Abstract
In response to demand from industry for microorganisms with auspicious biotechnological potential, a worldwide interest has developed in bacteria and fungi isolation. Microorganisms of interesting metabolic properties include non-pathogenic bacteria of the genus Clostridium, particularly C. acetobutylicum, C. butyricum and C. pasteurianum. A well-known property of C. butyricum is their ability to produce butyric acid, as well as effectively convert glycerol to 1,3-propanediol (38.2 g/L). A conversion rate of 0.66 mol 1,3-propanediol/mol of glycerol has been obtained. Results of the studies described in the present paper broaden our knowledge of characteristic features of C. butyricum specific isolates in terms of their phylogenetic affiliation, fermentation capacity and antibacterial properties.
Keywords: 1, 3-propanediol; Clostridium spp.; C. butyricum; glycerol; metabolic activity
Introduction
Microbiology and industrial biotechnology extensively utilize the immense potential of microorganisms. An important role is ascribed to bacteria of the genus Clostridium as they are microorganisms with numerous applications in several branches of industry (Zhang et al., 2009; Wang et al., 2011; Wilknes et al., 2011; Kaur et al., 2012; Metsoviti et al., 2012). The genus Clostridium is one of the largest in the Kingdom of Procaryota. These are anaerobic heterogenic bacteria, typically Gram-positive. Their distinctive feature is a cylindrical shape. Cells of bacteria of the genus Clostridium are ciliated, mobile and capable of forming endospores (Bahl and Dürre, 2001). For most species, the optimum growth temperature falls within the 30–40 °C range, while the optimum pH is contained in the 6.5–7.5 range. Guanine and cytosine contents in DNA amount to 24–54% mol. Commonly found in nature, the main habitats of Clostridium genus bacteria are soil, river sludge, active sludge, animal excrements, etc. (Schlegel, 1993; Bahl and Dürre, 2001).
These bacteria are characterized by intensive fermentation metabolism. They may use numerous organic compounds as sources of carbon and nitrogen. Products of their metabolism include CO2, H2, as well as organic compounds (butyric, lactic, acetic and succinic acids) and solvents (butanol, acetone, isopropanol) (Ezji et al., 2007; Ren et al., 2007; Skonieczny and Yargeau, 2009; Song et al., 2011; Wang et al., 2011). Bacteria of the genus Clostridium, e.g., C. butyricum, C. pasteurianum, C. diols, C. butylicum and C. perfingens also participate in biotransformation processes (Colin et al., 2000; Hao et al., 2008). The above mentioned bacterial strains are capable of effectively converting glycerol to 1,3-propanediol (Kubiak et al., 2012). Another species of great value for industry is the C. thermocellum strain, which is an anaerobic thermophile capable of converting waste cellulose into ethanol (Colin et al., 2000; Demain et al., 2005; Ezeji et al., 2007; Ren et al., 2007; Leja et al., 2011; Song et al., 2011; Kubiak et al., 2012). By fermenting pectins, pectinolytic species of the genus Clostridium (e.g. C. pectinovorum) loosen plants’ tissue structure and facilitate a rapid separation of cellulose fibers, which appears particularly attractive in terms of biomass conversion(Perry, 2008). By fermenting sugars (glucose, sucrose, lactose), C. acetobutylicum and C. butyricum are capable of producing hydrogen (Oh et al., 2009; Beckers et al., 2010). An increasing number of reports on the probiotic potential of C. butyricum have been published (Takahashi et al., 2000, 2004; Zhang et al., 2002; Seki et al., 2003; Araki et al., 2004; Shimbo et al., 2005). An interesting example is the C. butyricum MIYAIRI 588 strain, which exhibits probiotic properties and is recommended as an additive to animal feeds (Shimbo et al., 2005). Bacteria of the C. butyricum and C. pectinovorum species have also found applications in ecology, as they play a considerable role in soil mineralization processes and conversion of organic matter.
Besides a sizeable representation of the genus Clostridium characterized by significant positive potential for industrial application, there also exist several species exhibiting adverse effects on foodstuffs, as well as on human and animal health. Mesophilic proteolytic species such as C. putrefaciens frequently cause food spoilage. C. botulinum and C. tetani species are among the most dangerous bacteria, and their pathogenic character manifests itself in their capacity to produce very strong exotoxins. On the other hand, the botulin toxin produced by C. botulinum is used in medicine (to alleviate neurological symptoms) and in cosmetology, as an agent effectively reducing mimic wrinkles (Schlegel, 1993; Ting and Freiman, 2004).
The primary application of C. butyricum bacteria is the production of butyric acid. The aim of the present study was to indicate other potential areas of industrial application.
Materials and Methods
Microorganisms
The examined microorganisms (original environmental isolates) came from the Department of Biotechnology and Food Microbiology (Poznan University of Life Sciences, Poland).
Microbiological media
The Reincorfed Clostridial Medium RCM (BIOCORP, Poland) was used as the proliferation medium for the bacteria of the genus Clostridium.
The production medium (Rich Medium) was prepared according to Himmi et al. (1999) and supplemented with crude glycerol (Wratislawia, Poland) at a concentration of 70 g/L. Glycerol feedstock had the following composition (in w/w): 86% glycerol; 8–10% water; 5–6% NaCl; free fatty acids, and < 0.01% methanol.
Bacteria identification
Complete DNA of the analyzed microorganisms was isolated using a Genomic Mini AX Bacteria Kit (A&A Biotechnology, Gdansk, Poland), following incubation with lysozyme (Sigma-Aldrich, Poland) at a concentration of 50 mg/mL for 1 h at 37 °C. For amplification of the small rRNA, subunit primers SDBact0008aS20 and SUniv1492bA21 were used (Suau et al., 1999). PCR products were purified with the use of Clean-up Kit (A&A Biotechnology, Poland), and next sequenced (Genomed, Poland). Obtained sequences of 16S rRNA of the analyzed strains were compared with the sequences available at the GenBank using the BLAST program.
