scophthalmus maximus) from a mariculture farm (Xiao et al., 2009).
The ability of E. tarda to invade fish epithelial cells and tissues has been reported as virulence factors (Marques et al., 1984; Janda & Abbott 1993; Ling et al., 2001). Despite these reports and the description of genes required to cause infection in fish and the role of type III secretion system in E. tarda virulence (Tan et al., 2005), little is known about the pathogenic characteristics of E. tarda strains isolated from different sources, especially from marine mammals.
The aim of this study was to establish the ability of 17 E. tarda strains isolated from SASL fecal material to adhere to and invade HEp-2 cells in vitro.
Material and Methods
The E. tarda strains were isolated on MacConkey and XLD agars (Oxoid) and identified according to their biochemical characteristics with the assesment of the Vitek AutoMicrobic System (bioMérieux). All the adherence and invasion tests were carried out twice in duplicate.
Adhesiveness
The adhesive capacity was determined infecting HEp-2 cells with the strains under study following the protocol from Skaletsky et al. (1984). In brief, HEp-2 cells were cultured overnight on coverslips (20 × 8 mm), in Leighton tubes containing minimum essential medium with 10% fetal calf serum (MEM-10%), at 37 °C under 5% CO2 atmosphere. After being washed three times with phosphate-buffered saline (PBS), the medium was replaced with 1 mL E. tarda suspension (6 × 108 colony forming units) in MEM-10% and incubated for 30 min at 37 °C (infection period). The tubes were then washed with phosphate buffered saline six times, and 1 mL of MEM with 10% fetal calf serum was added to each tube. The tubes were then incubated for 3 h at 37 °C (multiplication period). After this period, cover slips were washed three times with PBS, and the cells were fixed with methanol and stained with May-Grünwald stain. The stained cover slips were washed with water, then removed from the Leighton tubes, dried, mounted on glass slides, and examined under a light microscope (x400 and x1,000). An adherent Escherichia coli isolate was included as positive control, whereas un-inoculated cell lines were used as negative controls. Adhesion results were expressed as percentage of HEp-2 cells (adhesion percentage) showing adhering bacteria and the number of bacteria (± SD) adhered to cells was determined. The adhesion percentage expressed the number of invaded cells/total cells examined × 100 and it was determined by counting at least 200 cells. Strains were considered as adherent if at least 20% of HEp-2 cells showed one or more adhered bacteria (Fernández et al., 2010). Adherence was also demonstrated by scanning electronic microscopy.
Invasiveness
The acridine orange-crystal violet stain (Fernández et al., 1997) was used to assess invasiveness. HEp-2 cells were cultured overnight on coverslips (20 × 8 mm), in Leighton tubes containing MEM-10%, at 37 °C in 5% CO2 atmosphere. After washing three times with PBS, the medium was replaced with 1 mL E. tarda suspension (6 × 108 colony forming units) in MEM-10%. Cells were incubated for 2 h at 37 °C and 5% CO2 (infection period), washed 10 times in PBS and reincubated for 3 h with 1 mL MEM-10% (multiplication period). Then, coverslips were washed three times with PBS and, without fixing, stained with 0.01% acridine orange in Gey’s solution for 45 s, rinsed with Hanks balanced salt solution and counterstained with 0.05% crystal violet in 0.15 N NaCl for 45 s, mounted on slides and sealed with colorless nail polish and then examined under epi-fluorescence microscopy at 400× magnification for screening and at 1000× magnification for quantitative evaluation. The invasion percentage (number of invaded cells/total cells examined × 100) was determined by counting at least 200 cells. The average number (± SD) of invading bacteria was estimated in a minimum of 35 invaded HEp-2 cells. Invasion was confirmed by using transmission electron microscopy as standard control.
