Abstract
The alpine plant Arabis alpina (alpine rock-cress) is a thoroughly studied species in the fields of perennial plant flowering regulation, phylogeography, and adaptation to harsh alpine climatic conditions. We report the complete A. alpina chloroplast genome sequence obtained through de novo assembly of Illumina paired-end reads produced by total DNA sequencing. The A. alpina cp circular genome is 152,866 bp in length and built of two inverted repeats of 26,933 bp separated by unique regions: a large single copy of 82,338 bp and a small single copy of 17,938 bp. The genome contains 131 genes, some of them being duplicated in the inverted repeats. Seventy-nine unique protein-coding genes were annotated, together with 29 tRNA genes and 4 ribosomal RNA genes. Sequencing and mapping of 23 additional A. alpina DNA samples enabled to gain insight into the intraspecies polymorphism of the sequenced cp genome. Genetic variability among genomes was detected as 44 indels, most of them being located in noncoding regions, and 130 single-nucleotide polymorphisms, 37 of them corresponding to mutations in coding regions. A. alpina chloroplast genome sequence will be helpful in population studies or investigations of chloroplast functions of this alpine plant species.
Keywords: Alpine rock-cress, Plant, Plastome, Genetic diversity
Highlights
-
•
We report the complete A. alpina chloroplast genome sequence through de novo assembly.
-
•
Arabis alpina plastome is 152,866 in length and harbors 131 genes.
-
•
Intraspecies polymorphism analysis was investigated among 24 samples.
-
•
We detected 44 indels and 130 single nucleotide polymorphisms.
1. Introduction
Chloroplasts are organelles specific to the green lineage where photosynthesis takes place. They provide essential energy for plants and algae (Gray, 1989; Howe et al., 2003). Some essential metabolic pathways are also located in this cellular compartment (Tetlow et al., 2009). Chloroplasts are derived from an ancestral endosymbiosis event between a cyanobacteria and a nonphotosynthetic host cell (Dyall et al., 2004). This organelle has its own genome, the plastome, which has evolved from the genome of the bacterial ancestor. The chloroplast genome still encodes part of the plastid protein components; the rest of the genes have been transferred to the nucleus during evolution (Martin et al., 1998). The plastome is a circular double-stranded DNA molecule present in multiple copies per organelle (Green, 2011). Its structure is conserved in higher plants and built most of the time of two inverted repeats (IRA and IRB of 20 to 28 kb in size) separated by two unique sequences named small single-copy (SSC, 16–27 kb) region and large single-copy (LSC, 80–90 kb) region. A comparison of available higher plant plastome sequences reveals a significant conservation in terms of gene order, gene content (~ 130 genes) and genome organization (Palmer and Stein, 1986); GC content (30%–40%); and genome size, with few exceptions (120–160 kb) (Chumley et al., 2006). Chloroplast genome sequencing has contributed to the study of the primary function of this compartment, the photosynthetic process (Leister and Schneider, 2003). Chloroplast DNA sequences are also widely used in the reconstruction of evolutionary relationships among plants and population genetics. These studies include phylogeny (Nie et al., 2012) and phylogeography (Pouget et al., 2013; Fan et al., 2013; Hodel and Gonzales, 2013) that look at the geographical distribution of species in relation with their genealogy (Hickerson et al., 2010).
The alpine plant Arabis alpina (alpine rock-cress) is a thoroughly studied species in the fields of perennial plant flowering regulation (Wang et al., 2009; Bergonzi et al., 2013), phylogeography (Koch et al., 2006; Assefa et al., 2007; Ehrich et al., 2007; Ansell et al., 2008, 2011; Karl et al., 2012), and adaptation to harsh alpine climatic conditions (Manel et al., 2010; Poncet et al., 2010; Zulliger et al., 2013). A. alpina is an arctic–alpine plant of the Brassicaceae family. Arctic–alpine plants are species growing naturally in the tundra of arctic regions and in mountains at southern latitudes. These regions represent a similar environment where plants have to face harsh climatic conditions. A. alpina has been recently studied looking for nuclear regions involved in adaptation to such an environment (Poncet et al., 2010; Zulliger et al., 2013). Alpine plants are often exposed to stress in the forms of UV, high light, wind, dryness, etc., and need to be adapted in order to maintain, for example, their photosynthetic activity in such environmental conditions (Körner, 2003). The photosynthetic apparatus is built of multiproteic complexes whose components are encoded by the nucleus and the plastome. A coordinate expression of nuclear and chloroplast genes is essential for the photosynthetic process to occur in chloroplasts. The A. alpina plastome sequence has not been reported so far. Five chloroplast genomes from plants of the Brassicaceae family are available in sequence databases (Arabidopsis thaliana, Arabis hirsuta, Brassica napus, Capsella bursa-pastoris, Draba nemorosa), but few data have been deposited for A. alpina. The only plastome sequences available for this species came from phylogeographic studies (Koch et al., 2006; Ansell et al., 2011; Karl et al., 2012), where the trnL–trnF region of the chloroplast genome was amplified by polymerase chain reaction (PCR) and sequenced in order to acquire phylogenetic data about A. alpina populations.