Phylogenetic analysis
A phylogenetic analysis of newly discovered strains makes it possible to determine the species affiliation, as well as assess affinity of the new strains to those already described in literature.
16S rRNA sequences (obtained at the identification stage) of DSP1, DSP5, DO14 and DO16 C. butyricum strains were compared with the Clostridium strains available at the GenBank. Clustal program implemented in BioEdit (ver. 7.0.9) was used to fit the DNA sequences (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The phylogenetic tree was constructed in MEGA 4.0 program (Tamura et al., 2007). The neighbor joining method and the p-distance model were used.
Bioreactor culture
The first stage of culture consisted in proliferation of bacteria in the Hungate test tube in anaerobe culture chamber (Whitley MG500 by Scientific). After the adopted incubation time (24 h), the preinoculum was transferred by means of a sterile syringe to a bottle (Duran®) integrated with a 30 L bioreactor (Sartorius Stedim Ltd., Germany). The bottle was placed in a water bath (32 °C) and incubated for 24 h to proliferate bacterial biomass. After incubation, the contents of the bottle was pumped to a bioreactor with a working capacity of 10 L (total capacity of 30 L) with the use of a peristaltic pump. Incubation was repeated at 32 °C for 24 h. Then 10 L proliferated bacterial biomass was used to inoculate 90 L production medium. Prior to inoculation the medium was sterilized for 30 min. at a temperature of 121 °C (internal sterilization) in a bioreactor (New Brunswick Scientific, USA). Fermentation was run for 34 h at 36 °C and constant agitation rate of 60 rpm. The pH value of 7.0 was maintained by the addition of 20% NaOH.
Biochemical properties
Metabolic potential of the analyzed microorganisms was assessed using biochemical ANAEROtest 23® (Pliva-Lachema Diagnostica, Czech Republic). The ANAEROtest 23® kit is typically used for routine identification of anaerobic bacteria found in clinical material and foodstuffs. The kit makes it possible to determine 23 characteristic features of a given microorganism. For identification of a strain, assays were placed in wells on microtitration plates, at three rows with eight wells. Biochemical traits were determined based on chemical reactions occurring in the specifically composed growth media (found in plate wells). Macroscopic readings of test results included observations of growth, observations of changes in medium color occurring as a result of microbial metabolism, activation of color reaction by means of introducing a reagent reacting with the metabolite produced by a given microorganism or gas release.
Comparison of isolated C. butyricum strains with indicator microorganisms
In order to identify antagonism of the tested C. butyricum strains in relation to indicator microorganisms, analyses were conducted, including preparation of culture media of C. butyricum DSP1 and C. butyricum DO14 strains, separation of culture media into fractions (the supernatant and precipitate), preparation of indicator microorganisms, as well as analyses of activity of the obtained culture medium and the supernatant by the well method.
Preparation of culture liquid media
Antibacterial activity was determined using 24 h cultures (on RCM) of C. butyricum DSP1 and C. butyricum DO14. Cultures were run under strictly anaerobic conditions at a temperature of 32 °C.
Separation of the culture liquid media into fractions
In order to obtain supernatant (S) cultures, the analyzed strains were centrifuged (5000 g; 10 min.). Analyses of the supernatant aimed to determine the activity of bacterial exocellular metabolites.
Preparation of indicator microorganisms
Indicator microorganisms (Pseudomonas aeruginosa, Listeria monocytogenes, Salmonella enteritidis, Staphylococcus aureus, Bacillus subtilis, Escherichia coli) were transferred to test tubes containing 10 mL broth medium with an addition of 2% glucose (to proliferate biomass). Cultures were run at 37 °C for 24 h. Next, in order to obtain a distinct confluent layer, the liquefied agar medium was inoculated with 10% (v/v) 24 h indicator culture and poured onto Petri dishes.
Liquid culture medium and supernatant fraction antibacterial activity analyses
After solidification of the broth medium inoculated with indicator microorganisms, wells were made using a cork borer. Each well was supplemented with 150 μL liquid culture medium and 150 μL supernatant fraction of the analyzed strain. Next, diameters of growth inhibition or reduction of indicator bacteria were measured. Growth inhibition of the indicator microorganism, which manifested itself in complete lightening around the place where the liquid culture/supernatant was transferred, indicated bactericidal activity of the tested strain. Bacteriostatic properties were determined by measuring the diameter of the growth inhibition zone (growth limitation of indicator strain). An increase in the density of confluent layer around the well indicated a stimulatory action of the tested culture/supernatant in relation to the applied indicator. Growth inhibition of the indicator microorganism, manifested in a complete lightening around the place where the supernatant fraction was transferred, indicated antibacterial activity of exocellular metabolites. Bacteriostatic properties or stimulatory action of the supernatant were determined in the same way as with the liquid culture medium.
Analytical methods
1,3-PD, glycerol and organic acids were assayed by high performance liquid chromatography (HPLC) on an Agilent Technologies 1200 series chromatograph (the set included automatic sample feeder G1329B, and double G1312B pump with a G1362A refractometric detector). Assays were performed on a Rezex ROA column using 0.005 N H2SO4 as the mobile phase, at a flow rate of 0.6 mL/min at 40 °C. Samples were transferred on a column at 10 μL. Quantitative and qualitative identifications were performed by the external standard method based on peak areas (measurements and computer integration using ChemStation for LC 3D systems, Agilent).