Results and Discussion
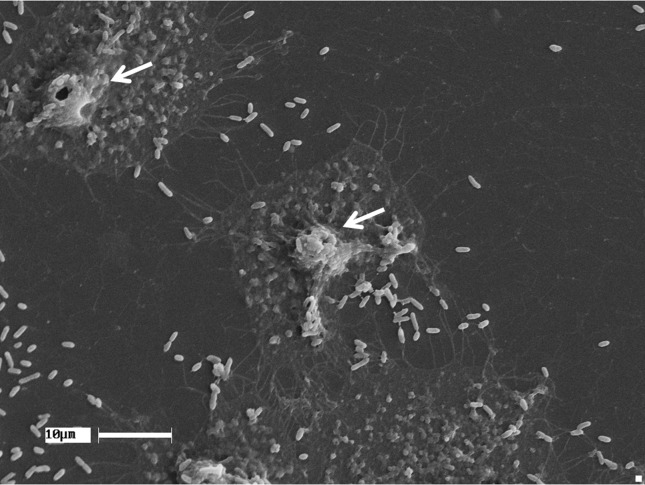
All the strains were adherent, with adhesion percentages ranging from 56 to 100% and the number of adhered bacteria per cell ± SD ranging from 2.88 ± 2.46 to 20.5 ± 8.05 (Table 1). The adhesion pattern observed using light microscopy was similar to the enteroaggregative pattern described by Nucci et al. (2002) in E. tarda strains (data not shown). Scanning microscopy confirmed the actual adhesion of E. tarda to HEp-2 cells in vitro showing rounded ends and straight bacilli adhering to HEp-2 cells, creating an arrangement that resembled a microcolony (Figure 1).
Table 1.
Adherence to and invasion of HEp-2 cells by Edwarsiella tarda studied with light microscopy and acridine orange-crystal violet staining and controlled by scanning electronic microscopy (SEM) and transmission electronic microscopy (TEM), respectively.
| Strains | Adhesion percentage (%) | N° of adhered bacteria/cell ± SD | SEM adherence control | Invasion percentage (%) | N° of invading bacteria/cell ± SD | TEM invasion control |
|---|---|---|---|---|---|---|
| 1 | 70 | 2.88 ± 2.46 | + | 74 | 1.68 ± 1.30 | + |
| 3 | 56 | 3.76 ± 1.76 | + | 53 | 1.32 ± 1.07 | + |
| 4 | 86 | 6.6 ± 2.59 | ND | 58 | 2.92 ± 1.00 | ND |
| 7 | 92 | 5.38 ± 2.89 | ND | 47 | 2.76 ± 0.93 | ND |
| 8 | 78 | 5.26 ± 3.76 | ND | 54 | 2.24 ± 1.22 | ND |
| 9 | 93 | 12.1 ± 7.66 | ND | 37 | 3.62 ± 1.22 | ND |
| 10 | 98 | 17.22 ± 6.36 | + | 46 | 5.32 ± 1.71 | + |
| 11 | 96 | 9.4 ± 6.48 | + | 42 | 3.28 ± 1.75 | + |
| 15 | 94 | 8.22 ± 6.19 | ND | 48 | 3.94 ± 1.07 | ND |
| 17 | 100 | 9.52 ± 4.01 | + | 39 | 4.12 ± 1.72 | + |
| 20 | 99 | 13.66 ± 7.01 | ND | 30 | 4.42 ± 0.70 | ND |
| 22A | 90 | 5.96 ± 3.12 | ND | 21 | 2.30 ± 0.81 | ND |
| 22B | 93 | 14.62 ± 8.30 | + | 39 | 4.74 ± 1.19 | + |
| 23 | 98 | 20.5 ± 8.05 | + | 50 | 5.84 ± 1.33 | + |
| 25 | 96 | 15.5 ± 9.01 | + | 61 | 4.56 ± 1.38 | + |
| 26 | 94 | 8.08 ± 4.26 | ND | 59 | 2.12 ± 1.08 | ND |
| 27 | 90 | 7.84 ± 4.46 | + | 51 | 2.88 ± 1.11 | + |
SD = standard deviation; SEM = scanning electron microscopy; ND = not done.
TEM = transmission electron microscopy.
Figure 1.
Scanning electronic microphotography showing E. tarda adhered to HEp-2 cell and arrangements resembling microcolonies (arrows).
E. tarda infects a broad range of freshwater and marine life including fish, amphibians, reptiles, birds, and mammals; while it is also the only species of the genus Edwarsiella that is pathogenic to humans (Leung et al., 2012).
In order to cause an infection, E. tarda may attach to the host’s epithelial cells. Fish and human cells models have been used to assess the adherence of E. tarda. Ling et al. (2001), demonstrated through a fish model that the main sites of attachment of E. tarda were the gastrointestinal tract epithelia, gills, and body surface. On the other hand, the adherence of an E. tarda strain isolated from a mariculture farm to an epithelioma papulosum cyprini cell line was determined by Xiao et al. (2009).
