Abstract
Classical research has suggested that early palate formation develops via epithelial-mesenchymal interactions, and in this study we reveal which signals control this process. Using Fgf10–/–, FGF receptor 2b–/– (Fgfr2b–/–), and Sonic hedgehog (Shh) mutant mice, which all exhibit cleft palate, we show that Shh is a downstream target of Fgf10/Fgfr2b signaling. Our results demonstrate that mesenchymal Fgf10 regulates the epithelial expression of Shh, which in turn signals back to the mesenchyme. This was confirmed by demonstrating that cell proliferation is decreased not only in the palatal epithelium but also in the mesenchyme of Fgfr2b–/– mice. These results reveal a new role for Fgf signaling in mammalian palate development. We show that coordinated epithelial-mesenchymal interactions are essential during the initial stages of palate development and require an Fgf-Shh signaling network.
Introduction
Isolated cleft palate is a common congenital craniofacial birth defect, occurring once in every 1,000 births. Cleft palate as an isolated malformation is distinct from cleft lip with or without cleft palate, which has a higher incidence (1:700) (1). Cleft palate associated with other abnormalities as part of a syndrome account for 55% of cases (2). Higher incidences of cleft palate are reported in offspring of individuals with cleft palate as part of a syndrome than in those in families with no obvious syndrome (3). Mutations causing clefting have been found in a variety of genes including transcription factors, growth factor receptors, extracellular matrix components, and cell surface adhesion molecules (4). Mutations in FGF receptor 2 (FGFR2) and in TWIST, a transcription factor involved in FGF signaling, cause the craniosynostosis syndromes Apert and Saethre-Chotzen, respectively (4–6). Both of these syndromes are characterized by cleft palate; Kreiborg and Cohen (1992) have described 76% of Apert syndrome patients exhibiting cleft palate. Almost all patients had a Byzantine arch–shaped palate (7).
The mammalian secondary palate forms from outgrowths of the maxillary processes, which start to develop at embryonic day 12 (E12) in the mouse. These bilateral projections form shelves that grow downward between the developing tongue and the lower jaw. At E14 they elevate to a horizontal position above the tongue, where they approximate and fuse (8). They also fuse with the primary palate and thereby form a barrier between the oral and nasal cavities, which allows breathing and feeding to continue at the same time. Failure of any of these processes can result in a hole or cleft between the mouth and nose.
Soluble growth factors play key roles in reciprocal and reiterative signaling, which control the development of many organs through tissue-tissue interactions (9). Classical experiments suggest that epithelial-mesenchymal interactions play key roles during palate development. However, experimental studies have focused mostly on the events of palatal fusion (10, 11). A recent publication by Zhang et al. (12) showed that bone morphogenetic protein/Msx1 (Bmp/Msx1) signaling has a role in early palate development. Despite members of the Fgf gene family playing important roles in the development of embryonic outgrowths, including the early facial primordia, no investigations have been conducted on Fgf signaling during palatal shelf development (13).
In order to study the role of Fgf signaling in early palate development, we investigated Fgfr2b (IIIb isoform)–null mice and also mice lacking Fgf10, which is a ligand for Fgfr2b (14). In the absence of Fgfr2b, Fgf10 could bind to Fgfr1b. Not only has binding affinity been demonstrated, but activation of Fgfr1b by Fgf10 has also been shown to stimulate mitogenesis (15). We report that, in addition to Fgfr2b–/– mice (16), Fgf10–/– mice also exhibited a cleft palate. Both mutants showed a disruption in palatogenesis during the initial stages of development prior to shelf elevation. Here we show that Fgfr2b was expressed in the developing palatal epithelium and that Fgf10 transcripts were localized in the adjacent underlying mesenchyme, indicating that epithelial-mesenchymal interactions are involved in growth control during normal palatogenesis.
We also show that the epithelium of both mutants was thin due to a severe reduction in cell proliferation. There was also a reduction in cell proliferation in the mesenchyme. As the Fgf10/Fgfr2b signaling affects the epithelium, the proliferation defects in the mesenchyme were secondary to the lack of reciprocal epithelial signals. Sonic hedgehog (Shh) expression in the epithelium was downregulated in both Fgfr2b–/– and Fgf10–/– mice. In vitro experiments showed that recombinant FGF10 protein induced both proliferation as well as Shh expression in the palatal epithelium and that it also induced the hedgehog receptor Patched 1 (Ptc1) in the adjacent underlying mesenchyme. We suggest that Shh acts downstream of Fgf10/Fgfr2b signaling and stimulates mesenchymal proliferation. In support of this hypothesis, we showed that exogenous Shh peptide induced proliferation in isolated palatal mesenchymal explants and that targeted inactivation of Shh in the oral epithelium led to a similar cleft phenotype to that of Fgfr2b–/– and Fgf10–/– mutants. Interestingly, lack of Smoothened (Smo) in the epithelium did not disturb palatal development. As Shh signals through Smo, this confirms that the target tissue of Shh is not the epithelium but the mesenchyme.
Taken together, our results provide molecular evidence that reciprocal epithelial-mesenchymal interactions are essential in the growth and morphogenesis of the developing palate and that these interactions are under the control of Fgf10/Fgfr2b signaling.