Results and Discussion
Phylogenetic analysis
One of the first stages in the examination of the new environmental isolates was to determine their species affiliation based on biochemical tests or 16S rRNA sequences. The capacity of the isolated strains of the C. butyricum species to synthesize 1,3-PD was presented in subchapter “Synthesis of 1,3-PD from crude glycerol in batch culture”, and in a study by Szymanowska-Powalowska et al. (2013). In order to present interdependencies between strains of C. butyricum DSP1, C. butyricum DSP5 and C. butyricum DO16, as well as other strains of the genus Clostridium, a phylogenetic tree recreated on the basis of the nucleotide sequence of the 16S rRNA coding gene was constructed. In case of C. butyricum DO14 strain, the sequencing quality did not make it possible to include this strain in the graphic illustration of interstrain dependencies.
In most cases, strains selected from the data base for the purpose of phylogenetic analyses are capable of using glycerol as a source of carbon (Biebl and Spröer, 2002). Some of the compared microorganisms exhibit capacity to synthesize 1,3-PD (C. butyricum VIP 3266, C. butyricum DSM 2478, C. butyricum DSM 523, C. diolis DSM 5430, C. diolis DSM 5431, C. diolis E5) (Biebl and Spröer, 2002). The phylogenetic tree presented in Figure 1 shows the relationship between the new C. butyricum isolates and the strains described in literature as auspicious to 1,3-PD producers.
Figure 1.
A phylogenetic tree constructed on the basis of the 16S rRNA nucleotide sequence showing evolutionary relationships between C. butyricum DSP1, DSP5 and DO16, and other strains from the genus Clostridium.
Biochemical properties of the genus Clostridium isolates
A major stage in the characteristics of the new strains, particularly those revealing industrial applicability potential, was an examination of their biochemical properties (such as the capacity to assimilate, ferment or decompose chemical compounds). Assimilation consists in conversion of a nutrient to compounds used as building blocks for the organism and to energy in the form of ATP. Fermentation was defined as anaerobic enzymatic conversion of organic compounds, particularly carbohydrates, to simpler products, mainly alcohols, and to energy in the form of ATP. Different types of microorganisms were characterized by specific enzymatic systems active in the processes of assimilation, fermentation, or decomposition. Identification of microorganisms’ capacity to assimilate, ferment and decompose different substrates determined their potential application in different branches of industry.
Table 1 presents data recorded as a result of biochemical tests conducted on the novel bacterial strains of C. butyricum species (DSP1, DSP5, DO14 and DO16). A total of 23 biochemical traits were investigated. All isolates exhibited capacity to ferment mono- (six and five carbon) and disaccharides, which was also reported for other stains, particularly with reference to the metabolism of glucose into butyric acid (Quilaguy-Ayure et al., 2010). The capacity of C. butyricum bacteria to produce butyric acid using simple sugars is the basic trait used in the chemical industry. Strains analyzed in this study did not exhibit capacity to metabolize hexahydroxyl alcohols, such as sorbitol or mannitol, while being capable of using glycerol as a source of carbon, as evidenced in subchapter “Synthesis of 1,3-PD from crude glycerol in batch culture,” and in the study by Szymanowska-Powalowska et al. (2013). Conversion of glycerol to 1,3-PD (the primary product of glycerol metabolism) is an essential characteristics of significant industrial potential. 1,3-PD is used as a monomer in the production of polyesters, lubricants and organic solvents. Moreover, growing demand for polyesters and polypropylenes (PTT) has made it necessary to increase production of 1,3-PD used in the synthesis of polyurethanes. Biotechnological conversion of glycerol to 1,3-PD using C. butyricum bacteria can become an attractive alternative to the chemical synthesis, which is not only costly, but also brings harmful environmental consequences (Metsoviti et al., 2012; Kaur et al., 2012; Kubiak et al., 2012; Venkataramanan et al., 2013). Biochemical tests have proved that the analyzed isolates are capable of fermenting certain trisaccharides. Rafinose may serve as an example. DSP1, DSP5, DO14 and DO16 strains of C. butyricum exhibited β-glucosidase activity, which is associated with the ability to ferment cellobiose and can be applied in the process of plant biomass conversion. The strains did not show ability to metabolize N-acetyl-βglucosamine, a polysaccharide constituting a source of carbon and nitrogen for bacteria of the genera Flavobacterium, Bacillus, and Pseudomonas, as well as fungi of the genera Mucor or Aspergillus. Similar results of biochemical tests of new C. butyricum isolates were also obtained by other authors (Petidemange et al., 1995). One of the best isolates of the investigated strains (in terms of 1,3-PD synthesis) showed a comparable biochemical characteristics to C. butyricum E5 strain. Polymers, such as starch, xylene and pectins found in plant biomass, may constitute important sources of substrates for wild bacterial strains of the genus Clostridium. These bacteria were capable of degrading enzymes or enzymatic complexes, which makes biomass available for microorganisms (C. thermocellum, C. cellulovorans, C. celluloticum)(Biebl and Spröer, 2002). Similar effects were achieved when compounds like sucrose and xylose were used as sources of carbon; this was confirmed for isolates described both in the present study and in other reports (Taguchi et al., 1995; Biebl and Spröer, 2002; Yung-Chung et al., 2010). Besides cellulose and lignin, plant biomass contains hemicellulose. Xylose, in turn, is a monosaccharide contained in hemicellulose, and accouns for 30% of its composition. Reports confirm the existence of numerous representatives bacteria of the genus Clostridium capable of synthesizing xylanases (Bahl and