The first published cp genome sequence was from Nicotiana tabaccum (Shinozaki et al., 1986). Time-consuming procedures were required at that time to purify plastome DNA and to sequence generated fragments (Sato et al., 1999; Lee et al., 2006; Yukawa et al., 2006). PCR-based approaches using conserved primers to amplify plastome DNA regions have been subsequently developed, and these helped to gain new sequenced plastid genomes (Doorduin et al., 2011). These strategies have evolved recently with the emergence of next-generation sequencing (NGS) technologies opening the possibility to sequence chloroplast genomes more easily and at a lower cost (Yang et al., 2013; Wang and Messing, 2011). In order to gain a complete plastome sequence, genome assembly has been performed through, for example, contig building followed by PCR finishing (Zhang et al., 2011; Uthaipaisanwong et al., 2012; Pan et al., 2012) and/or assisted assembly based on a reference genome (Zhang et al., 2012; Nie et al., 2012; Huotari and Korpelainen, 2012; Wu et al., 2012; Hand et al., 2013).
We report the sequencing and de novo assembly of the complete A. alpina chloroplast genome. In this approach, an assembly software builds plastome fragments from sequencing reads only according to their sequence identity and paired-end relationships, if available. Detection of A. alpina plastome polymorphisms was performed through the study of 24 complete cp genomes sequenced from field-sampled individuals.
2. Materials and methods
2.1. Sampling
For chloroplast genome assembly, a reference A. alpina leaf sample was collected in the French Alps (individual 22 in Table 1). For polymorphism detection, 23 additional A. alpina leaf samples were collected in the French Alps from 12 locations (Table 1). Fresh leaves were immediately dried in silica gel. Plant material was stored in silica gel until DNA extraction was performed.
Table 1.
Location of sampling sites.
| Site | Latitude | Longitude | Individuals |
|---|---|---|---|
| 1 | 44.91643 | 6.41438 | 1,2 |
| 2 | 45.06387 | 6.38575 | 3,4 |
| 3 | 45.02676 | 6.38934 | 5,6 |
| 4 | 44.68557 | 6.98228 | 7,8 |
| 5 | 45.32995 | 5.84870 | 9,10 |
| 6 | 45.38354 | 5.81684 | 11,12 |
| 7 | 45.28696 | 5.78151 | 13,14 |
| 8 | 45.39815 | 5.89182 | 15,16 |
| 9 | 44.89122 | 5.43102 | 17,18 |
| 10 | 45.15070 | 5.61149 | 19,20 |
| 11 | 44.87973 | 5.52271 | 21,22 |
| 12 | 45.01837 | 5.57010 | 23,24 |
2.2. DNA extraction and sequencing
For each plant sample, DNA was extracted from 20 mg dried material ground into a fine powder with the Gentra Puregene Tissue Kit (Qiagen) according to the manufacturer's instructions. Two rounds of purification were performed to obtain pure DNA suitable for sequencing (A260/A280 > 1.8). DNA quality was checked using agarose gel electrophoresis, and quantification was performed using a NanoDrop Spectrophotometer (Thermo Fischer Scientific Inc.).
Total DNA samples were sequenced using Illumina technology on HiSeq2000 sequencer by DNAVision (www.dnavision.com). DNA quantities were estimated using PicoGreen (Invitrogen). Shearing of DNA was performed using a Covaris ultrasonicator (Covaris Inc.) to produce 300 bp fragments, and fragmentation quality was checked on a Bioanalyzer 2100 (Agilent Technologies). Paired-end libraries were prepared using the Mate Pair Library Preparation Kit (Illumina Inc.) and sequenced as 100 base reads. Libraries were sequenced using multiplexing on HiSeq2000 flow cell lanes (Illumina Inc.).
2.3. Chloroplast genome assembly
Prior to contig assembly of reads from the reference individual, sequence filtering was performed, and only paired-end sequences were selected at each step. After multiplexing tag removal of raw sequences, quality filtering was performed. Sequences with average Phred quality lower than 25 were discarded, and low-quality bases were trimmed from extremities (Phred score < 20). Reads shorter than 75 bases were removed in line with the size limit of the assembler used.
Reads were aligned on available Brassicaceae chloroplast genomes using BLAST software (Altschul et al., 1990), and each sequence giving a positive hit was retained from the data set as paired-end reads. The pool of reads enriched in plastome sequences was then submitted to a filtering step to reduce read redundancy. Clustering was performed using SEED 1.4.1 (Bao et al., 2011) with no mismatch, and three overhanging residues set up. The resulting processed data set was subjected to DNA fragment assembly using the WGS 6.1 assembler (wgs-assembler.sourceforge.net) (Myers et al., 2000). The default overlapper ovl was used. Software's default error rate gave good results with our data set. UtgErrorLimit was set up to 2.5 for Illumina sequences as suggested by the WGS developers.
2.4. Genome annotation and sequence alignments
Annotation of A. alpina chloroplast genome was performed on the Dual Organellar GenoMe Annotator (DOGMA) website (http://dogma.ccbb.utexas.edu) (Wyman et al., 2004). Each annotated gene was manually checked for start and stop codons or intron junctions to correct errors and ensure accurate annotation of the genome. The annotated GenBank format sequence file was used to draw the circular map using GenomeVx (http://wolfe.ucd.ie/GenomeVx) (Conant and Wolfe, 2008). A complete chloroplast genome comparison was performed with mVISTA program in LAGAN mode (http://genome.lbl.gov/vista/mvista) (Dubchak and Ryaboy, 2006) using GenBank format files as input. ClustalW2 enabled the alignment of complete chloroplast genome sequences (http://www.ebi.ac.uk/Tools/msa/clustalw2), and the tree output file was drawn using the R (R Development Core Team, 2011) package Pegas (http://cran.r-project.org/) (Paradis, 2010).