Results
Fgfr2b–/– and Fgf10–/– mice exhibit cleft palate
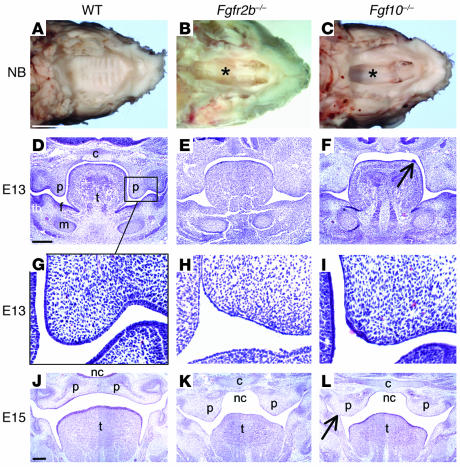
A total of 51 Fgfr2b–/– and 33 Fgf10–/– mice between the ages E11 and newborn (NB) were compared with a similar number of WT littermates. All Fgfr2b–/– and Fgf10–/– mutants studied exhibited a complete, wide cleft of the secondary palate (Figure 1, A–C). Abnormalities in the palate were first noted at E13. The mutant palates were short and squared in shape in comparison to WT and did not have “finger-like” palatal projections lateral to the tongue with a characteristic bend between the medial edge and the developing molar tooth (Figure 1, D–F). In the mutants, the palatal epithelium was thin and lacked stratification (Figure 1, G–I). In some instances, the epithelial covering at the tip of the palatal projection (medial epithelial edge, MEE) had disintegrated, and underlying mesenchymal cells had broken through into the oral cavity (data not shown). Partial ankylosis of the tongue was also evident, with defective epithelialization between the floor of the mouth and the tongue (Figure 1, J–L). Also, epithelial fusions were noted between the nasal surface of the palate and the tongue, as well as the oral surface of the palate and the mandible (Figure 1L). More evidence of a disordered epithelium in the mutant was exemplified by the occurrence of isolated epithelial outgrowths on the dorsal surface of the tongue. These were more prevalent in Fgf10–/– mice (Figure 1F). By E15 the palatal shelves were positioned above the tongue and had fused in the WT mice, but they were widely spaced in both mutants (Figure 1, J–L). At birth, the palatal shelves appeared to have regressed in size (Figure 1, A–C). Due to their abnormal morphology and size, the palatal shelves had no possibility of meeting and fusing. In addition, the mutant palatal shelves lacked rugae (Figure 1, A–C).
Figure 1.
Palatal abnormalities in Fgfr2b_/_ and Fgf10_/_ mice. (A_C) View of the palate with mandible removed from NB mice. (A) WT palate was completely closed with clear symmetrical rugae. (B and C) Fgfr2b_/_ and Fgf10_/_ mutants exhibited a similar wide cleft of the secondary palate, with a view into the nasal cavity (asterisks). (D_L) E13 and E15 hematoxylin stained coronal sections through the oral cavity. (G_I) Higher-magnification views of D_F. By E13 the epithelium of WT mice had begun to thicken and stratify into a squamous pattern. That of both mutants was thin and lacked organization. Occasional patches of thicker epithelium did form, notably in Fgf10_/_ mice either at the MEE (I) or as outgrowths from the dorsum of the tongue (F, arrow). However, cells in these patches underwent apoptosis (see Figure 4). (K and L) Palatal shelves were positioned above the tongue but neither met nor fused. (L) Epithelial fusion between the palatal shelf and mandible (arrow). C, cranial base; f, floor of the mouth; m, Meckel’s cartilage; nc, nasal cavity; p, palatal shelf; t, tongue; tb, molar tooth bud. Scale bars: A, 2 mm (A_C, same magnification); D, 200 ∝m (D_F, same magnification); and J, 200 ∝m (J_L, same magnification).
Expression of Fgfr2b, Fgf10, and Fgf7 transcripts during palate development
Fgfr2b transcripts were detected in the developing oral epithelium (E12–14) particularly in the palate, floor of the mouth, and the tongue (Figure 2, A, D, and G). At E13 Fgfr2b mRNA was also detected in the mesenchyme of the nasal side of the palate, but at a much lower level (Figure 2D). Fgf10 transcripts were localized from E12 to E14 in the mesenchyme of the palate, floor of mouth, and tongue. These locations were directly adjacent to the epithelium, where Fgfr2b was expressed, suggesting epithelial-mesenchymal interactions (Figure 2, B, E, and H). In the palate, Fgf10 was mainly expressed in mesenchyme adjacent to the MEE and the oral epithelial surface. Fgf7 displayed a similar expression pattern to that of Fgf10, though in the palate it was mainly localized on the nasal rather than oral aspect (Figure 2, C, F, and I). Interestingly, Fgf10 and Fgf7, both ligands of Fgfr2b, were colocalized in the region of the “bend” between the palatal shelf and the cranial base (Figure 2, E and F). Fgf3, another ligand of Fgfr2b, was not detected in the developing palate (data not shown).
Figure 2.