Dürre, 2001; Wang et al., 2011; Yung-Chung et al., 2010). Metabolic capacity associated with decomposition and fermentation of biomass is particularly important in the production of 2nd generation biofuels (ethanol from cellulose) (Maki et al., 2009; Williams et al., 2013). Sucrose and xylose are used as sources of carbon in the microbiological synthesis of hydrogen by the CGS2 C. butyricum strain (Yung-Chung et al., 2010). The use of hydrogen carries great potential for such applications, as production of nitrogen fertilizers, or crude oil processing. It can also be used in fat industry, metallurgy, and, as fuel, in the power industry (Urbaniec et al., 2012). Wang et al. (2011) investigated the effect of carbohydrate substrates on the growth of C. butyricum TK2 strain. Glucose, sucrose and lactose were selected as carbon sources. An increase in the number of microorganisms was observed in all the examined cases: 5 × 108 cfu/mL (glucose), 2.5 × 108 cfu/mL (lactose) and 2.4 × 108 cfu/mL (sucrose). Identification of bacteria’s metabolic capacity, particularly this associated with decomposition of complex compounds, may be a sign of enzymatic activity. Decomposition of starch polysaccharides, which indicates the presence of the amylolytic enzyme profile by a given microorganism, may serve here as an example. The ability to synthesize amylases by Clostridium spp. bacteria was also reported by other authors (Annous and Blaschek, 1990). Important biochemical indexes also included the ability to decompose tryptophane to indol, and to reduce nitrates (reduction of nitrates (V) to nitrates (III) or nitrogen). These are main characteristic features to determine bacteria’s affiliation to the family Enterobacteriaceae. For the new isolates of the genus Clostridium, which constitute the subject of the present study, the result for these indexes was negative. Within the scope of the present study, no capacity was found in these strains to decompose urea (to ammonia and carbon dioxide), whereas this trait was characteristic of the bacteria from the genera Klebsiella or Proteus, and had even found certain applications in the area of environment protection (ammonification) (Bahl and Dürre, 2001).
Table 1.
Biochemical properties of new Clostridium isolates in comparison to reference strains
| Biochemical properties | New C. butyricum isolates | Reference C. butyricum strains | |||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| DSP1 | DSP5 | DO14 | DO16 | IP 3044* | E5** | DSM 5431*** | |
| Indole | − | − | − | − | − | − | NA |
| Glucose | + | + | + | + | + | + | + |
| Maltose | + | + | + | + | + | + | NA |
| Fructose | + | + | + | + | + | + | NA |
| Galactose | + | + | + | + | + | + | + |
| Lactose | + | + | + | + | + | + | + |
| Sucrose | + | + | + | + | + | + | NA |
| Trehalose | + | + | + | + | NA | NA | + |
| Ramnose | − | − | + | − | + | + | NA |
| Mannose | + | + | + | − | + | + | NA |
| Rafinose | + | + | + | + | NA | NA | − |
| Cellobiose | + | + | + | + | W | W | + |
| Xylose | + | + | + | + | + | + | + |
| Arabinose | − | − | + | + | + | + | W |
| Melecitose | − | − | + | − | NA | NA | W |
| Salicin | + | + | + | + | NA | + | + |
| Sorbitol | − | − | + | − | NA | NA | W |
| Glycerol | + | + | + | + | + | + | + |
| Mannitol | − | − | + | − | + | + | + |
| Esculine | + | + | + | + | + | W | W |
| N-acetyl-β glucosamiase | − | − | − | − | NA | NA | NA |
| b-Glucosidase | + | + | − | − | NA | NA | NA |
| Urease | − | − | − | − | NA | NA | NA |
| Nitrates | − | − | − | − | NA | NA | NA |
(+) - positive result; (−) - negative result; (NA) - not analyzed; (W) - weak reaction, weak acidification.
IP 3044 - Clostridum butyricum from Institut Pasteur Collection (Paris, France) (Petidemange et al., 1995).
E5 - Clostridum butyricum (Petidemange et al., 1995).
DSM 5431*** - Clostridium butyricum (Biebl and Spröer, 2002).
Synthesis of 1,3-PD from crude glycerol in batch culture
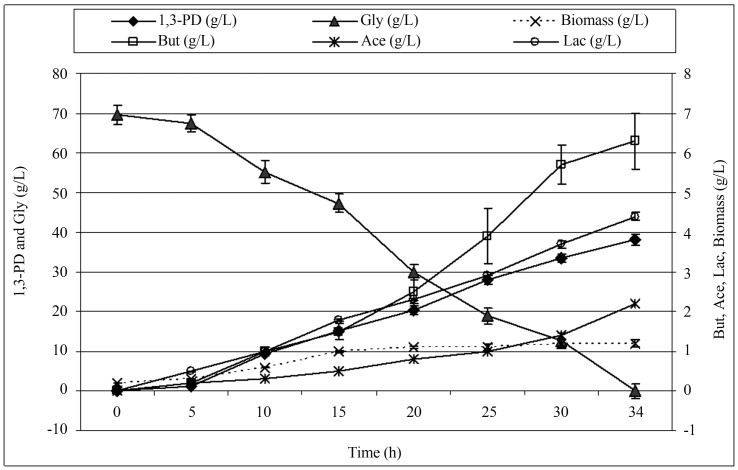
Glycerol obtained in the course of biofuel production is a crude product that is very difficult to utilize. However, it needs to be stressed that this crude product with a very limited utilization potential contains several contaminants which adversely affect microorganisms. Contaminations found in crude glycerol determine its potential for use in industry. According to sources, a 6% concentration of methanol inhibits microbial growth by as much as 90%. Ions of heavy metals retained in glycerol after the process of biodiesel production may have a negative effect on the viability of microbial cells. Moreover, high concentrations of salts contained in crude glycerol have an inhibitory effect on the growth of Clostridium species’ cells. On the other hand, in some investigations with yeast high concentration of salt stimulated production of erythritol by selected strain of Yarrowia lipolytica (Rywinska et al., 2013) or had no effect on biosynthesis of citric acid by Y. lipolytica on media containing crude glycerol (Rymowicz et al., 2010). Unsaturated free fatty acids also influence microbial metabolism and viability. Another problem, particularly in commercial production conditions, is the variability of the raw material (especially its quality), which may have adverse effect on process yields (Bahl and Dürre, 2001; Kaur et al., 2012; Kubiak et al., 2012). A potential method of the crude product utilization may be its biotechnological conversion to 1,3-PD. The biological process of glycerol conversion to diols involves microorganisms, e.g. bacteria of the species C. butyricum (Kaur et al., 2012; Kubiak et al., 2012). Figure 2 and Table 2 show analytical results obtained during the process of 1,3-PD synthesis from crude glycerol in a batch culture. Batch fermentation was run in a bioreactor of 100 L operating capacity. The initial glycerol concentration equaled 70 g/L. Conducted analyses made it possible to obtain a 1,3-PD concentration of 38.2 g/L. At the assumed glycerol concentration of 70 g/L and its complete conversion, the efficiency of 1,3-PD synthesis was 0.66 mol/mol glycerol.