2.5. Polymorphism detection and analysis
Sequences obtained from DNA samples were processed as mentioned above for quality. Reads shorter than 50 bases were filtered and discarded. Processed sequences were then mapped on the A. alpina plastome using BWA 0.5.9 as paired-end sequences (Li and Durbin, 2009). The maximum insert size between paired-end read extremities was fixed to 500, and only the best match was selected. Variant calling was performed using SAM tools 0.1.13 (Li et al., 2009). Single-nucleotide polymorphisms (SNP) were filtered according to the following criteria: biallelic sites were selected, only variants supported by more than 95% of mapped reads were retained, and mapping quality equal to or higher than 50 (Phred score). For each individual mapping, significant indels were selected to represent the majority at the mapped position.
3. Results and discussion
3.1. A. alpina chloroplast genome assembly
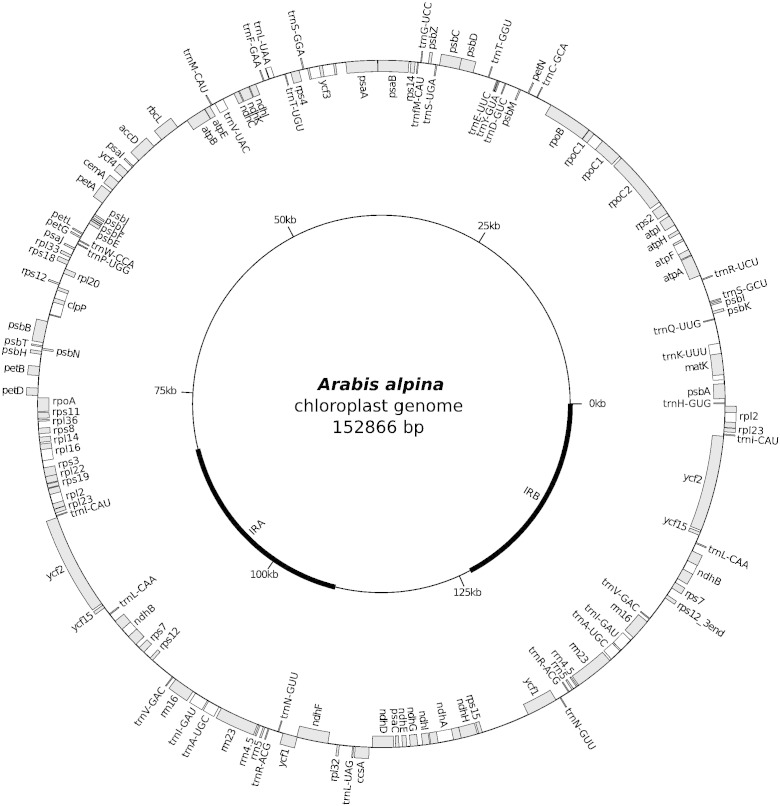
The high number of chloroplasts per leaf cell, together with the multiple copies of plastome per chloroplast, leads to a significant proportion of cpDNA in total leaf DNA. In the sample used in this work, plastome reads represented 11.4% of the high-quality reads available, which themselves corresponded to 77% of the raw sequences. The obtained filtered data set of 9.5.106 cp sequences was submitted to genome assembly but led to several contigs only partially covering the genome (81 kb). An additional step to reduce read redundancy by selecting unique sequences led a pool of 75,704 cp sequences corresponding to a 47 × coverage of the A. alpina cp genome. Assembly of these reads generated 135 kb of contigs, therefore significantly enhancing contig assembly. Contig alignment and scaffolding based on paired-end data enabled to gain a complete circular A. alpina cp genome sequence (Fig. 1) (GenBank accession number: HF934231).
Fig. 1.
Arabis alpina chloroplast genome map. Genes drawn outside of the outer circle are transcribed clockwise, while genes drawn inside the outer circle are transcribed counterclockwise. Gray boxes correspond to exons and white boxes to introns. The thick lines of the inner circle indicate the inverted repeats (IRA and IRB) separating the unique sequences SSC and LSC. GenomeVx was used to draw the map (Conant and Wolfe, 2008).
Having sequenced total DNA from 23 other A. alpina samples, a similar approach was conducted with sequences obtained from three other individuals from different locations. Independent de novo assembly for the three samples (Table 1, individuals 6, 9, and 24) led to a similar genome sequence assembly, except for some individual polymorphisms (see below). The assembly process is therefore reproducible from one individual to the other. The organization of the assembled genome is conserved when compared with other Brassicaceae cp genome using whole genome alignment (Fig. 1). Annotation of the A. alpina cp genome was successfully performed (Fig. 1). The assembled sequence enabled the consistent identification of start and stop codons or splicing junctions in agreement with data available from other plant species. Altogether, these results are in favor of an accurate A. alpina genome assembly using WGS 6.1. Other assemblers such as SOAP (Luo et al., 2012) or MIRA (http://sourceforge.net/apps/mediawiki/mira-assembler) generated partial genome assembly with contigs similar to what have been obtained with WGS 6.1 (data not shown). However, the overall assembly efficiency was lower in the conditions used.