Detection of Fgfr2b, Fgf10, and Fgf7 mRNA during palatogenesis by in situ hybridization. (A) At E12, Fgfr2b was expressed throughout the oral epithelium. (B) Fgf10 was detected in mesenchyme immediately adjacent to the epithelium in the early palatal outgrowths, the tongue, and the floor of the mouth. (D, E, G, and H) This pattern continued at E13 and E14, with Fgfr2b being strongly expressed in the epithelium of the palate and floor of the mouth. Fgf10 was mainly expressed in mesenchyme underlying the MEE and the oral epithelial surface of the palate. (D) Fgfr2b (weakly), (E) Fgf10, and (F) Fgf7 transcripts were also detected in the mesenchyme in the bend region between the palatal shelf and the cranial base (arrows) (arrowhead indicates the anterior-posterior groove between the palatal process and the body of the maxilla). (C and F) At E12 and E13 Fgf7 was mainly expressed in the palatal mesenchyme immediately adjacent to the future nasal epithelium. (I) After shelf elevation, Fgf7 was also expressed in the mesenchyme adjacent to the oral epithelium. f, floor of the mouth. All images are the same magnification; scale bar: 200 ∝m.
Cell proliferation is reduced in the developing palate of Fgfr2b–/– and Fgf10–/– mice
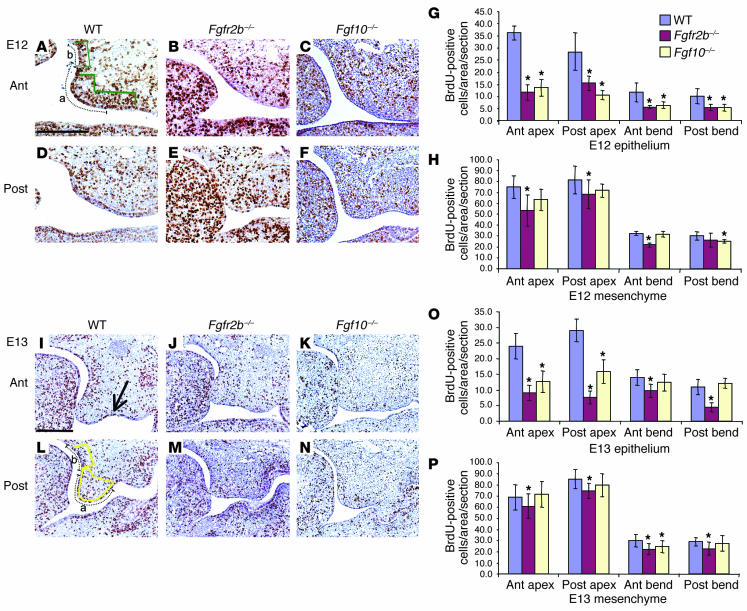
We further investigated the pathogenesis of the cleft palates in Fgfr2b–/– and Fgf10–/– mice by performing a detailed survey of cell turnover by BrdU incorporation and TUNEL staining. We chose two stages to investigate: E12, when palatal shelf growth has just begun, and E13, just prior to shelf elevation but when the shelves are still elongating. It has been reported that some of the genes involved in palatal development are differentially expressed in an anterior-posterior fashion (12). Therefore, sections from both anterior and posterior (at the level of the first lower molar tooth) regions were analyzed. Our in situ hybridization analysis showed that Fgf10 and Fgf7 were colocalized to a region of mesenchyme directly below the epithelium at the curved junction between nasal surface of the palatal shelf and the cranial base (Figure 2, E and F). This bend region was therefore assayed in addition to the apex of the palatal shelf itself.
Comparison of anterior and posterior palatal cell proliferation in WT mice.
At E12, cell proliferation was greater in the posterior apex epithelium (mean = 36.0) compared to the anterior apex epithelium (mean = 28.3, P = 0.07). At E13, proliferation was significantly greater (P < 0.0005) in the posterior apex epithelium (mean = 29.1) compared with the anterior apex (mean = 24.0), assessed by the mean number of BrdU-positive cells in the 20 sections assayed for each region (Figure 3O, column 1 vs. column 4). This was reflected in the mesenchyme with proliferation in the posterior apex (mean = 85.4) being greater (P < 0.0005) than the anterior (mean = 68.7) (Figure 3P, column 1 vs. column 4). In the bend region, epithelial proliferation was greater (P < 0.0005) in the anterior region (mean = 14.0) compared with the posterior (mean = 11.0) (Figure 3O, column 7 vs. column 10). In the bend mesenchyme no difference was recorded.
Figure 3.
Cell proliferation in the developing palate in WT, Fgfr2b_/_, and Fgf10_/_ mice. (A_H) BrdU analysis of E12 palate. (A_F) Histological sections of BrdU staining. Compared to that in WT animals, proliferation in Fgfr2b_/_ and Fgf10_/_ mutants was reduced in all areas analyzed. This was significant in all epithelial readings, and in general the reduction was greater in Fgfr2b_/_ compared with Fgf10_/_ mutants. (I_P) BrdU analysis of E13 palate. (I_N) Histological sections of BrdU staining. Proliferation rates were significantly reduced in all epithelial areas analyzed except in Fgf10_/_ mutants in the bend region at the junction of the nasal aspect of the palatal shelf and the cranial base. Mesenchymal proliferation rates were also reduced in both mutants, though not to the degree seen in the epithelium. (I) BrdU incorporation was particularly noticeable in the groove between the maxilla and the palatal process (arrow). All sections are the same magnification; scale bar, 200 ∝m. The y axes in G, H, O, and P indicate the mean values of BrdU incorporation of each area assayed. Error bars represent standard deviation; asterisk denotes a significant finding (P < 0.005) compared with the WT value. In A and L, dotted lines indicate length of epithelium measured, and green and yellow areas indicate the mesenchymal areas assayed in the apex (a) and bend (b) regions. Ant, anterior; Post, posterior.