Figure 2.
Kinetics of 1,3-PD synthesis from crude glycerol by C. butyricum DSP1. Fermentation conditions: initial glycerol concentration: 70 ± 1.0 g/L, temperature 36 °C; pH 7.0; culture vessel: bioreactor of 100 L. The experiment was conducted in two replications.
Table 2.
Basic parameters of the fermentation process.
| Microorganisms | Max.1 3-PD (g/L) | Utilization of glycerol (%) | Max.biomass (g/L) | Yield (mol/mol) | Productivity (g/Lh) |
|---|---|---|---|---|---|
| C. butyricum DSP1 | 38.2 ± 1.5 | 100 | 1.2 ± 0.2 | 0.66 | 1.12 |
Max.1,3-PD - maximum concentration of 1,3-propanediol; Max.biomass - maximum biomass concentration.
Literature of the subject presents a growing amount of data on the synthesis of 1,3-PD using crude glycerol, as it is only application of this type of substrate that is auspicious in terms of potential industrial use. In a batch culture, Metsoviti et al. (2012) obtained approx. 32 g/L 1,3-PD from 55 g/L crude glycerol and a very similar profile of waste metabolites in terms of their quantities and quality. In a fed-batch culture of AKR102a C. butyricum (at a temperature of 32 °C), Wilkens et al. (2011) obtained 1,3-PD concentrations of 93.7 and 76.2 g/L from for pure and crude glycerol, respectively. In their study, Chatzifragkou et al. (2011) also described the effect of crude glycerol on the parameters of 1,3-PD synthesis for the C. butyricum VPI 1718 strain. The efficiency of 1,3-PD synthesis for pure and crude glycerol was identical, and amounted to 0.67 mol/mol glycerol.
Literature of the subject, as well as the results presented in the current study, demonstrated the potential applicability of C. butyricum bacteria in the conversion of crude glycerol to synthesize 1,3-PD. Substitution of anhydrous glycerol with crude product would considerably lower the process cost, as well as facilitate development of an integrated process in which crude product from one process (glycerol obtained in the production of biodiesel) would become the primary raw material (the only source of carbon and energy) for another process, this of microbiological conversion of crude glycerol to 1,3-propanediol.
Antibacterial activity
Bacteria of the genus Clostridium are frequently associated with dangerous pathogens which pose a threat to health or even life of humans and animals. Novel strains of microorganisms exhibiting antibacterial properties constitute an interesting prospect, particularly for the feed industry. The Fodders Act of 22 July 2006 imposed a ban on the use of antibiotic growth stimulators in the feed industry. An alternative solution is the use of probiotics (besides acidifiers, enzymes, and plant extracts). According to WHO definition, probiotics are live microorganisms, which, when administered in adequate quantities, have a positive effect on the host organism. One of probiotics’ characteristic features is their operation against harmful microflora. It manifests itself, for example, in the inhibition of pathogen growth. This is the result of the microorganisms’ capacity to secrete organic acid (mainly lactic acid), hydrogen peroxide, and bacteriocins (i.e. small proteins of specific bactericidal qualities). The best known microorganisms which are attributed characteristic probiotic properties are the lactic fermentation bacteria. However, the greatest amount of information available on C. butyricum bacteria concentrates on their pathogenic qualities (Tanaka et al., 2000; Takahashi et al., 2004; Shimbo et al., 2005; Zhang et al., 2009).
A further stage in the study of characteristic qualities revealed by the new strains of C. butyricum was to investigate their operation against the most frequent human and animal pathogens. Table 3 shows the results of the study on adverse operation of C. butyricum bacteria (DSP1, DSP5, DO14, DO16) upon pathogenic microorganisms of Pseudomonas aeruginosa, Listeria monocytogenes, Salmonella enteritidis, Staphylococcus aureus, Bacillus subtilis, and Escherichia coli species.
Table 3.
