Abstract
The objective of this report is to describe the protocols for comparing the microRNA (miRNA) profiles of human induced-pluripotent stem (iPS) cells, retinal pigment epithelium (RPE) derived from human iPS cells (iPS-RPE), and fetal RPE. The protocols include collection of RNA for analysis by microarray, and the analysis of microarray data to identify miRNAs that are differentially expressed among three cell types. The methods for culture of iPS cells and fetal RPE are explained. The protocol used for differentiation of RPE from human iPS is also described. The RNA extraction technique we describe was selected to allow maximal recovery of very small RNA for use in a miRNA microarray. Finally, cellular pathway and network analysis of microarray data is explained. These techniques will facilitate the comparison of the miRNA profiles of three different cell types.
Keywords: Molecular Biology, Issue 88, microRNA, microarray, human induced-pluripotent stem cells, retinal pigmented epithelium
Introduction
Stem cells have the capacity to replicate without limit and the potential to differentiate into any somatic cell type. The development of techniques to reprogram somatic cells into pluripotent stem cells has elicited great excitement in the research community and among clinicians, as the advent of personalized tissue regeneration is on the horizon1. Induced-pluripotent stem (iPS) cells exhibit the same features of unlimited replicative potential and pluripotency as embryonic stem (ES) cells while circumventing the ethical dilemmas associated with ESCs. In addition, patient-derived stem cells will not stimulate an immune response, greatly increasing the probability of successful therapeutic applications2-3. Under specific culture conditions, iPS cells have been shown to differentiate into several different cell types in vitro, including cardiomyocytes, neurons, pancreatic beta cells, hepatocytes, and retinal pigmented epithelium (RPE)4-12.
The RPE is a specialized layer of pigmented epithelial cells located at the back of the retina that performs several functions that are essential for visual health and function such as absorption of stray light, phagocytosis of the photoreceptor outer segments, and processing of retinoids for the production of visual chromophore. Dysfunction of the RPE due to damage or disease profoundly affects photoreceptor health and visual function as evidenced by the blinding diseases that are the result of underlying RPE pathology such as age-related macular degeneration (AMD), Stargardt’s disease, and retinitis pigmentosa (RP)13. Truly effective treatments that can restore vision have not been achieved, and replacement of diseased RPE with healthy RPE may be the best option to prevent loss of vision14-15. RPE derived from iPS (iPS-RPE) is a possible source of cells to replace the damaged RPE. iPS-RPE expresses characteristic RPE proteins LRAT, CRALBP, PEDF, and RPE65; displays the classical highly pigmented hexagonal RPE morphology; and performs RPE functions such as phagocytosis, retinoid processing, and secretion of 11-cis retinal5,16. However, before iPS-RPE can be used therapeutically, the iPS-RPE must be thoroughly characterized. Understanding the factors that govern RPE differentiation is necessary to improve the yield and purity of the cells to be used for clinical applications.
Cell differentiation is the result of highly regulated gene expression. Epigenetic remodeling of the genome and coordination of transcription factors are required for cell fate decisions that occur during differentiation and development17. Regulation of message RNA (mRNA) translation by microRNA (miRNA) presents yet another level of regulation that affects cell fate1. MiRNAs are short, ~22 nt, lengths of nucleotides that either suppress translation by binding to the 3’ UTR of mRNA or target the mRNA for degradation. MiRNAs have been detected in virtually all tissues and to date over 2,000 unique human microRNAs have been registered in the miRBase database. Since miRNAs require only partial complementarity to bind to the target mRNA, a single miRNA can potentially bind to tens or hundreds of targets, and vice versa; i.e. a single mRNA can be targeted by several different miRNAs. This promiscuous binding characteristic dramatically increases the level of complexity of regulation as well as the level of difficulty of determining the functions of individual miRNAs and the role each plays in cellular functions18-21. Nevertheless, studies have shown that miRNA refines gene expression during differentiation by affecting DNA methylation status17. In a study more specific to the RPE, miR-204/211 was shown to promote the epithelial phenotype of the RPE22. Another group analyzed the miRNA profile of RPE during differentiation from ES cells and revealed distinct sets of miRNA are expressed during the differentiation process23. In fact, miRNA profiles can unambiguously distinguish cell types, including ES cells, precursor cells, and terminally differentiated cells24,25. Over 250 miRNAs are expressed in the retina. Based upon these studies, we hypothesize that miRNAs play an important role during the differentiation of RPE from iPS.