3.2. Features of A. alpina chloroplast genome
The A. alpina plastome is 152,866 bp in length and has a GC content of 36.45% (Table 2). Available Brassicaceae cp genomes are in the range of 152,860 to 154,490 bp, and the A. alpina plastome size is therefore consistent with data from plants of the same family. The average GC content in Brassicaceae plastomes is 36.4 ± 0.1%, very close to the value measured for A. alpina. As in other angiosperms, the A. alpina cp genome is a circular DNA molecule with quadripartite organization (Fig. 1): two identical inverted repeats (IRA and IRB) of 26,933 bp (35%) and two unique sequences (the SSC region is 17,938 bp long (11%), and the LSC region is 82,338 bp long (54%)). A. alpina cp genome contains a total of 131 predicted genes (Table 3). Among them, 87 correspond to protein-encoding genes, 8 being duplicated in the IR region (ndhB, rpl2, rpl23, rps7, rps12, ycf1, ycf2, ycf15). A set of 29 tRNA genes was detected, covering all families of amino acids. Seven of them are present in two copies because of their duplication in the IR regions. Genes encoding tRNA are spread throughout the genome: 20% in IRs, 3% in SSC, and 77% in LSC. Four genes encoding rRNA were identified in the IRs regions (rrn4.5, rrn5, rrn16, rrn23) and are therefore duplicated in the genome. Ribosomal RNA genes are located in the IR in all plastomes sequenced so far. Out of the 131 genes, 18 are interrupted by introns: 11 protein-coding genes and 7 tRNA genes. The clpP and ycf3 genes contain two introns. Trans-splicing occurs for the rps12 gene. Its 5′ exon is located in the LSC region, and the 3′ exon is present in two copies because of its localization in the IR regions. An A. alpina cp genome analysis revealed that protein coding sequences, tRNA and rRNA represent 58% of the genome, divided in 86.7%, 3.1%, and 10.2%, respectively. Intergenic spacers, introns, and pseudogenes correspond to 42% of the genome.
Table 2.
General features of Arabis alpina plastome.
| Features | Chloroplast |
|---|---|
| Genome size (bp) | 152866 |
| GC content (%) | 36.45 |
| Coding sequences (%) | 58 |
| Nb of protein-coding gene | 87 |
| Nb of ribosomal RNAs | 8 |
| Nb of tRNA genes | 36 |
| Nb of genes with introns | 18 |
Table 3.
Genes present in the Arabis alpina chloroplast genome.
| Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Cytochrome b6/f | petA, petB, petD, petG, petL, petN |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI |
| Rubisco | rbcL |
| NADH oxidoreductase | ndhA, ndhB (2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Large subunit ribosomal proteins | rpl 2 (2), rpl14, rpl16, rpl20, rpl22, rpl 23 (2), rpl 32, rpl33, rpl36 |
| Small subunit ribosomal proteins | rps2, rps3, rps4, rps7 (2), rps8, rps11, rps12 (2), rps14, rps15, rps18, rps19 |
| RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 |
| Other proteins | matK, accD, cemA, clpP, ccsA |
| Proteins of unknown function | ycf1 (2), ycf2 (2), ycf3, ycf4, ycf15 (2) |
| Ribosomal RNAs | rrn4.5 (2), rrn5 (2), rrn16 (2), rrn23 (2) |
| Transfer RNAs | trnA-UGC (2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-UCC, trnH-GUG, trnI-CAU (2), trnI-GAU (2), trnK-UUU, trnL-CAA (2), trnL-UAA, trnL-UAG, trnfM-CAU, trnM-CAU, trnN-GUU (2), trnP-UGG, trnQ-UUG, trnS-GCU, trnS-UGA, trnT-GGU, trnR-UCU, trnR-ACG (2), trnS-GGA, trnT-UGU, trnV-UAC, trnV-GAC (2), trnW-CCA, trnY-GUA |
3.3. Comparison with other Brassicaceae cp genomes
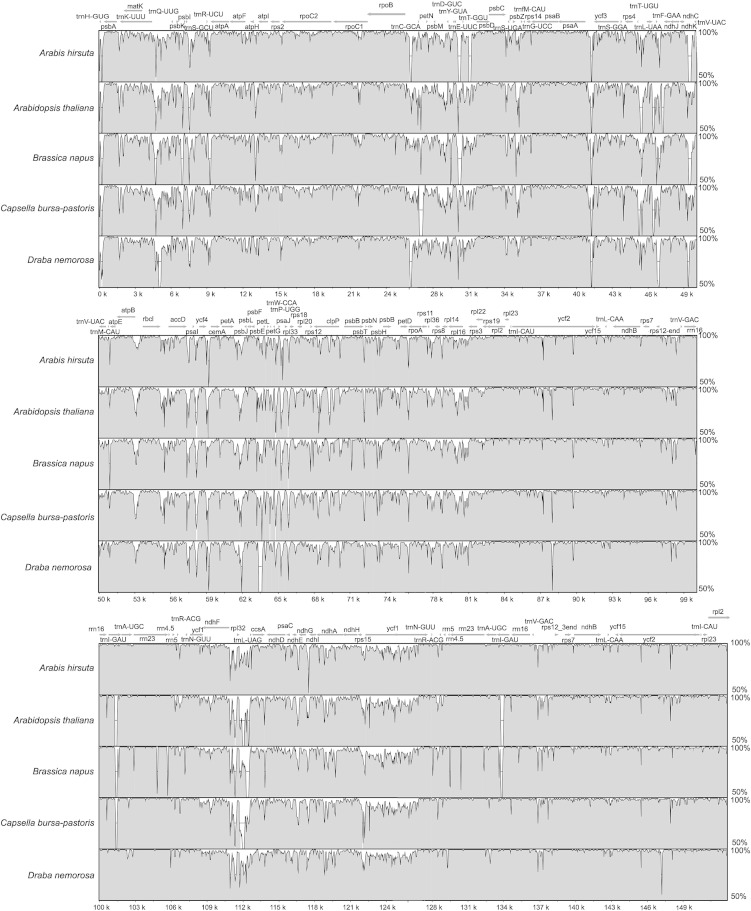
The availability of multiple complete Brassicaceae cp genomes provided the opportunity to compare the A. alpina plastome with these genomes (Fig. 2). Alignments and global sequence identity in a range of 93%–97% indicated a high conservation of cp genome sequences in this plant family. As shown in Fig. 2, IR appeared more conserved than unique sequences, as detected previously in other plant families (Doorduin et al., 2011). Variations were more abundant in noncoding regions than in coding regions. In size, the A. alpina cp genome is one of the smallest genomes of the Brassicaceae. Genome alignments revealed two deletions in the A. alpina plastome. The rps16 gene was not detected during the annotation step, which is also the case for D. nemorosa. The chloroplast protein synthesis apparatus is very similar to the bacterial machinery. Many ribosomal proteins are homologous to those of Escherichia coli. The RPS16 protein is essential for E. coli viability (Persson et al., 1995) and is involved in 30S ribosomal subunit assembly (Held