Antibacterial activity of C. butyricum DSP1 and C. butyricum DO14 against selected pathogens.
| Indicator bacteria | Antibacterial activity (mm) | |||
|---|---|---|---|---|
|
|
||||
| Well method | ||||
|
|
||||
| C. butyricum DO14 | C. butyricum DSP1 | |||
|
|
|
|||
| P | S | P | S | |
| Pseudomonas aeruginosa | 12(bg) | 10(bd) | 10(bd) | 10(bd) |
| Listeria monocytogenes | NF | NF | NF | NF |
| Salmonella enteritidis | NF | NF | 12(bd) | 12(bd) |
| Staphylococcus aureus | NF | NF | NF | NF |
| Bacillus subtilis | NF | NF | 16(S) | 16(S) |
| Escherichia coli | 12(bg) | 10(bd) | 12(bg) | 12(bg) |
P - culture liquid, S - supernatant, bd - bacteriostatic activity (reduction density); bg - bacteriostatic activity (complete growth inhibition); S - growth stimulation; NF - not found.
The highest level of bactericidal and bacteriostatic properties against Pseudomonas aeruginosa, Salmonella enteritidis and Escherichia coli was found with the C. butyricum DSP1 strain. No such activity, in turn, was observed with any of the analyzed isolates against Listeria monocytogenes and Staphylococcus aureus strains. Numerous reports concentrate on the inhibitory effect of C. butyricum on the pathogenic bacteria Helicobacter pylori (Tanaka et al., 2000; Takahashi et al., 2004; Shimbo et al., 2005). Takahashi et al. (2004) described similar properties of C. butyricum MIYAIRI 588 against Escherichia coli O157:H7. In their opinion, what determines the antibacterial potential of the Clostridium genus bacteria is their capacity to synthesize organic acids. The inhibitory effect of lactic acid and butyric acid of the tested C. butyricum MIYAIRI 588 strain was observed at neutral and low pH, which is significant information with view to its applicability potential (e.g. tolerance to gastrointestinal juices). Antibacterial properties of lactic acid were also discussed by other authors (Ammor et al., 2006). It needs to be stressed that C. butyricum bacteria also exhibit antagonistic interactions with Candida albicans, C. difficile, Klebsiella spp., Salmonella spp., Vibrio spp. (Chen et al., 1987; Fujita and Takashi, 1987; Kuroiwa et al., 1990; Takahashi et al., 2004).
EU Commission’s implementing regulation no. 373/2011 admitted C. butyricum FERM-BP 2789 preparation for use as a feed additive for birds and pigs. Use of the bacterial strain specified in the regulation is intended to increase weight gains and improve feed conversion rates in the above mentioned species. Imase et al. (2008) also described the effect of C. butyricum CMB 588 bacteria on the pathogenic strain Helicobacter pylori. Additionally, these authors observed an increase in the level of beneficial intestinal microflora (e.g. bacteria of the genus Bifidobacterium) during administration of an antibiotic therapy containing bacteria of the genus Clostridium. Other authors analyzed the probiotic effect of C. butyricum MIYAIRI 588 on a dangerous pathogen of the same species, the C. difficile(Woo et al., 2011).
Conclusions
In conclusion, the non-pathogenic clostridia reveal a large potential for industrial application. The study was designed to present new interesting potential applications of C. butyricum. The biochemical profile of the novel C. butyricum isolates indicates their capacity to ferment a wide range of sugars (simple and complex). The C. butyricum DSP1 strain was capable of synthesizing 1,3-PD from crude glycerol in the batch culture with an efficiency of 0.66 mol/mol glycerol. In addition, the C. butyricum DSP1 and C. butyricum DO14 isolates exhibit antibacterial qualities against Pseudomonas aeruginosa and Escherichia coli.
Acknowledgments
The paper was prepared within the framework of project PO IG 01.01.02-00-074/09 co-funded by The European Union from The European Regional Development Fund within the framework of the Innovative Economy Operational Programme 2007–2013.
References
- Ammor S, Tauveron G, Dufour E, Chevallier I. Antibacterial activity of lactic acid bacteria against spoilage and pathogenic bacteria isolated from the same meat small-scale facility. Screening and characterization of the antibacterial compounds. Food Control. 2006;1:454–461. [Google Scholar]
- Annous BA, Blaschek HP. Regulation and localization of amylolytic enzymes in Clostridium acetobutylicum ATTC 824. Appl Environ Microbiol. 1990;56:2559–2561. doi: 10.1128/aem.56.8.2559-2561.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Andoh A, Takizawa J, Takizawa W, Fujiyama Y. Clostridium butyricum, a probiotic derivative, suppresses dextran sulfate sodium-induced experimental colitis in rats. Int J Mol Med. 2004;13:577–580. [PubMed] [Google Scholar]
- Bahl H, Dürre P. Clostridia Biotechnology and Medical Applications. WV Press; Weinheim: 2001. [Google Scholar]
- Beckers L, Hiligsmann S, Hamilton Ch, Masset J, Thonart P. Fermentative hydrogen production by Clostridium butyricum CWBI1009 and Citrobacter freundii CWBI952 in pure and mixed cultures. Biotechnol Agron Soc Environ. 2010;14:541–548. [Google Scholar]