Comparison of epithelial cell proliferation in Fgfr2b–/–, Fgf10–/– and WT mice.
Cell proliferation was significantly reduced in all epithelial areas surveyed in Fgfr2b–/– palates when compared with WT littermates at both E12 and E13. This ranged from a 30% to a 74% reduction (Figure 3, B, E, G, J, M, and O). Epithelial cell proliferation was also significantly reduced in all areas in Fgf10–/– palates, except for the bend region in E13 mice (Figure 3 C, F, G, K, N, and O).
Comparison of mesenchymal cell proliferation in Fgfr2b–/–, Fgf10–/– and WT mice.
In Fgfr2b–/– mice, mesenchymal proliferation was reduced at all locations compared with WT littermates at both E12 and E13 (12–32% reduction). The reduction was less than that seen in the epithelium (Figure 3, B, E, H, J, M, and P). This suggests that the epithelium is supporting mesenchyme proliferation and that when the epithelial receptor is blocked, a mitogenic downstream signal from the epithelium to the mesenchyme is blocked. This is evidence that, in addition to there being signaling from the mesenchyme to the overlying epithelium, signaling also occurs from the epithelium back to the mesenchyme. In Fgf10–/– mice, mesenchymal proliferation was reduced at E12 and to a lesser extent at E13 (Figure 3, C, F, H, K, N, and P).
Fgf10/Fgfr2b signaling acts as an epithelial survival factor
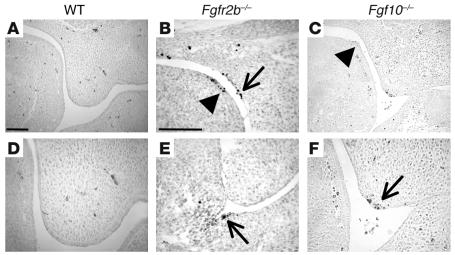
Prior to E14, TUNEL-positive cells were not detected in the developing posterior palatal epithelium of WT mice. However at E13, in both Fgfr2b–/– and Fgf10–/– mutants, we detected isolated areas of TUNEL-positive cells (Figure 4). These were in patches of thickened epithelium, notably in the MEE and bend region of the palate, in the dorsum of the tongue, and in the floor of the mouth (Figure 4, B, C, E, and F). This indicated that Fgfr2b/Fgf10 signaling could act not only as a proliferative factor but also as a cell survival factor. Areas known to contain apoptotic cells, the MEE at E14 and the enamel knot of the developing tooth, were used as positive controls (data not shown). No difference in TUNEL staining was detected in the palatal mesenchyme between WT and mutant samples.
Figure 4.
Detection of TUNEL-positive cells in the posterior oral cavity of WT, Fgfr2b_/_, and Fgf10_/_ mice at E13. (A and D) TUNEL-positive cells were not detected in WT oral epithelium. (B, C, E, and F) TUNEL-positive cells were detected in the dorsum of the tongue (B and C, arrowheads), in the palatal bend (B, arrow), in the floor of the mouth (E, arrow), and in the MEE (F, arrow) in both Fgfr2b_/_ and Fgf10_/_ mice. TUNEL-positive cells were also occasionally detected in the palatal mesenchyme (B). D and F are high magnifications of the palatal shelf apex in A and C, respectively. A and C are the same magnification, as are B and E; scale bars: 100 ∝m.
Palates from Fgfr2b–/– mice fuse in vitro
Although the expression pattern of Fgfr2b and the morphology of Fgfr2b–/– mice suggested that the defect in palatogenesis was in early palatal growth, we investigated the possibility that there may be a defect in the fusion process. Palates from both E12.5 and E13.5 Fgfr2b–/– mice were dissected free and placed side by side in Trowell-type culture dishes. The explants were placed close to each other in the same anterior-posterior orientation and were cultured for 96 hours. Hematoxylin and eosin–stained tissue sections were examined in order to assess whether the explants were fused. Explants of palates from Fgfr2b–/– mice were compared with those of heterozygote (Het) and WT littermates (E12: 7 mutants, 13 WT or Het mice; E13: 5 mutants, 9 WT or Het mice). After 48 hours in culture, all explants had started to fuse. After 96 hours, all explants had fused in a similar fashion, such that the mesenchymal tissue from opposing shelves had become continuous (data not shown). No differences were observed in fusion between explants cultured in media containing serum or in chemically defined media.