The objective of this report is to describe the protocols for the differentiation of RPE from IMR90-4 iPS cells, collection of RNA for analysis by microarray, and the analysis of microarray data to identify miRNAs that are differentially expressed among three cell types, iPS cells, iPS-RPE and fetal RPE. Total RNA was extracted from cultures of each cell type and hybridized to a miRNA microarray, containing probes specific for 1,205 human miRNAs and 144 viral miRNAs. The microarray results were compared to determine which miRNAs were differentially expressed among the different cell types. miRNAs with 2 fold or greater fold change in expression were selected for further analysis. A miRNA analysis software program was used to identify potential targets of the differentially expressed miRNAs and to generate cellular networks regulated by the selected miRNAs.
Protocol
1. Preparation of Culture Reagents and Culture Plates
mTeSR1 media: Prepare mTeSR1 media according to the manufacturer’s directions. Thaw 100 ml of mTeSR1 5x supplement overnight at 4 °C. Add the 100 ml of mTeSR1 5x supplement to 400 ml of mTeSR1 basal media and mix well. This media is stable at 4 °C for up to 2 weeks and at -20 °C for 6 months.
Differentiation media: Prepare differentiation media in DMEM/F12 to yield 0.1mM β-mercaptoethanol, 0.1 mM nonessential amino acids, 2.0 mM L-glutamine, 10% knockout serum, and 10 µg/ml gentamicin.
iPS-RPE media: Prepare iPS-RPE media in MEM to yield 1x N1 supplement, 0.1 mM nonessential amino acids, 250 µg/ml taurine, 13 ng/ml triiodo-L-thyronine sodium salt, 20 ng/ml hydrocortisone, 5 µg/ml gentamicin, and 15% FBS26.
Triiodo-L-thyronine sodium salt stock solution. To prepare stock solution dissolve 1.0 mg of triiodo-L-thyronine sodium salt in 1 N sodium hydroxide by gently swirling. Next add 49.0 ml of base media to make 50.0 ml of 20 µg/ml of triiodo-L-thyronine sodium salt. Prepare aliquots and freeze at -20 °C until needed. Use appropriate volume to achieve desired concentration in final culture media.
Hydrocortisone stock solution: To prepare a hydrocortisone stock solution, solubilize 1 mg of hydrocortisone in 1 ml of absolute ethanol by gentle agitation. Next add 19.0 ml of base media for 20 ml hydrocortisone stock solution of 50 µg/ml. aliquot and freeze at -20 °C until needed. Use appropriate volume to achieve the desired concentration in the final culture media.
Fetal RPE growth medium. Prepare the growth media by mixing all reagents in the RTEGM kit in the basal media, according to the manufacturer directions except for the FBS.
Fetal RPE plating media. Prepare plating media by transferring a small amount of growth media to a sterile container and add FBS to a final concentration of 2%.
Dispase: Prepare working solution of dispase of 1 mg/ml in DMEM/F12.
Matrigel coated plates: Prepare matrigel in DMEM/F12 to 0.08 mg/ml.
Coat each well of a 6-well plate with 1.0 ml of the matrigel solution.
Incubate the coated plates at room temperature for 1.0 hr.
After the 1.0 hr incubation, aspirate the excess DMEM/F12.
Add 0.5 ml of mTeSR1 media to avoid drying. The matrigel coated plates are now ready for use.
2. iPS Culture
Prior to iPS cell seeding have all tubes, warm mTESR1 media to room temperature, and the matrigel coated plates ready.
Quickly thaw the IMR90-4 iPS cells in a 37 °C water bath. Then remove the cryovial from the water bath and sterilize using 70% ethanol.
Transfer the cells to a 15 ml conical tube using a 2 ml pipette to minimize breaking of cell aggregates.
Drop-wise add 5 ml of warm mTeSR1 media to the cells and gently mix. Centrifuge the cells at 300 x g for 5 min at room temperature and remove the supernatant.
Carefully resuspend the cell aggregates in 2 ml of mTeSR1 media and seed the cells onto one well of a six well plate.
Place the plate into a 37 °C incubator and culture with 5% CO2, 95% humidity.