The adherence of E. tarda to human epithelial cells was previously described by Nucci et al. (2002). These authors found that all of the strains they studied were able to adhere to HeLa cells, independently of their human or fish origin. Moreover, they described for all their isolates an adherence pattern similar to enteroaggregative adherence, which had been previously described in E. coli by Scaletsky et al. (1984). This kind of adhesive pattern could probably be responsible for the microcolony-like arrangement that was observed on the HEp-2 cells through scanning electron microscopy. Similar arrangements have been described in adhesive Arcobacter butzleri strains (Fernández et al., 2010). Since enteroaggregative E. coli forms biofilms on the intestinal mucosa (Sheikh et al., 2001), it may be possible that the enteroaggregative pattern of adhesion observed in the E. tarda strains could be related to the capacity of this bacterium to form biofilm as described by He et al. (2011). After the attaching process, E. tarda can invade or can be internalized into the cells (Wang et al., 2010) assisted by invasion-like proteins and/or the participation of a type III secretion system recently described (Tan et al., 2005).
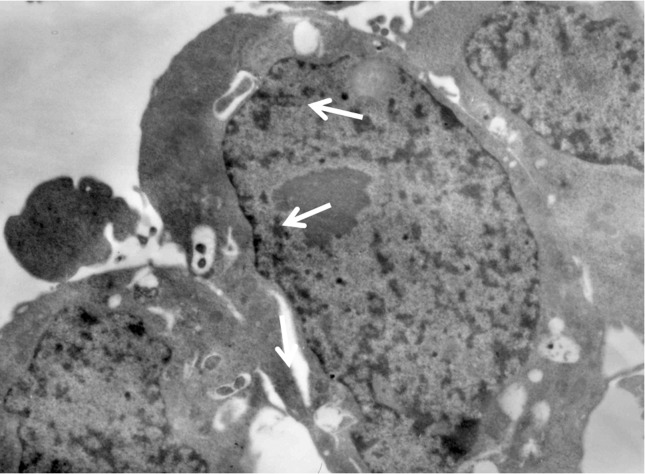
All the strains were able to invade HEp-2 cell with invasion percentages ranging from 21 to 74% while the number of internalized bacteria per cell ± SD ranged from 1.32 ± 1.07 (Table 1). Internalized bacteria were seen inside vacuoles through epi-fluorescence microscopy, this was later corroborated by using transmission electron microscopy (Figure 2).
Figure 2.
Transmission electron microphotography of HEp-2 cells showing E. tarda enclosed within endocytic vacuoles (arrows).
The in vitro invasive property of E. tarda was first described by Marques et al. (1984) in human strains and by Janda & Abbott (1993) in clinical, fish and environmental strains. However, the latter authors also found non-invasive strains. Later, Nucci et al. (2002) reported that invasion was a feature common to all isolates of E. tarda analyzed, independently of their source. Similarly to the findings reported by Marques et al. (1984) and Nucci et al. (2002), all our strains were able to invade HEp-2 cells with invasion percentages ranging from 21 to 74%. Examination by means of transmission electron microscopy allowed us to confirm the intracellular presence of E. tarda, with the bacteria being enclosed within endocytic vacuoles (Figure 2). Similar observations were previously reported by Marques et al. (1984) and Janda & Abbott (1993).
E. tarda seems to be a frequent inhabitant of the intestinal tract of the SASL (González et al., 2010). As shown in this study, this bacteria species can express adherence and invasion in epithelial cells, properties that are considered as pathogenic factors. However, the role played by this bacterium as primary pathogen in this and in other species of pinnipeds is unknown. Several infectious diseases have been reported in different species of sea lions and other pinnipeds, but they were all secondary processes associated to previous predisposing clinical conditions like traumas, starvation, dystocia, neoplasia or primary infectious diseases (Coles et al., 1978; Stroud and Roffe, 1979). These findings suggest that E. tarda could be an opportunistic invader in sick or injured marine mammals. Further studies are needed in order to clarify the real pathogenic role that E. tarda could play in these animals.
Acknowledgments
The authors would like to thank Dr. Hans G. Richter (Department of Anatomy, Histology and Pathology, Universidad Austral de Chile) for his advice and critical review. This study was financially supported by Grant DID-UACh SE-01-99.