FGF10 induces proliferation in the palatal epithelium
To confirm that the reduction in proliferation described in Fgfr2b–/– and Fgf10–/– mutants was a direct result of Fgf10/Fgfr2b signaling, we tested the effects of recombinant FGF10 protein on the developing palate. FGF10-impregnated beads were placed on WT mouse palatal explants, cultured for 48 hours, pulsed with BrdU, and analyzed. FGF10 stimulated palatal epithelial proliferation and increased epithelial thickness (5/5 explants) (Figure 5A). Control BSA-coated beads had no effect (Figure 5B).
Figure 5.
Shh is a target of Fgf10/Fgfr2b signaling. (A) FGF10-impregnated bead stimulates palatal epithelial proliferation (arrow). (B) BSA-impregnated bead had no effect on proliferation. (C) Shh bead stimulated proliferation in isolated palatal mesenchymal explants. (D) FGF10 induced Shh in vitro, in the oral side of WT palatal epithelium (arrow), but not in Fgfr2b_/_ mutant mice (E). (F) FGF10 also induced Ptc1 in the mesenchyme immediately adjacent to the bead in palatal explants (arrows). Ptc1 is a known target of Shh and an indicator of the level of hedgehog signaling. (G_L) In situ hybridization to detect Shh mRNA. Shh was expressed in the MEE and the oral side of the palatal epithelium, as well as in the dorsum of the tongue. (H, I, K, and L) In Fgfr2b_/_ and Fgf10_/_ mutants, Shh expression was reduced and concentrated to discrete patches of palatal epithelium. Shh remained expressed throughout the tongue dorsum epithelium (arrow). (M and N) Frontal histological sections through the posterior oral region of NB WT (M) and K14-Cre;Shhc/n (N) mice, stained with Ladewig’s trichrome. (M) In the WT mouse, the palatal shelves had elevated and fused in the midline forming a barrier between the oral cavity and the nasopharynx (asterisk). (N) The palatal shelves of K14-Cre;Shhc/n mutants failed to develop beyond rudimentary processes. (O) In situ hybridization to detect Smo mRNA at E13. Transcripts were detected in the mesenchyme of the palate (arrow). G_I, L and O, same magnification; scale bars: 200 ∝m. J and K: original magnification, ∞12.5.
FGF10 induces Shh and Ptc1, Shh has mitogenic activity, and its mRNA expression is altered in Fgfr2b–/– and Fgf10–/– mutants
As Fgfr2b is expressed in the palatal epithelium, the reduced proliferation rate in the palatal mesenchyme of Fgfr2b–/– mutants suggests that a signal from the epithelium may stimulate proliferation in the underlying mesenchyme. We therefore sought to establish the nature of this potential signal. Shh is expressed in the oral epithelium (Figure 5, G and J). It possesses mitogenic properties, and in other developing organs it acts as a signaling molecule over both short and long ranges, often downstream of Fgf signaling (17). To test whether Shh could induce proliferation in the palatal mesenchyme, we performed experiments in which exogenous Shh peptide was applied to mesenchymal palatal explants. Shh induced proliferation in the palatal mesenchyme as assayed by BrdU (Figure 5C). Significantly, we found the expression pattern of Shh to be altered in Fgfr2b–/– and Fgf10–/– mutants (Figure 5, H, I, K, and L). At E11, before the palatal shelves had started to elongate, Shh was expressed in a similar pattern in both WT and mutant mice (data not shown). In the anterior region, transcripts were localized to the oral epithelia in the midline of the developing maxilla and mandible and also more laterally in the maxillary epithelium. Further posteriorly, Shh was expressed widely in the oral and tongue epithelia, though the intensity may have been reduced in the Fgfr2b–/– mutants (data not shown). However, at E13 in both the anterior and posterior regions of the palate, Shh expression was restricted to isolated patches of epithelia in Fgfr2b–/– and Fgf10–/– mutants, whereas Shh was highly expressed in the MEE and the palatal oral epithelium in WT mice (Figure 5, G–L). The expression pattern of Shh in the dorsum of the tongue in the mutants was similar to that of WT mice. In addition, FGF10 induced Shh in the epithelium in E12.5 WT palatal explants (4/5 explants) but not in the palatal explants from Fgfr2b–/– littermates (0/3 explants) (Figure 5, D and E). Furthermore, FGF10 induced the downstream target of Shh, Ptc1 in the underlying palatal mesenchyme (4/4 explants) (Figure 5F.
We observed that in WT mice, the MEE and oral epithelium were thicker than the epithelium on the nasal aspect of the palate (Figure 1G and Figure 2, D–F). This thickening was most notable in the groove that runs in an anterior-posterior direction between the palatal shelf and the body of the maxilla (Figure 2F). In this groove area, groups of BrdU-positive cells were noted (Figure 3, I and L). Fgfr2b–/– and Fgf10–/– mice showed neither generalized epithelial thickening of the oral epithelium nor thickening in the groove area. Interestingly, in WT mice Shh expression was restricted to the MEE and oral surfaces (Figure 5, G and J). During the development of other organs, regions of the epithelium thicken and exhibit differential gene expression patterns. Examples of this include the apical ectodermal ridge (AER) in the limb and the enamel knot in the tooth (9, 18). These regions act as signaling centers and are important in determining identity and patterning during morphogenesis. It is tempting to speculate that the oral surface of the palatal epithelium, particularly the groove region, may have a similar role.