Change the iPS cell culture media daily. Undifferentiated colonies will appear 5-7 days after seeding. Check for undifferentiated colonies that are ready for passage (colonies with a dense center).
3. Passaging of iPS Cells
Prior to beginning to passage the iPS cells, have all tubes, dispase solution, DMEM/F12 and mTERS1 media warmed to room temperature, and the matrigel coated plates ready.
Before passaging iPS cells, remove any areas of differentiation by scraping the area and aspirating the media. Differentiation should remain below 20% of the well for high quality cultures. Carefully rinse the iPS cell-containing well with 2 ml DMEM/F12 and add 1 ml of 1 mg/ml dispase to each well.
Incubate at 37 °C for 7 min. Aspirate the dispase and gently rinse the cell colonies with 2 ml of DMEM/F12 twice.
After rinsing, add 2 ml of mTSER1 to each well.
Using a 5 ml pipette gently scrape the colonies of the plate to form aggregates.
Transfer the detached colonies to a 15 ml conical tube. Add sufficient mTeSR1 media to seed the next passage of cells.
4. Differentiation of iPS Cells to iPS-RPE
Plate the iPS cells on to a newly coated plate in mTESR1 media.
Allow iPS cell colonies to expand for 4-5 days in culture.
Substitute mTeSR1 media with 4 ml differentiation media. Change one half of the media every 3 days.
Allow pigmented foci to grow large enough to be manually dissected out of the culture (40-50 days).
Use a 200 µl pipette tip to cut around and lift the pigmented colonies.
Collect and pool the dissected colonies in a 15 ml conical tube.
Then add 5 ml of DMEM/F12 to the pooled RPE cells and centrifuge at 300 x g for 5 min twice.
Prepare a single cell solution by incubating the pooled iPS-RPE with 0.25% trypsin EDTA.
Rinse the iPS-RPE cells with iPS-RPE media and plate the collected cells on a fresh matrigel-coated well in 4 ml of iPS-RPE media.
Culture the cells for 3 weeks prior to passage. Change the media every 3 days. For the experiment, plate iPS-RPE at 100,000 cells/cm2. Collect cells on Day 17. Count the day of seeding as Day 0.
5. Sample Collection
- iPS Cell Collection
- Culture the iPS cells until undifferentiated colonies are observed to be dense at the center.
- Rinse the colonies in DMEM/F12.
- Dissociate into single cells using accutase. Collect the cells into a 15 ml conical tube and centrifuge at 300 x g for 5 min. Aspirate supernatant. Resuspend in 2 ml of DMEM/F12.
- Fetal RPE Collection
- Rinse Fetal RPE cell cultures with DMEM/F12 and dissociate into single cells using 0.25% trypsin EDTA.
- Collect the cells in a 15 ml conical tube. Centrifuge at 300 x g for 5 min. Aspirate supernatant. Resuspend in 2 ml of DMEM/F12.
- iPS-RPE Cell Collection
- When the iPS-RPE reach passage 3 and passage 5, collect the cells on Day 17 after passage. Rinse iPS-RPE twice in PBS and prepare a single cell suspension by incubation with 1.0 ml of 0.25% trypsin.
- Neutralize trypsin with 2 ml of ice cold iPS-RPE media.
- Collect the cells in a 15 ml conical tube and centrifuge at 300 x g for 5 min. Aspirate supernatant.
- Resuspend the cell suspension in 3 ml of staining buffer from microbead kit.
6. Negative Selection of iPS-RPE to Remove Undifferentiated Cells
Keep all reagents at 4 °C until use, but do not work on ice. Centrifuge cell suspension from step 5.3.4 at 300 x g for 5 min. Aspirate supernatant.
Resuspend cells in 100 μl of staining buffer from microbead kit per 2 x 106 cells.
Add 10 μl of Anti-TRA-1-60-PE antibody per 2 x 106 cells.
Mix well and incubate at 4 °C for 10 min.
Wash cells in 1-2 ml of buffer per 2 x 106 cells. Centrifuge for 10 min at 300 x g. Aspirate supernatant.
Resuspend cells in 80 μl of staining buffer per 2 x 106 cells. Add 20 μl of anti-PE labeled microbeads (from kit) per 2 x 106 cells. Mix well. Incubate for 15 min at 4 °C.