and Nomura, 1975). This gene is also absent from the Populus alba and Medicago truncatula cp genomes (Ueda et al., 2008). In these species, Ueda et al. (2008) have shown a dual targeting of the nuclear RPS16 product to the mitochondria and the chloroplast. Such a feature is not specific to these species since in A. thaliana, for example, whose plastome contains the rps16 gene, the nuclear rps16 gene encodes a protein with the ability to be targeted into the two plant organelles (Ueda et al., 2008). It is therefore likely that in A. alpina, the absence of cp rps16 gene is rescued by the import into the chloroplast of a nuclear rps16 gene product. Another deletion of 444 bp in the A. alpina plastome was detected in comparison with the A. thaliana, C. bursa-pastoris, and B. napus cp genomes. The deletion is shared by the D. nemorosa and A. hirsuta plastomes and is located in the intergenic region between the psbE and petL genes.
Fig. 2.
Graphic view of the alignment of the six available Brassicaceae chloroplast genomes. VISTA identity plot of sequence identity between the Arabis alpina, Arabidopsis thaliana (NC_000932), Arabis hirsuta (NC_009268), Draba nemorosa (NC_009272), Brassica napus (NC_016734), Capsella bursa-pastoris (NC_009270) plastomes. Sequence identity varying between 50% and 100% are drawn on the y axis of the plot. The x axis corresponds to the coordinates on the Arabis alpina chloroplast genome. Arrows indicate the annotated genes and their transcriptional direction.
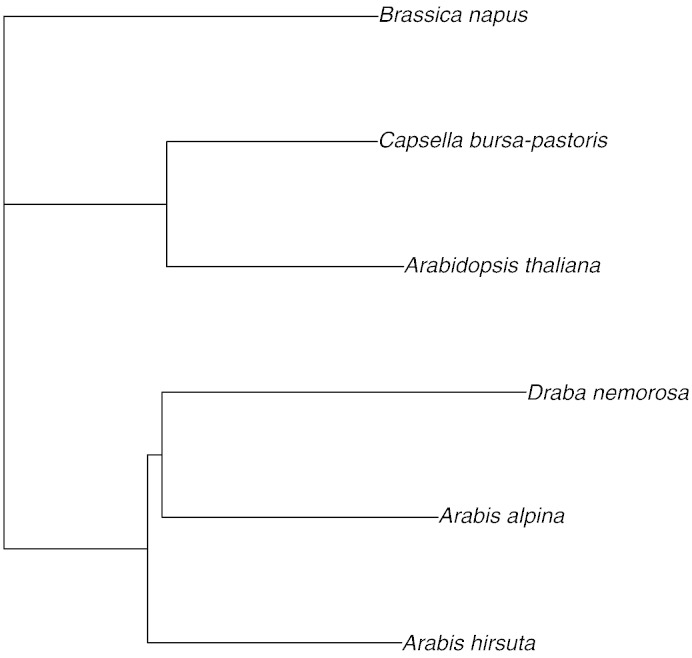
A phylogenetic tree based on the whole Brassicaceae cp genome sequence alignment indicated a phylogenetic position of A. alpina in a group together with A. hirsuta and D. nemorosa (Fig. 3) that is closer to this latter species, which shares the rps16 gene deletion.
Fig. 3.
Complete chloroplast genome phylogeny of Brassicaceae. Plastome sequences were aligned using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2), and a tree was drawn using the R package Pegas (Paradis, 2010).
3.4. Polymorphic loci detection
In order to investigate intraspecies polymorphisms, 23 additional A. alpina total DNA samples were sequenced, and reads were mapped on the assembled A. alpina plastome as a reference. These samples represent local variability analysis of plants sampled in the French Alps in an area of approximately 80 km2. Significant coverage throughout the genome was obtained for each individual; the lowest coverage for a SNP in this study was 53 ×. Forty-four indels were detected: only one was located in a putative coding region (ycf1 position 126648). The 24 individuals' genome mapping also yielded a total of 130 SNPs, 37 of them being located in protein coding regions. These SNPs were shared by at least two individuals. Out of the 130 SNPs detected, 32% corresponded to transitions and 68% to transversions. The transition–transversion ratio (R) was therefore 0.47, indicating no bias between transition and transversion for the detected mutations. The transition–transversion bias has been documented in Drosophila and mammals sequence analysis (Chen et al., 2009; Seplyarskiy et al., 2012). It was considered as being a consequence of the chemical basis of mutations. However, Keller et al. (2007) showed that this bias does not apply, for example, to grasshopper pseudogenes. Some transition–transversion bias has been detected in chloroplast genes (Morton et al., 1997; Guhamajumdar and Sears, 2005) but was not detected in cp genomes from the Lemnoideae family (Wang and Messing, 2011) or A. alpina in this work. Mutations detected in the A. alpina cp genome were mainly located in the unique regions, LSC and SSC, with an average of one SNP per 808 bp. As mentioned above for interspecies sequence comparison, IR regions appeared more conserved than the unique region of the cp genome. SNPs are more abundant in noncoding regions in agreement with a higher evolution rate in such sequences. Table 4 summarizes the localization of SNPs that were found in coding regions. Out of the 37 positions detected, 35% correspond to synonymous substitutions. A significant rate of nonsynonymous mutations was therefore measured, leading to functional variants of different chloroplast proteins. All mutations in genes encoding photosynthetic proteins were synonymous (Table 4), which could reflect a selective pressure on these sequences encoding functional polypeptides of photosystems. Nonsynonymous substitutions occur, for example, in ribosomal protein or RNA polymerase subunit genes.