Keratin 14Cre;Shhc/n mice exhibit cleft palate but K-14Cre;Smoc/n mice do not
Although Shh-null mice do survive until birth, they exhibit such severe disruption of the head and face that identifiable nasal, maxillary, and mandibular structures fail to form, thus making it difficult to study palatogenesis (19). Using Cre-recombinant technology we generated mice deficient in either Shh or Smo under the Keratin 14 (K-14) promoter (20, 21). In these transgenic mice, Shh and Smo expression is inactivated only in the epithelium (20, 22). Eighty-five percent of K-14Cre;Shhc/n mice exhibited a cleft palate with rudimentary palatal shelves spaced widely apart, demonstrating that epithelial Shh is critical for palate development (Figure 5, M and N). To test whether the effect of Shh is on the epithelium or the underlying mesenchyme, we analyzed the palates of K-14Cre;Smoc/n mice. These were indistinguishable from those of their WT littermates (data not shown). Thus, blocking the downstream targets of Shh in the epithelium had no effect on palatogenesis. Therefore, Shh protein must be exerting its effect on the adjacent mesenchyme. In addition, we found that Smo was expressed in the mesenchyme of the palate at E13 (Figure 5O).
Discussion
In this study we have found that when either Fgf10 or its receptor Fgfr2b is knocked out, mice develop cleft palate. Development is affected early, with the palatal processes failing to grow and to make the morphological change from a bud into a normal palatal extension. This disruption is similar to abnormalities seen in the development of both Fgf10–/– and Fgfr2b–/– mice limb buds; although these are initiated, outgrowth fails to occur (16, 23). In Fgfr2b–/– mutants, the skin is thin and shows a low proliferation rate in the basal layer, and in the developing limb, the AER exhibits an increase in apoptosis (16, 24). Consistent with this, the epithelium was found to be thin and cell proliferation reduced in the palates of both Fgf10–/– and Fgfr2b–/– mice. Where the palatal epithelium did show signs of thickening, the cells underwent apoptosis, indicating that Fgf10/Fgfr2b signaling has both cell proliferation and cell survival functions during palatal development. We found the change in cell proliferation and cell death to be of a similar magnitude to that found in other developmental systems affected in Fgf10–/– and Fgfr2b–/– mice. These included the limb, skin, and thymus, all of which showed gross abnormalities (16, 24, 25). A similar degree of change in cell turnover was detected in the palatal mesenchyme of Msx1–/– mice, and this accounted for the cleft palate. Interestingly, in Msx1–/– mutants, the timing of the proliferation defect seemed to be critical in that defective proliferation was detected only at E12 and not at E13 (12). In Fgf10–/– and Fgfr2b–/– mice, there is defective cell turnover from E12 onward. It thus seems that not only can the degree of change, but also the timing of the defect, in cell turnover account for the wide cleft palate seen in NB mice.
Epithelial-mesenchymal interactions regulate early palate development.
Vertebrate limbs and teeth develop by a series of interactions with signals from the developing mesenchyme passing to the overlying epithelium, causing it to thicken. This thickened epithelium signals back to the underlying mesenchyme to direct growth and patterning. We have previously shown that in both limbs and teeth, Fgf10/Fgfr2b signaling plays important roles in initial budding and in maintaining growth (24, 26). Here we provide evidence that Fgf10 signals from the palatal mesenchyme to its receptor in the palatal epithelium and that signals pass back to the mesenchyme. Furthermore, we show that during palatal shelf budding and growth, the epithelium on the oral aspect of the palate thickens and that Fgf10 induces this thickening. We therefore demonstrate that signals operating in the developing palate are similar to those in the developing tooth and limb.
Previous studies have shown that the palatal mesenchyme can maintain cell proliferation and apoptosis in the epithelium through the action of epidermal growth factor on its receptor, which is located in the epithelium (27, 28). Also, in vitro tissue dissociation and recombination studies have shown that the mesenchyme can specify the palatal epithelium into its different regions: the oral, nasal, and medial edge epithelia (11). In this study we show that Fgf10 signals from the mesenchyme to the epithelium to regulate Shh which signals back to the underlying mesenchyme. We propose that this signaling network leads to changes in palatal size and shape.
Shh is a target of Fgf10/Fgfr2b signaling.
We propose that Shh is a downstream effector of Fgf10/Fgfr2b signaling, as Shh expression was downregulated in the palatal epithelium of both Fgf10–/– and Fgfr2b–/– mutants. Altered Shh expression has previously been demonstrated to occur in the limb bud and lung endoderm of Fgf10–/– mice (23). In the developing lung, Shh is also known to negatively regulate Fgf10, and removal of the Shh inhibitor Hip results in a downregulation of Fgf10 and lung hypoplasia (29, 30). In the developing limb, Shh is expressed in the zone of polarizing activity in the mesenchyme, and epithelial Fgf’s regulate its expression. It has a key role in determining anterior-posterior identity, and an analogous role in the palate may be postulated, with Shh exhibiting an asymmetric expression pattern; i.e., only on the oral aspect and the MEE (Figure 5, D and G). Interestingly, in the limb, Shh is not only dependent on Fgf10 but can also maintain the expression of Fgf10 in the progress zone in the mesenchyme (31). Thus, in both the developing limb and lung, Fgf10 regulates Shh and vice versa.