Add 1-2 ml of staining buffer per 2 x 106 cells. Centrifuge for 10 min at 300 x g. Aspirate supernatant.
Resuspend cells in 500 μl of staining buffer.
- Magnetic cell separation:
- Place column in magnetic cell sorter. Rinse column with 3 ml of staining buffer.
- Apply cell suspension from 6.8 onto column.
- Place a tube at the negative port to collect the TRA-1-60 negative cells.
7. Total RNA Extraction
Lyse the cells by running the samples through a commercially available microcentrifuge homogenizer mini spin column designed for nucleic acid minipreps.
Extract total RNA from iPS cells, iPS-RPE, and fetal RPE with the commercially available RNA extraction kit designed to preserve small RNA molecules27.
Determine total RNA concentrations using a Nanodrop spectrophotometer.
8. Bioanalyzer and Microarray Assay
Check the RNA integrity with Bioanalyzer in conjunction with commercially available RNA quality kit.
Incubate 100 ng of total RNA with calf intestinal phosphatase at 37 °C for 30 min
Denature the RNA using 100% DMSO at 100 °C for 5 min.
Label the samples with pCp-Cy3 using T4 ligase by incubation at 16 °C for 1 hr.
Hybridize on an 8_x_15K format human miRNA array.
Wash the arrays according to manufacturer's instructions and scan at a resolution of 5 µm using a scanner.
Acquire data using microarray feature extraction software compatible with the miRNA microarray.
Process the miRNA signals to determine the relative level of expression in each sample.
Compare the expression levels of each miRNA between the sample groups. Select miRNAs with at least two-fold differential expression for further analysis.
Using commercially available software for data analysis, follow the instructions to generate heat maps for visualization of differentially expressed miRNAs28.
9. Pathway Analysis
Save data from the miRNAs selected in 8.9 in Excel format.
Log into the online RNA pathway analysis program. Upload the Excel file. Select “Flexible Format” for file format and select “Agilent” from drop down list for identifier type.
Under raw data heading, assign “ID” to probe ID column and assign “Observation 1” and “log ratio” to the log ration column. Select “ignore” for all other columns.
Select the parameters for analysis as follows: Confidence: experimentally observed only; include direct and indirect relationships; and include endogenous chemicals; Select the Ingenuity Knowledge base as the reference set. Choose the database of reference molecules that includes data from all species, all cell lines and tissues, and all mutations.
Run network and pathway analyses.
Representative Results
iPS cells (Figure 1A) were grown in differentiation conditions to induce differentiation into iPS-RPE cells. The iPS-RPE exhibited classic RPE phenotype of hexagonal pigmented cell morphology (Figure 1B) similar to fetal RPE (Figure 1C).
To better understand the role that miRNA may play during the process of differentiation from iPS to iPS-RPE, microarray analysis of miRNA expression was conducted. Total RNA was collected from iPS cells, fetal RPE, and iPS-RPE using the mirVana miRNA isolation kit to ensure maximal retention of small RNAs. The RNA was quantified and tested for purity before hybridization to the microarray labchip. Probes representing 1,205 human and 144 viral miRNAs were used to analyze miRNA expression from each sample set. The signals were analyzed to determine the relative levels of expression of each miRNA from each sample. The data was used to compare the miRNA expression profile of iPS-RPE to iPS and fetal RPE. Heat maps were generated to enable visualization of differential miRNA expression between the groups (Figure 2A). 83 miRNAs that were differentially expressed in the iPS compared to iPS-RPE, with 47 miRNAs upregulated in the iPS compared to iPS-RPE, and 36 downregulated in the iPS (Figure 2B). 110 miRNAs were differentially expressed in fetal RPE compared to iPS-RPE, with 61 miRNAs upregulated in the fRPE, and 49 downregulated in the fRPE compared to iPS-RPE (Figure 2C). Pathway analysis with Ingenuity IPA software indicated the differentially expressed miRNAs target transcripts for genes involved in cellular development, growth, proliferation and survival, cell cycle, and cellular movement. MiRNAs expressed in ocular tissues in vivo were found to be enriched in iPS-RPE and fetal RPE compared to iPS (Figures 3 and 4).