Table 4.
List of SNPs identified in Arabis alpina plastome genes.
| Position | Protein | Base change | Amino acid | Function |
|---|---|---|---|---|
| 6543 | psbK | T/C | A/A | Photosystem II |
| 11245 | atpF | G/T | H/Q | ATP synthase |
| 14350 | rps2 | C/G | E/D | Small subunit ribosomal protein |
| 15306 | rpoC2 | A/G | F/C | RNA polymerase |
| 16295 | G/A | N/N | ||
| 16474 | G/A | P/A | ||
| 17307 | A/C | I/R | ||
| 20512 | rpoC1 | T/C | L/L | RNA polymerase |
| 22005 | C/T | G/S | ||
| 24729 | rpoB | T/C | K/K | RNA polymerase |
| 34855 | psbZ | C/T | F/F | Photosystem II |
| 39586 | psaA | C/T | L/L | Photosystem I |
| 47964 | ndhK | A/C | S/A | NADH oxidoreductase |
| 58892 | ycf4 | G/A | A/T | Protein of unknown function |
| 59442 | cemA | G/T | W/L | Chloroplast envelope membrane protein |
| 60485 | petA | C/A | V/V | Cytochrome B6/f |
| 66051 | rps18 | A/C | I/I | Small subunit ribosomal protein |
| 66267 | G/T | T/T | ||
| 66674 | rpl20 | G/T | L/I | Large subunit ribosomal protein |
| 66794 | T/C | S/G | ||
| 69066 | clpP | A/G | F/F | Protease |
| 71867 | psbB | C/T | S/S | Photosystem II |
| 76906 | rpoA | C/T | R/K | RNA Polymerase |
| 78533 | rps8 | C/T | D/N | Small subunit ribosomal protein |
| 81021 | rps3 | G/T | G/G | Small subunit ribosomal protein |
| 81699 | rpl22 | C/T | C/Y | Large subunit ribosomal protein |
| 81890 | G/T | F/L | ||
| 85215 | ycf2 | G/T | C/F | Proteins of unknown function |
| 108838 | ndhF | T/A | I/M | NADH oxidoreductase |
| 110679 | A/C | Y/D | ||
| 113079 | ccsA | T/G | F/L | Cytochrome c biogenesis protein |
| 118680 | ndhA | G/A | N/N | NADH oxidoreductase |
| 121810 | rps15 | T/C | R/G | Small subunit ribosomal protein |
| 125941 | ycf1 | G/A | S/L | Protein of unknown function |
| 125954 | C/T | E/K | ||
| 126332 | T/C | T/A | ||
| 149991 | ycf2 | C/A | C/F | Protein of unknown function |
4. Conclusion
Using NGS, the A. alpina chloroplast genome was sequenced, revealing the structure and organization of the plastome of this alpine plant. Through complete chloroplast genome sequencing, new potential markers, including indels and SNPs, were identified. These data will represent a valuable source of markers in future studies about A. alpina populations. We found more polymorphic sites in noncoding regions that also contained most of the indels, with selective pressure being usually higher in coding regions. Polymorphism data are therefore available for genome regions having different evolutionary rates that could help to analyze both recent and more ancient diversifications. In addition, complete cp genome sequence also provides data about functional protein variability in chloroplasts.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Christelle Melodelima, Email: christelle.melo-de-lima@ujf-grenoble.fr.