We show that exogenous FGF10 induced Shh in WT palatal epithelium and that this was not the case in Fgfr2b–/– mice. Thus, the altered expression pattern of Shh in Fgf10–/– and Fgfr2b–/– mutants is not secondary to defective epithelium. We demonstrate the essential role of Shh in palatogenesis by showing that K-14Cre;Shhc/n mice exhibit cleft palate. We also provide evidence that Shh acts on the palatal mesenchyme, as K-14Cre;Smoc/n mice, in which Shh signaling is blocked in the epithelium, exhibited normal palatogenesis.
Fgf signaling during palatogenesis.
We have shown that Fgf10/Fgfr2b signaling regulates Shh during palatogenesis. Fgfr2b is predominately in the ectoderm of most developing organs, and in many it is required for cell proliferation. In addition, it appears that Fgf10/Fgfr2b signaling is important for the expression of the subsequent signals regulating mesenchymal development. Thus, in the limb bud, Fgf10, signaling through Fgfr2b, regulates the expression of other members of the Fgf family in the epithelium (reviewed in 18). The expression of members of the Fgf gene family in the palatal epithelium has not been reported. Interestingly, Fgf18-null mice have been reported to exhibit cleft palate (32, 33).
Tissue recombination experiments in the chick facial primordia have demonstrated that the epithelium is required to maintain FGFR2 expression levels in the underlying mesenchyme. Also, Fgf2, which is present in the facial epithelium, can partially compensate for the removal of the epithelium (34–36). Nevertheless, it is the mesenchyme that controls the shape of the facial primordia (37).
Fgf10 and Fgf7have different but overlapping expression domains. We show that Fgf10 is mainly expressed in the oral side of the palatal mesenchyme and Fgf7 expression is more restricted to the nasal aspect. Their expression domains overlap at the palatal apex and in the bend region (Figure 6A). This suggests that they may have differential, combined, or compensatory actions. We found that the effects on both epithelial and mesenchymal cell proliferation in Fgf10–/– mice were less severe in the bend region than those in Fgfr2b–/– mice. As Fgf7 is known to bind to Fgfr2b and has been suggested to play a role in epithelial-mesenchymal interactions in other organs including the lung and limb (29, 38), Fgf7 may partially compensate for the loss of Fgf10 in this region. Palatal abnormalities in Fgf7–/– mice have not been reported (39). We also suggest that as Fgf10 and Fgf7 are differentially expressed in the oral and nasal aspects of the developing palate, they may not only have distinct effects on epithelial proliferation and palatal morphogenesis, but also different effects on determining whether the epithelium differentiates into nasal, medial, or oral type. The abnormal epithelial fusions observed between the palatal processes, the tongue, and the mandible, in both Fgf10–/– and Fgfr2b–/– mutants (Figure 1L), may have resulted from abnormalities in epithelial differentiation. Loss of Fgf10/Fgfr2b function may also affect mesenchymal cell differentiation. Thus, differentiation abnormalities in either the palatal epithelium or the mesenchyme could result in a morphological change in the developing palatal shelves, manifesting in the formation of a cleft. To test this hypothesis we analyzed the immunolocalization of α-actin and detected no difference between WT, Fgf10–/–, and Fgfr2b–/– palates (unpublished data). However, a more extensive survey of other differentiation markers could still reveal defects.
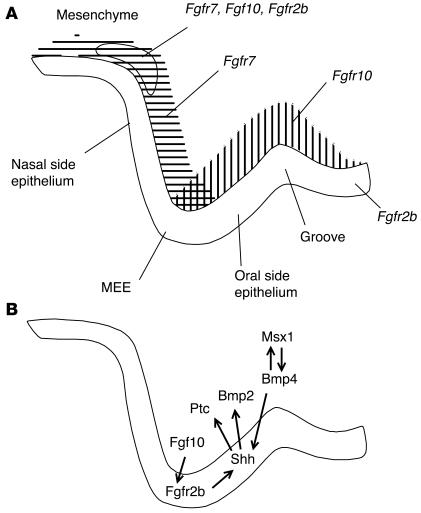
Figure 6.
Molecular control of early palate development. (A) Schematic diagram showing the mRNA expression of Fgfr2b, Fgf10, and Fgf7 in the E13 mouse palate. Fgfr2b was expressed in the oral epithelium and at a low level in the mesenchyme in the bend area between the cranial base and palatal shelf. Fgf10 and Fgf7 were also expressed at this site. In addition, Fgf10 was mainly expressed in the mesenchyme on the oral side of the developing palate and Fgf7 on the nasal aspect. In the mesenchyme adjacent to the MEE, Fgf10 and Fgf7 expression domains overlapped. “Groove” indicates the anterior-posterior groove between the palatal process and the body of the maxilla. (B) Schematic diagram illustrating the proposed Fgf10/Fgfr2b epithelial-mesenchymal interactions and possible downstream signaling in the developing palate.