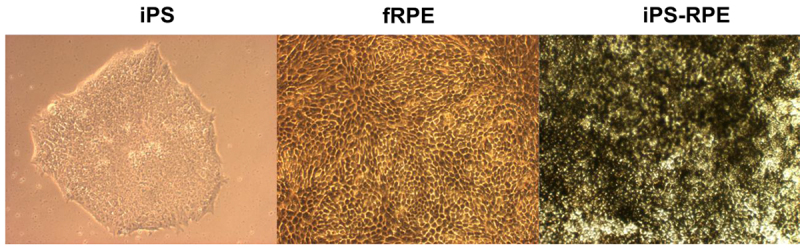 Figure 1. Brightfield images of iPS, fetal RPE, and iPS-RPE. Brightfield images (100X) of iPS cells prior to differentiation, fetal RPE, and RPE derived from iPS. Both the fetal RPE and iPS-RPE display classical RPE morphology of hexagonal shape and pigmentation. Please click here to view a larger version of this figure.
Figure 1. Brightfield images of iPS, fetal RPE, and iPS-RPE. Brightfield images (100X) of iPS cells prior to differentiation, fetal RPE, and RPE derived from iPS. Both the fetal RPE and iPS-RPE display classical RPE morphology of hexagonal shape and pigmentation. Please click here to view a larger version of this figure.
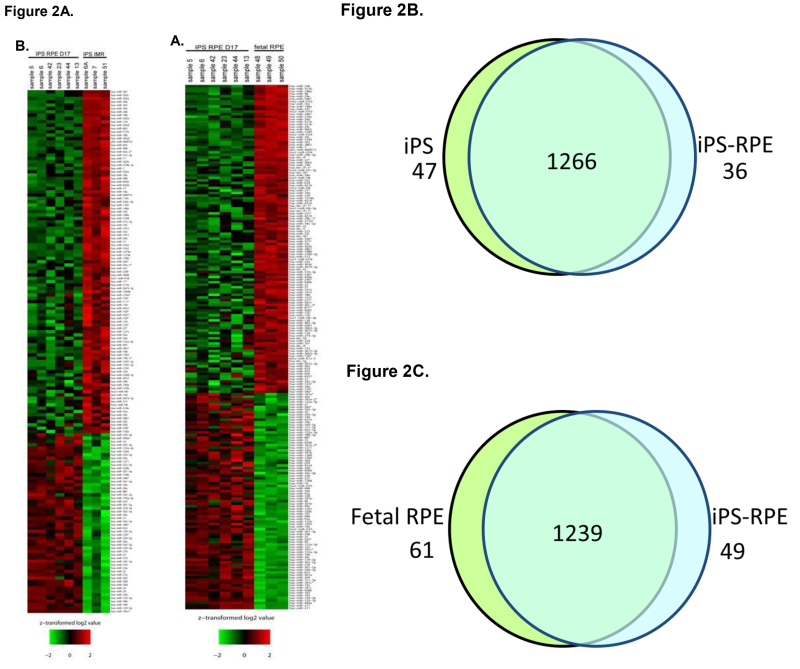 Figure 2. miRNA expression profile from iPS-RPE compared to iPS and fetal RPE.A) Heat maps depict the relative degree of differential expression of miRNAs. The color scale represents the expression levels of the miRNA as above (red), below (green), or at the mean expression level (black) across all samples. B) Comparison of miRNA profiles of iPS-RPE to iPS cells. C) Comparison of miRNA profiles of iPS-RPE to fetal RPE. Please click here to view a larger version of this figure.
Figure 2. miRNA expression profile from iPS-RPE compared to iPS and fetal RPE.A) Heat maps depict the relative degree of differential expression of miRNAs. The color scale represents the expression levels of the miRNA as above (red), below (green), or at the mean expression level (black) across all samples. B) Comparison of miRNA profiles of iPS-RPE to iPS cells. C) Comparison of miRNA profiles of iPS-RPE to fetal RPE. Please click here to view a larger version of this figure.