- Biebl H, Spröer C. Taxonomy of the glycerol fermenting Clostridia and description of Clostridium diolis sp. nov. Syst Appl Microbiol. 2002;25:491–497. doi: 10.1078/07232020260517616. [DOI] [PubMed] [Google Scholar]
- Chatzifragkou A, Papanikolau S, Dietz D, Doulgeraki AI, Nychas GJE, Zeng AP. Production of 1,3-propanediol by Clostridium butyricum growing on biodiesel - derived crude glycerol through a non-sterilized fermentation process. App Microbiol Biotechnol. 2011;91:101–112. doi: 10.1007/s00253-011-3247-x. [DOI] [PubMed] [Google Scholar]
- Chen HY, Kaneda S, Mikami Y, Arai T, Igarashi K, Saito M, Miyoshi T, Akira F. Protection activity induced by the bacterial vaccine, heat-killed Clostridium butyricum against Candida albicans infection in mice. Jap J Med Mycol. 1987;28:262–269. [Google Scholar]
- Colin T, Bories A, Moulin G. Inhibition of Clostridium butyricum by 1,3-propanediol and diols during glycerol fermentation. Appl Microbiol Biotechnol. 2000;54:201–205. doi: 10.1007/s002530000365. [DOI] [PubMed] [Google Scholar]
- Demain AL, Newcomb M, Wu JHD. Cellulase, Clostridia and ethanol. Microbiol Mol Biol. 2005;69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezeji TCL, Qureshi N, Blaschek HP. Bioproduction of butanol from biomass: from genes to bioreactors. Curr Opin Biotechnol. 2007;18:220–227. doi: 10.1016/j.copbio.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Fujita I, Takashi K. Studies on the anti-diarrheal activity of Clostridium butyricum Miyairi 588. Effect of Clostridium butyricum Miyairi 588 on the enterotoxicity cholera enterotoxin. Jap J Pharm. 1987;15:3249–3255. [Google Scholar]
- Hao J, Lin R, Zheng Z, Liu H, Liu D. Isolation and characterization of microorganisms able to produce 1,3-propanediol under aerobic conditions. World J Microbiol Biotechnol. 2008;9:1731–1740. [Google Scholar]
- Himmi EL, Bories A, Barbirato F. Nutrient requirements for glycerol conversion to 1,3-propanediol by Clostridium butyricum. Bioresource Technol. 1999;67:123–128. [Google Scholar]
- Imase K, Takahashi M, Tanaka A, Tokunaga K, Sugano H, Tanaka M, Ishida H, Kamiya S, Takahashi S. Efficacy of Clostridium butyricum preparation concomitantly with Helicobacter pylori eradication therapy in relation to changes in the intestinal microbiota. Microbiol Immunol. 2008;52:156–161. doi: 10.1111/j.1348-0421.2008.00026.x. [DOI] [PubMed] [Google Scholar]
- Kaur G, Srivastava AK, Chand S. Simple strategy of repeated batch cultivation for enhanced production of 1,3-Propanediol using Clostridium diolis. Appl Biochem Biotechnol. 2012;167:1061–1068. doi: 10.1007/s12010-012-9715-7. [DOI] [PubMed] [Google Scholar]
- Kubiak P, Leja K, Myszka K, Celinska E, Spychala M, Szymanowska-Powalowska D, Czaczyk K, Grajek W. Physiological predisposition of various Clostridium species to synthetize 1,3-propanediol from glycerol. Process Biochem. 2012;47:308–1319. [Google Scholar]
- Kuroiwa T, Kobari K, Iwanaga M. Inhibition of enteropathogens by Clostridium butyricum MIYAIRI 588. Jpn J Infect Dis. 1990;64:257–263. doi: 10.11150/kansenshogakuzasshi1970.64.257. [DOI] [PubMed] [Google Scholar]
- Leja K, Myszka K, Kubiak P, Wojciechowska J, Olejnik-Schmidt AK, Czaczyk K, Grajek W. Isolation and identification of Clostridium spp. from natural samples that performs effective conversion of glycerol to 1,3-propanediol. Acta Sci Pol Biotechnol. 2011;10:25–34. [Google Scholar]
- Maki M, Leung KT, Qin WS. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int J Biol Sci. 2009;5:500–516. doi: 10.7150/ijbs.5.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsoviti M, Paramithiotis S, Drosinos EH, Galiotou-Panayotou M, Nychas GJE, Zeng AP, Papanikolaou S. Screening of bacterial strains capable of converting biodiesel-derived raw glycerol into 1,3-propanediol, 2,3-butanediol and ethanol. Eng Life Sci. 2012;1:57–68. [Google Scholar]
- Oh SE, Zuo Y, Zhang H, Guiltinan MJ, Logan BE, Regan JM. Hydrogen production by Clostridium acetobutylicum ATCC 824 and megaplasmid-deficient mutant M5 evaluated using a large headspace volume technique. Int J Hyd Energy. 2009;34:9347–9353. [Google Scholar]
- Perry DA. Pectolytic Clostridium spp. in soils and rhizospheres of carrot and other arable crops in east Scotland. J Appl Microbiol. 2008;3:403–408. [Google Scholar]
- Petitdemange E, Durr C, Andaloussi SA, Raval G. Fermentation of raw glycerol to 1,3-propanediol by new strains of Clostridium butyricum. J Ind Microbiol. 1995;15:498–502. [Google Scholar]
- Quilaguy-Ayure DM, Montoya-Solano JD, Suárez-Moreno ZR, Bernal-Morales JM, Montoya-Castaño D. Analysing the dhaT gene in Colombian Clostridium sp. (Clostridia) 1,3-propanediol-producing strains. Univ Sci. 2010;15:17–26. [Google Scholar]
- Ren Z, Ward TE, Logan BE, Regan JM. Characterization of the cellulolytic and hydrogen-producing activities of six mesophilic Clostridium species. J Appl Microbiol. 2007;103:2258–2266. doi: 10.1111/j.1365-2672.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- Schlegel HG. General Microbiology. CU Press; Cambridge: 1993. [Google Scholar]