Stéphane Lobréaux, Email: stephane.lobreaux@ujf-grenoble.fr.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ansell S.W., Grundmann M., Russell S.J., Schneider H., Vogel J.C. Genetic discontinuity, breeding-system change and population history of Arabis alpina in the Italian Peninsula and adjacent Alps. Mol. Ecol. 2008;17:2245–2257. doi: 10.1111/j.1365-294X.2008.03739.x. [DOI] [PubMed] [Google Scholar]
- Ansell S.W., Stenøien H.K., Grundmann M., Russell S.J., Koch M.A., Schneider H., Vogel J.C. The importance of Anatolian mountains as the cradle of global diversity in Arabis alpina, a key arctic–alpine species. Ann. Bot. 2011;108:241–252. doi: 10.1093/aob/mcr134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefa A., Ehrich D., Taberlet P., Nemomissa S., Brochmann C. Pleistocene colonization of afro-alpine ‘sky islands’ by the arctic–alpine Arabis alpina. Heredity. 2007;99:133–142. doi: 10.1038/sj.hdy.6800974. [DOI] [PubMed] [Google Scholar]
- Bao E., Jiang T., Kaloshian I., Girke T. SEED: efficient clustering of next-generation sequences. Bioinformatics. 2011;27:2502–2509. doi: 10.1093/bioinformatics/btr447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergonzi S., Albani M.C., Loren Ver, van Themaat E. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science. 2013;340:1094–1097. doi: 10.1126/science.1234116. [DOI] [PubMed] [Google Scholar]
- Chen J.Q., Wu Y., Yang H., Bergelson J., Kreitman M., Tian D. Variation in the ratio of nucleotide substitution and indel rates across genomes in mammals and bacteria. Mol. Biol. Evol. 2009;26:1523–1531. doi: 10.1093/molbev/msp063. [DOI] [PubMed] [Google Scholar]
- Chumley T.W., Palmer J.D., Mower J.P. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24:861–862. doi: 10.1093/bioinformatics/btm598. [DOI] [PubMed] [Google Scholar]
- Doorduin L., Gravendeel B., Lammers Y., Ariyurek Y., Chin-A-Woeng T., Vrieling K. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 2011;18:93–105. doi: 10.1093/dnares/dsr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak I., Ryaboy D.V. VISTA family of computational tools for comparative analysis of DNA sequences and whole genomes. Methods Mol. Biol. 2006;338:69–89. doi: 10.1385/1-59745-097-9:69. [DOI] [PubMed] [Google Scholar]
- Dyall S.D., Brown M.T., Johnson P.J. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- Ehrich D., Gaudeul M., Assefa A. Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Mol. Ecol. 2007;16:2542–2559. doi: 10.1111/j.1365-294X.2007.03299.x. [DOI] [PubMed] [Google Scholar]
- Fan D.M., Yue J.P., Nie Z.L., Li Z.M., Comes H.P., Sun H. Phylogeography of Sophora davidii (Leguminosae) across the ‘Tanaka-Kaiyong Line’, an important phytogeographic boundary in Southwest China. Mol. Ecol. 2013;22:4270–4288. doi: 10.1111/mec.12388. [DOI] [PubMed] [Google Scholar]
- Gray M.W. The evolutionary origins of organelles. Trends Genet. 1989;5:294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- Green B.R. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66:34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]
- Guhamajumdar M., Sears B.B. Chloroplast DNA base substitutions: an experimental assessment. Mol. Genet. Genomics. 2005;273:177–183. doi: 10.1007/s00438-005-1121-1. [DOI] [PubMed] [Google Scholar]
- Hand M.L., Spangenberg G.C., Forster J.W., Cogan N.O. Genes Genomes Genetics; 2013. Plastome Sequence Determination and Comparative Analysis for Members of the Lolium–Festuca Grass Species Complex. (pii, g3.112.005264v1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held W.A., Nomura M. Escherichia coli 30S ribosomal proteins uniquely required for assembly. J. Biol. Chem. 1975;250:3179–3184. [PubMed] [Google Scholar]
- Hickerson M.J., Carstens B.C., Cavender-Bares J., Crandall K.A., Graham C.H., Johnson J.B., Rissler L., Victoriano P.F., Yoder A.D. Phylogeography's past, present, and future: 10 years after Avise, 2000. Mol. Phylogenet. Evol. 2010;54:291–301. doi: 10.1016/j.ympev.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Hodel R.G., Gonzales E. Phylogeography of Sea Oats (Uniola paniculata), a dune-building coastal grass in Southeastern North America. J. Hered. 2013;104:656–665. doi: 10.1093/jhered/est035. [DOI] [PubMed] [Google Scholar]
- Howe C.J., Barbrook A.C., Koumandou V.L., Nisbet R.E., Symington H.A., Wightman T.F. Evolution of the chloroplast genome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:99–106. doi: 10.1098/rstb.2002.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari T., Korpelainen H. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene. 2012;508:96–105. doi: 10.1016/j.gene.2012.07.020. [DOI] [PubMed] [Google Scholar]
- Karl R., Kiefer C., Ansell S.W., Koch M.A. Systematics and evolution of Arctic–Alpine Arabis alpina (Brassicaceae) and its closest relatives in the eastern Mediterranean. Am. J. Bot. 2012;99:778–794. doi: 10.3732/ajb.1100447. [DOI] [PubMed] [Google Scholar]
- Keller I., Bensasson D., Nichols R.A. Transition–transversion bias is not universal: a counter example from grasshopper pseudogenes. PLoS Genet. 2007;3:e22. doi: 10.1371/journal.pgen.0030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Kiefer C., Ehrich D., Vogel J., Brochmann C., Mummenhoff K. Three times out of Asia Minor: the phylogeography of Arabis alpina L. (Brassicaceae) Mol. Ecol. 2006;15:825–839. doi: 10.1111/j.1365-294X.2005.02848.x. [DOI] [PubMed] [Google Scholar]
- Körner C. Springer; Heidelberg: 2003. Alpine Plant Life: Functional Plant Ecology of High Mountain Ecosystems. [Google Scholar]