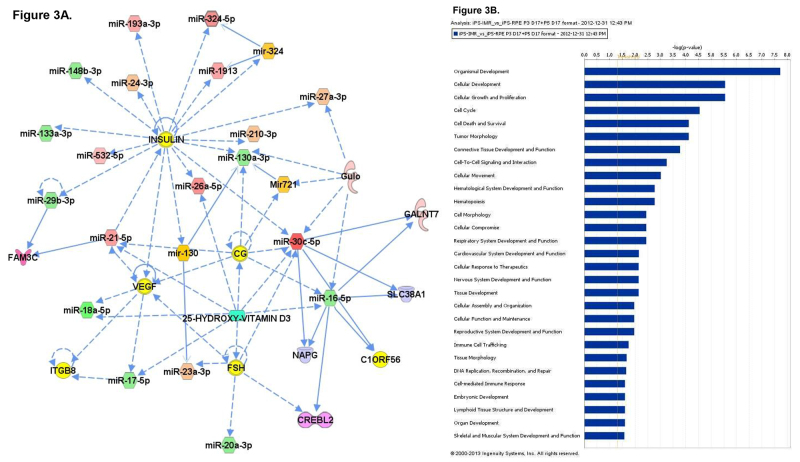 Figure 3. A) Network analysis of the miRNA expression profiles of the parental iPS cells versus the iPS-derived RPE. The top 4 networks are shown separately (left) and merged (center). B) Pathway analysis of differentially expressed miRNAs. Please click here to view a larger version of this figure.
Figure 3. A) Network analysis of the miRNA expression profiles of the parental iPS cells versus the iPS-derived RPE. The top 4 networks are shown separately (left) and merged (center). B) Pathway analysis of differentially expressed miRNAs. Please click here to view a larger version of this figure.
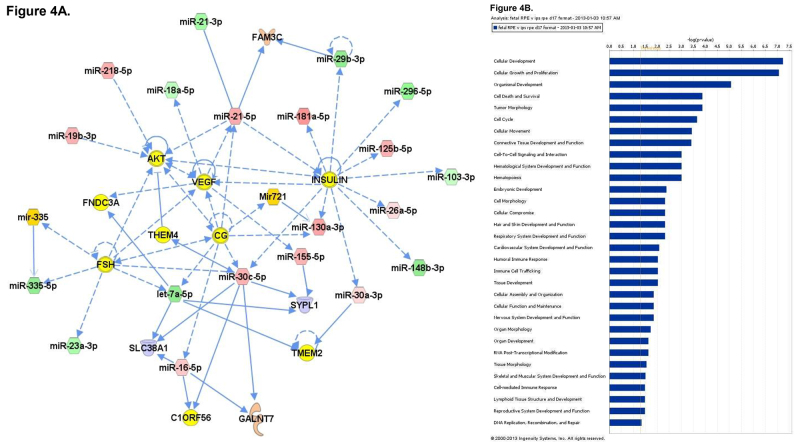 Figure 4. A) Network analysis of the miRNA expression profiles of fetal RPE versus the iPS-derived RPE. The top 7 networks are shown separately (left) and merged (center). B) Pathway analysis of differentially expressed miRNAs. Please click here to view a larger version of this figure.
Figure 4. A) Network analysis of the miRNA expression profiles of fetal RPE versus the iPS-derived RPE. The top 7 networks are shown separately (left) and merged (center). B) Pathway analysis of differentially expressed miRNAs. Please click here to view a larger version of this figure.
Discussion
In conclusion, this report describes the methods used to culture iPS cells, iPS-RPE, and fetal RPE. RPE derived from iPS are morphologically and functionally similar to fetal RPE. The iPS-RPE also expresses characteristic RPE genes including RPE65, CRALBP, PEDF, and LRAT16. To further characterize these cells, RNA was extracted and used to perform microarray analysis to identify differentially expressed miRNA. Analysis of the signal intensities revealed that the greatest number of differentially expressed miRNAs was detected in the fetal RPE versus iPS-RPE comparisons, suggesting the iPS-RPE may be more mature. Future studies are planned to validate these results with RT-PCR.
There are several critical points during this experiment that must be carefully performed to ensure the success of the assay and acquisition of accurate data. The first key step occurs during the culture of the iPS cells. iPS cells must remain in a pluripotent state to maintain their stemness. The cells must be carefully inspected daily for signs of differentiation. Undifferentiated iPS cells grow as compact multicellular colonies. The cells should have a high nuclear to cytoplasm ratio and prominent nucleoli. The iPS colonies are characterized by a distinct border, with several layers of cells at the center. Signs of differentiation include loss of defined colony borders, non-uniform cell morphology, and the appearance of obvious cell types, such as neurons and fibroblasts. Single cells that have differentiated can be removed by dispase and rinsing, however, colonies with these characteristics must be manually removed from the culture29.