References
- Coles BM, Stroud RK, Sheggeby S. Isolation of Edwardsiella tarda from three sea mammals. J Wildlife Dis. 1978;14:339–341. doi: 10.7589/0090-3558-14.3.339. [DOI] [PubMed] [Google Scholar]
- Fernández H, Eller G, Freymuller E, Vivanco T. Detection of Campylobacter jejuni invasion of HEp-2 cells by acridine orange-crystal violet staining. Mem Inst Oswaldo Cruz. 1997;92:509–511. doi: 10.1590/s0074-02761997000400012. [DOI] [PubMed] [Google Scholar]
- Fernández H, Flores S, Inzunza F. Arcobacter butzleri strains Isolated from different sources display adhesive capacity to epithelial cells in vitro. Acta Scient Vet. 2010;38:287–291. [Google Scholar]
- González M, Latif F, Fernández F, Villanueva MP, Ulloa J, Fernández H. Species of the family Enterobacteriaceae in feces of South American sea lion Otaria flavescens settled in the Valdivia River. RBMO. 2010;45:331–334. [Google Scholar]
- González M, Villanueva MP, Debruyne L, Vandamme P, Fernández H. Campylobacter insulaenigrae: first isolation report from South American sea lion (Otaria flavescens, (Shaw, 1800) Braz J Microbiol. 2011;42:261–265. doi: 10.1590/S1517-83822011000100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Xu T, Han Y, Shi X, Zhang XH. Phenotypic diversity of Edwardsiella tarda isolated from different origins. Lett Appl Microbiol. 2011;53:294–299. doi: 10.1111/j.1472-765X.2011.03103.x. [DOI] [PubMed] [Google Scholar]
- Janda JM, Abbott SL. Infections associated with the genus Edwardsiella: the role of Edwardsiella tarda in human. Disease Clin Infect Dis. 1993;17:742–748. doi: 10.1093/clinids/17.4.742. [DOI] [PubMed] [Google Scholar]
- Leung KY, Siame BA, Tenkink BJ, Noort RJ, Mok YK. Edwardsiella tarda - Virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. 2012;14:26–34. doi: 10.1016/j.micinf.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Lima LC, Fernandes AA, Costa AAP, Velasco FO, Leite RC, Hackett JL. Isolation and characterizaton of Edwardsiella tarda from pacu Myleus micans. Arq Bras Med Vet Zootec. 2008;60:275–277. [Google Scholar]
- Ling SH, Wang XH, Lim TM, Leung KY. Green fluorescent change proteintagged for: protein-tagged Edwardsiella tarda reveals portal of entry in fish. FEMS Microbiol Lett. 2001;194:239–243. doi: 10.1111/j.1574-6968.2001.tb09476.x. [DOI] [PubMed] [Google Scholar]
- Marques LRMM, Toledo RF, Silva NP, Magalhães M, Trabulsi LR. Invasion of HeLa Cells by Edwardsiella tarda. Curr Microbiol. 1984;10:129–132. [Google Scholar]
- Nucci C, da Silveira WD, da Silva Correa S, Nakazatoa G, Bando SY, Ribeiro MA, Pestana de Castro AF. Microbiological comparative study of isolates of Edwardsiella tarda isolated in different countries from fish and humans. Vet Microbiol. 2002;89:29–39. doi: 10.1016/s0378-1135(02)00151-7. [DOI] [PubMed] [Google Scholar]
- Scaletsky ICA, Silva MLM, Trabulsi LR. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45:534–536. doi: 10.1128/iai.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J, Hicks S, Dall’Agnol M, Phillips AD, Nataro JP. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol Microbiol. 2001;41:983–997. doi: 10.1046/j.1365-2958.2001.02512.x. [DOI] [PubMed] [Google Scholar]
- Stroud RK, Roffe TJ. Causes of death in marine mammals stranded along the Oregon coast. J Wildlife Dis. 1979;15:91–97. doi: 10.7589/0090-3558-15.1.91. [DOI] [PubMed] [Google Scholar]
- Tan YP, Zheng J, Tung SL, Rosenshine I, Leung KY. Role of type III secretion in Edwardsiella tarda virulence. Microbiol. 2005;151:2301–2313. doi: 10.1099/mic.0.28005-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Q, Xiao J, Liu Q, Wu H, Zhang Y. Hemolysin EthA in Edwardsiella tarda is essential for fish invasion in vivo and in vitro and regulated by two-component system EsrA-EsrB and nucleoid protein HhaEt. Fish Shellfish Immunol. 2010;29:1082–1091. doi: 10.1016/j.fsi.2010.08.025. [DOI] [PubMed] [Google Scholar]
- Xiao J, Qin Liu Q, Wang X, Liu H, Zhang Y. Isolation and identification of fish pathogen Edwardsiella tarda from mariculture in China. Aquacult Res. 2009;40:13–17. [Google Scholar]