- Lee S.B., Kaittanis C., Jansen R.K., Hostetler J.B., Tallon L.J., Town C.D., Daniell H. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics. 2006;23:61. doi: 10.1186/1471-2164-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D., Schneider A. From genes to photosynthesis in Arabidopsis thaliana. Int. Rev. Cytol. 2003;228:31–83. doi: 10.1016/s0074-7696(03)28002-5. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Subgroup G.P.D.P. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y. SOAPdenovo2: an empirically improved memory-efficient short read de novo assembler. Giga Sci. 2012;1:1–18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel S., Poncet B.N., Legendre P., Gugerli F., Holderegger R. Common factors drive adaptive genetic variation at different spatial scales in Arabis alpina. Mol. Ecol. 2010;19:3824–3835. doi: 10.1111/j.1365-294X.2010.04716.x. [DOI] [PubMed] [Google Scholar]
- Martin W., Stoebe B., Goremykin V., Hapsmann S., Hasegawa M., Kowallik K.V. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- Morton B.R., Oberholzer V.M., Clegg M.T. The influence of specific neighboring bases on substitution bias in noncoding regions of the plant chloroplast genome. J. Mol. Evol. 1997;45:227–231. doi: 10.1007/pl00006224. [DOI] [PubMed] [Google Scholar]
- Myers E.W., Sutton G.G., Delcher A.L. A whole-genome assembly of Drosophila. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- Nie X., Lv S., Zhang Y., Du X., Wang L., Biradar S.S., Tan X., Wan F., Weining S. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora) PLoS One. 2012;7:e36869. doi: 10.1371/journal.pone.0036869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J.D., Stein D.B. Conservation of chloroplast genome structure among vascular plants. Curr. Genet. 1986;10:823–833. [Google Scholar]
- Pan I.C., Liao D.C., Wu F.H. Complete chloroplast genome sequence of an orchid model plant candidate: Erycina pusilla apply in tropical oncidium breeding. PLoS ONE. 2012;7(4):e34738. doi: 10.1371/journal.pone.0034738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E. Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- Persson B.C., Bylund G.O., Berg D.E., Wikstrom P.M. Functional analysis of the ffh–trmD region of the Escherichia coli chromosome by using reverse genetics. J. Bacteriol. 1995;177:5554–5560. doi: 10.1128/jb.177.19.5554-5560.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet B.N., Herrmann D., Gugerli F. Tracking genes of ecological relevance using a genome scan in two independent regional population samples of Arabis alpina. Mol. Ecol. 2010;19:2896–2907. doi: 10.1111/j.1365-294X.2010.04696.x. [DOI] [PubMed] [Google Scholar]
- Pouget M., Youssef S., Migliore J., Juin M., Médail F., Baumel A. Phylogeography Sheds Light on the Central-Marginal Hypothesis in a Mediterranean Narrow Endemic Plant. Ann. Bot. 2013;112:1409–1420. doi: 10.1093/aob/mct183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . 2011. R: A Language and Environment for Statistical Computing. (Vienna, Austria. URL http://www.R-project.org) [Google Scholar]
- Sato S., Nakamura Y., Kaneko T., Asamizu E., Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- Seplyarskiy V.B., Kharchenko P., Kondrashov A.S., Bazykin G.A. Heterogeneity of the transition/transversion ratio in Drosophila and Hominidae genomes. Mol. Biol. Evol. 2012;29:1943–1955. doi: 10.1093/molbev/mss071. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlow I.J., Rawsthorne S., Raines C., Emes M.J. Plastid Metabolic Pathways. In: Moller S.G., editor. Annual Plant Reviews, Plastids. vol. 13. Blackwell Publishing; 2009. pp. 60–125. [Google Scholar]
- Ueda M., Nishikawa T., Fujimoto M., Takanashi H., Arimura S., Tsutsumi N., Kadowaki K. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 2008;25:1566–1575. doi: 10.1093/molbev/msn102. [DOI] [PubMed] [Google Scholar]
- Uthaipaisanwong P., Chanprasert J., Shearman J.R., Sangsrakru D., Yoocha T., Jomchai N., Jantasuriyarat C., Tragoonrung S., Tangphatsornruang S. Characterization of the chloroplast genome sequence of oil palm (Elaeis guineensis Jacq.) Gene. 2012;500:172–180. doi: 10.1016/j.gene.2012.03.061. [DOI] [PubMed] [Google Scholar]
- Wang W., Messing J. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS One. 2011;6:e24670. doi: 10.1371/journal.pone.0024670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Farrona S., Vincent C. PEP1 regulates perennial flowering in Arabis alpina. Nature. 2009;459:423–427. doi: 10.1038/nature07988. [DOI] [PubMed] [Google Scholar]
- Wu J., Liu B., Cheng F., Ramchiary N., Choi S.R., Lim Y.P., Wang X.W. Sequencing of chloroplast genome using whole cellular DNA and solexa sequencing technology. Front. Plant. Sci. 2012;3:243. doi: 10.3389/fpls.2012.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S.K., Jansen R.K., Boore J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Yang J.B., Tang M., Li H.T., Zhang Z.R., Li D.Z. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013;13:84. doi: 10.1186/1471-2148-13-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukawa M., Tsudzuki T., Sugiura M. The chloroplast genome of Nicotiana sylvestris and Nicotiana tomentosiformis: complete sequencing confirms that the Nicotiana sylvestris progenitor is the maternal genome donor of Nicotiana tabacum. Mol. Genet. Genomics. 2006;275:367–373. doi: 10.1007/s00438-005-0092-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y.J., Ma P.F., Li D.Z. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae) PLoS One. 2011;6:e20596. doi: 10.1371/journal.pone.0020596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Fang Y., Wang X., Deng X., Zhang X., Hu S., Yu J. The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: insights into the evolution of plant organellar genomes. PLoS One. 2012;7:e30531. doi: 10.1371/journal.pone.0030531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulliger D., Schnyder E., Gugerli F. Are adaptive loci transferable across genomes of related species? Outlier and environmental association analyses in Alpine Brassicaceae species. Mol. Ecol. 2013;22:1626–1639. doi: 10.1111/mec.12199. [DOI] [PubMed] [Google Scholar]