Preparation of RNA for microarray analysis is the next critical step. Inconsistent RNA quality is a significant source of variability in microarray data. RNA extraction should yield high molecular weight and minimally degraded RNA for best results. Successful RNA extraction will yield total RNA with minimal degradation and free of any contaminating RNases. After determination of the RNA concentration by spectrophotometry at 260 nm, the purity of the sample should be determined at 230 and 280 nm to detect contamination with polysaccharides or protein. The 230:260:280 ratio for RNA should be 1:2:1 to indicate high quality RNA with no contamination30.
When planning the microarray experiment, the most critical step is the inclusion of negative controls to ensure the hybridization to the probes is specific, and to run each sample in triplicate to ensure that the signals detected are valid for that sample set. During the analysis of the microarray results, a key point is background correction, in which the background noise is subtracted from the observed signal to give the true signal intensity for each particular probe. This step will eliminate false positives, while highlighting the true positive signals.
Finally, during network and pathway analysis of the microarray results, the most important step is to establish the correct parameters and filters before running the analysis. This step will determine which databases will be used by the software to generate the networks and identify cellular pathways that are impacted by the miRNAs shown to be differentially expressed in the comparisons.
After acquisition of data using the methods described in this report, it is necessary to consider the limitations of these assays during interpretation of the results. Although unlikely, it remains possible that the RPE culture is not a homogenous population of RPE cells. The anti-TRA-1-60 sorting process is a negative selection process that removes the cells that still express stem cell markers; however it does not guarantee the remaining cells are pure RPE cells. Although morphologically the cells exhibit classic RPE morphology of pigmentation and polygonal cell shape, it is possible that not all cells are RPE.
Microarray experiments may produce false positives for the expression of RNA, particularly in the case of microRNA due to the very short sequences. Although the microarray system used for this experiment included multiple probes to target different regions of each target, the results should always be validated by qRT-PCR.
Analysis of microarray data by Ingenuity IPA is dependent upon the quantity and quality of information in the Ingenuity database. Much of the information on gene function and cellular pathways is derived from experiments conducted on transformed cell lines or in the context of cancer research. Therefore, the results of pathway and network analysis using this program may not always be relevant for primary cells or, in this case, stem cells.
Despite these limitations, the methods described in this report can be used for culture of iPS cells and fetal RPE, deriving RPE from iPS cells, and extraction of RNA for microarray analysis of miRNA. The data generated by these assays can be used to identify miRNAs involved in the differentiation process, as well as those that regulate RPE functions.
Although more miRNAs were found to be differentially expressed in the fetal RPE vs. iPS-RPE comparison than in the iPS vs. iPS-RPE comparison, there are many possible reasons for this. First of all, at the time this assay was performed, the miRNA microarray platform was capable of detecting 1,205 human and 144 viral miRNAs. Since that time, the platform has been expanded to include probes for over 2,000 human miRNAs. It is certainly possible that the miRNA differential expression profiles will change if more miRNAs are detected. Of the 1,349 miRNAs detected by this assay, most of miRNAs showed no difference in expression between the cell types (Figures 2B and 2C). More important than counting the overall number of miRNAs expressed will be determining the functions of these miRNAs and understanding which ones are regulating the process of differentiation and RPE functions. These are topics actively being investigated in our lab. The current report is intended to provide a description of the methods used to derive iPS-RPE from iPS and to perform miRNA analysis on these cell types. Finally, although this report is focused on miRNA, miRNAs are only a part of the process, as gene transcription, post-translational modifications, intracellular localization, and many other cellular events are critical regulators of differentiation.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense.
This research was performed while the authors Whitney A. Greene, Alberto Muñiz, and Ramesh R. Kaini held a National Research Council Postdoctoral Research Associateship at the USAISR.
Microarray assays were performed by the Greehey Children's Cancer Research Institute Microarray Core Facility and Bioinformatics Department at UT Health Science Center San Antonio.
This work was supported by U.S. Army Clinical Rehabilitative Medicine Research Program (CRMRP) and Military Operational Medicine Research Program (MOMRP).