- Rymowicz W, Fatykhova A, Kamzolova SV, Rywinska A, Morgunov IG. Citric acid production from glycerol-containing waste of biodiesel industry by Yarrowia lipolytica in batch, repeated batch and cell recycle regimes. Appl Microbiol Biotechnol. 2010;87:971–979. doi: 10.1007/s00253-010-2561-z. [DOI] [PubMed] [Google Scholar]
- Rywinska A, Juszczyk P, Wojtatowicz M, Robak M, Lazar Z, Tomaszewska L, Rymowicz W. Glycerol as a promising substrate for Yarrowia lipolytica biotechnological applications. Biomass Bioener. 2013;48:148–166. [Google Scholar]
- Seki H, Shiohara M, Matsumura T, Miyagawa N, Tanaka M, Komiyama A, Kurata S. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr Int. 2003;45:86–90. doi: 10.1046/j.1442-200x.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Shimbo I, Yamaguchi T, Odaka T, Nakajima K, Koide A, Koyama H, Saisho H. Effect of Clostridium butyricum on fecal flora in Helicobacter pylori eradication therapy. World J Gastroenterol. 2005;11:7520–7524. doi: 10.3748/wjg.v11.i47.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skonieczny MT, Yargeau V. Biohydrogen production by Clostridium beijerinckii: effect of pH and substrate concentration. Int J Hyd Energy. 2009;34:3288–3294. [Google Scholar]
- Song JH, Ventura JRS, Lee ChH, Jahng D. Butyric acid production from brown algae using Clostridium butyricum ATCC 25755. Biotechnol Bioprocess Eng. 2011;16:42–49. [Google Scholar]
- Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J. Direct analysis of genes encoding 16 S rRNA from complex communities reveals many novel molecular species within human gut. Appl Environ Microbiol. 1999;65:4799–4807. doi: 10.1128/aem.65.11.4799-4807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanowska-Powalowska D, Drozdzynska A, Remszel N. Isolation of new strains of bacteria able to synthesize 1,3-propanediol from glycerol. Adv Microbiol. 2013;3:171–180. [Google Scholar]
- Taguchi F, Mizukami N, Saito-Taki T, Hasegawa K. Hydrogen production from continuous fermentation of xylose during growth of Clostridium sp. strain No. 2. Can J Microbiol. 1995;41:536–540. [Google Scholar]
- Takahashi M, Taguchi H, Yamaguchi H, Osaki T, Kamiya S. Studies of the effect of Clostridium butyricum on Helicobacter pylori in several test models including gnotobiotic mice. J Med Microbiol. 2000;49:635–642. doi: 10.1099/0022-1317-49-7-635. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Taguchi H, Yamaguchi H, Osaki T, Komatsu A, Kamiya S. The effect of probiotic treatment with Clostridium butyricum on enterohemorrhagic Escherichia coli O157:H7 infection in mice. Immunol Med Microbiol. 2004;41:219–229. doi: 10.1016/j.femsim.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Miyagawa N, Tsutsumi Y. Preventive effect of Clostridium butyricum on the side effect of the Helicobacter pylori eradication. Chin J Gastroenterol. 2000;5:116–119. [Google Scholar]
- Ting PT, Freiman A. The story of Clostridium botulinum: from food poisoning to botox. Clin Med Res. 2004;4:258–261. doi: 10.7861/clinmedicine.4-3-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniec K, Huisingh D, Friedl A, Claassen P. Hydrogen for a sustainable global economy. J Clean Prod. 2012;18:81–83. [Google Scholar]
- Venkataramanan KP, Boatman JJ, Kurniawan Y, Taconi KA, Bothun GD, Scholz C. Impact of impurities in biodiesel-derived crude glycerol on the fermentation by Clostridium pasteurianum ATCC 6013. Appl Microbiol Biotechnol. 2013;93:1325–1335. doi: 10.1007/s00253-011-3766-5. [DOI] [PubMed] [Google Scholar]
- Wang HK, Li AD, Liu FF, Qi W. Determination of an economical medium for growth of Clostridium butyricum TK2 using orthogonal test. Afr J Microbiol Res. 2011;5:1773–1777. [Google Scholar]
- Wilkens E, Ringel AK, Hortig D, Willke T, Vorlop KD. High-level production of 1,3-propanediol from crude glycerol by Clostridium butyricum AKR102a. Appl Microbiol Biotechnol. 2011;5:232–230. doi: 10.1007/s00253-011-3595-6. [DOI] [PubMed] [Google Scholar]
- Williams K, Zheng Y, McGarvey J, Fan Z, Zhang R. Ethanol and volatile fatty acid production from lignocellulose by Clostridium cellulolyticum. ISRN Biotechnol. 2013;2013:1–7. doi: 10.5402/2013/137835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo TDH, Oka K, Takahashi M, Hojo F, Osaki T, Hanawa T, Kurata S, Yonezawa S, Kamiya S. Inhibition of the cytotoxic effect of Clostridium difficile in vitro by Clostridium butyricum MIYAIRI 588 strain. J Med Microbiol. 2011;60:1617–1625. doi: 10.1099/jmm.0.033423-0. [DOI] [PubMed] [Google Scholar]
- Yung-Chung L, Wei-Chung L, Chun-Yen C, Chang J. Dark fermentative hydrogen production from enzymatic hydrolysate of xylan and pretreated rice straw by Clostridium butyricum CGS5. Bioresource Technol. 2010;101:5885–5891. doi: 10.1016/j.biortech.2010.02.085. [DOI] [PubMed] [Google Scholar]
- Zhang SB, Cui YL, Wu SE, Li D, Wan FC. Inhibitory effect of Clostridium butyricum (CB) on bacteria. Zhongguo Xingyao Zazhi. 2002;11:322–324. [Google Scholar]
- Zhang CH, Ma YJ, Yang FX, Liu W, Zhang YD. Microbial production of 1,3-propanediol by Klebsiella pneumoniae using crude glycerol from biodiesel preparations. Bioresour Technol. 2009;100:134–139. doi: 10.1007/s10529-006-9154-z. [DOI] [PubMed] [Google Scholar]