References
- Yu Z, et al. miRNAs regulate stem cell self-renewal and differentiation. Front Genet. 2012;3 doi: 10.3389/fgene.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G, et al. A commentary on iPS cell biology: Potential applications in autologous tissue and cell transplantation, development of new models for the study of human pathological conditions and drug screening. J Cell Physiol. 2014;229:148–152. doi: 10.1002/jcp.24437. [DOI] [PubMed] [Google Scholar]
- Almeida PE, et al. Immunogenicity of pluripotent stem cells and their derivatives. Circ Res. 2013;112:549–561. doi: 10.1161/CIRCRESAHA.111.249243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz DE, et al. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427–2434. doi: 10.1002/stem.189. [DOI] [PubMed] [Google Scholar]
- Kokkinaki M, et al. Human induced pluripotent stem-derived retinal pigment epithelium (RPE) cells exhibit ion transport, membrane potential, polarized vascular endothelial growth factor secretion, and gene expression pattern similar to native RPE. Stem Cells. 2011;29:825–835. doi: 10.1002/stem.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti K, et al. The new paradigm: retinal pigment epithelium cells generated from embryonic or induced pluripotent stem cells. Pigment Cell Melanoma Res. 2011;24:21–34. doi: 10.1111/j.1755-148X.2010.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pera MF. Stem cells. A new year and a new era. Nature. 2008;451:135–136. doi: 10.1038/451135a. [DOI] [PubMed] [Google Scholar]
- Subba Rao M, et al. Thinking outside the liver: Induced pluripotent stem cells for hepatic applications. World J Gastroenterol. 2013;19:3385–3396. doi: 10.3748/wjg.v19.i22.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Yamanaka S. iPS cells: a source of cardiac regeneration. J Mol Cell Cardiol. 2011;50:327–332. doi: 10.1016/j.yjmcc.2010.10.026. [DOI] [PubMed] [Google Scholar]
- Lee KS, et al. Human sensory neurons derived from induced pluripotent stem cells support varicella-zoster virus infection. PLoS One. 2012;7 doi: 10.1371/journal.pone.0053010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipio Z, et al. Reversal of hyperglycemia in diabetic mouse models using induced-pluripotent stem (iPS)-derived pancreatic beta-like cells. Proc Natl Acad Sci U S A. 2010;107:13426–13431. doi: 10.1073/pnas.1007884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Cour M, Tezel T. The retinal pigment epithelium. Adv Organ Biol. 2005;10:253–272. [Google Scholar]
- Carr AJ, et al. Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SD, et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- Muniz A, et al. Retinoid uptake, processing and secretion in human iPS-RPE support the visual cycle. Invest Ophthalmol Vis Sci. 2014;55:198–209. doi: 10.1167/iovs.13-11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo TR, et al. The functions of microRNAs in pluripotency and reprogramming. Nat Cell Biol. 2012;14:1114–1121. doi: 10.1038/ncb2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora A, et al. Prediction of microRNAs affecting mRNA expression during retinal development. BMC Dev Biol. 2010;10 doi: 10.1186/1471-213X-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- Mallanna SK, Rizzino A. Emerging roles of microRNAs in the control of embryonic stem cells and the generation of induced pluripotent stem cells. Dev Biol. 2010;344:16–25. doi: 10.1016/j.ydbio.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adijanto J, et al. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J Biol Chem. 2012;287:20491–20503. doi: 10.1074/jbc.M112.354761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, et al. Identification of miRNA signatures during the differentiation of hESCs into retinal pigment epithelial cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KD, et al. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu P, et al. MicroRNA profiling reveals two distinct p53-related human pluripotent stem cell states. Cell Stem Cell. 2010;7:671–681. doi: 10.1016/j.stem.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Maminishkis A, et al. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest Ophthalmol Vis Sci. 2006;47:3612–3624. doi: 10.1167/iovs.05-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiagen miRNeasy Mini Handbook. Sample and Assay Technologies. 2013.
- Microarray System with miRNA Complete Labeling and Hyb Kit. Agilent Technologies. 2011.
- Maintenance of hESCs and hiPSCs in mTESR1 and mTESR2. Stem Cell Technologies. 2010.
- The Analysis of DNA or RNA using its wavelengths: 230 nm, 260 nm, 280 nm. About Biotechnology. 2013.