We have shown how Fgf10/Fgfr2b/Shh signaling is essential for normal mammalian palate development. Elucidation of this pathway allows the design of therapies where the downstream effects are treated. The model presented (Figure 6B) integrates well with the Msx1 and Pax9 loss of function models in mice, in which Bmp signaling is disrupted in early palatogenesis (12, 40). Analogously, loss of function mutations in MSX1 have been found in patients with cleft palate, and PAX9 has been associated with human clefting (41, 42). Mutant mice lacking Transforming growth factor β3 (Tgfβ3) exhibit an isolated cleft palate, and this is characterized by defects in the fusion process of the palatal shelves (43–45). Interestingly the expression patterns of Fgfr2b and Shh overlaps with that of Tgfβ3. We demonstrate that isolated palatal shelf explants from Fgfr2b–/– mice fused. The palatal phenotype in Fgfr2b–/– embryos occurs earlier than that in Tgfb3–/– embryos. The most notable difference in the phenotypes is that in Tgfβ3–/– animals, the palatal shelf growth is normal until fusion at E14.5, while Fgfr2b–/– animals lack the early palatal shelf outgrowth. Even though we show that a Fgf-Shh signaling network has a role during early palatogenesis, this does not exclude a role for either Fgf or Hedgehog signaling later during palatogenesis.
Growth factor signaling during early palatogenesis.
We have started to put together a signaling network operative during early palate morphogenesis (Figure 6B). We have shown that Fgf10 signals from the palatal mesenchyme to its receptor Fgfr2b in the overlying epithelium. Besides stimulating cell proliferation, this signaling activates other cues, at least Shh. This signaling network controls epithelial and mesenchymal cell turnover, which directs early palatal shelf morphogenesis and growth. Also, we suggest that differential ligand activity through Fgfr2b may influence the fate of the palatal epithelium, determining its differentiation into nasal, oral, or medial edge epithelium. This model ties in well with the work of Zhang et al. (12), in which the cleft palate found in Msx1-deficient mice is rescued by overexpressing the downstream effector of Msx1, BMP4. They show that Shh can induce Bmp2 in the tissue culture of anterior palatal mesenchyme and that Shh and BMP2 can induce mesenchymal proliferation. Shh is absent from the epithelium of Msx1–/– mice but is restored when the cleft phenotype is rescued by mesenchymally expressed BMP4. This indicates that Bmp4 can regulate Shh (12).
Taken together, we reveal a new role for Fgf signaling in mammalian secondary palate development, which controls epithelial and mesenchymal proliferation as well as early palatal shelf morphogenesis and growth.
Methods
Transgenic mice and tissue preparation
Fgfr2(IIIb)–/– and Fgf10–/– mice were obtained by crossing heterozygous breeding pairs. The generation and genotyping of these mice has been described previously (16, 46). The generation of K-14Cre;Shhc/n and K-14Cre;Smoc/n mice has been described previously (20, 21). Whole heads of mice aged between E11 and NB were dissected free under a stereomicroscope. The age of the embryos was determined by the day of the appearance of the vaginal plug (day 0) and by morphological criteria. Following overnight fixation in 4% paraformaldehyde (PFA) in PBS at 4°C, the tissues were dehydrated in an ethanol series and embedded in paraffin. Frontal sections of 7 ∝m in thickness were cut and mounted on Superfrost plus slides (VWR International Ltd., Poole, United Kingdom), dried overnight at 37°C, and stored at 4°C. WT littermates were used as controls in all experiments.
Organ culture
Palatal shelf cultures.
E12.5 and E13.5 palatal shelves were dissected free and placed side by side in a similar manner to that in Taya et al (44), though we used a Trowell-type organ culture system (47). Both DMEM (Sigma-Aldrich, Poole, United Kingdom) with 10% bovine calf serum and serum-free BGJ-b (Gibco-Life Technologies, Paisley, United Kingdom) media were used. Both media were supplemented with glutamax and penicillin/streptomycin (Sigma-Aldrich).
Palatal mesenchymal explants.
E12.5 palates were dissected out with the surrounding maxillary tissue, and epithelia were removed by dissection following treatment with pancreatin (2.25%) (Sigma-Aldrich) and trypsin (0.75%) (Sigma-Aldrich) solution on ice for 30 minutes, followed by 30 minutes in media on ice. Isolated palatal mesenchymal explants were cultured for 48 hours in DMEM with 10% bovine calf serum, glutamax, and penicillin/streptomycin.
Bead assays.
Heparin-coated acrylic beads (Sigma-Aldrich) were incubated in 50 ng/∝l recombinant human FGF10 (R&D Systems, Abingdon, United Kingdom) or BSA at 37°C for 40 minutes and stored at 4°C before being placed on the explant. Bead assays were cultured for 24–48 hours. Similarly, Affi-gel agarose beads (Biorad, Hemel Hempstead, United Kingdom) were impregnated with mouse Shh peptide (Sigma-Aldrich) (1mg/ml).
In situ hybridization
Preparation of Fgfr2b, Fgf3, Fgf7, Fgf10, Ptc1, and Shh 35S-UTP–labeled riboprobes, in situ hybridization, and image analysis have all been described previously (5, 26, 48–50). Antisense Smo probe (a kind gift from M.T. Cobourne, King’s College, London, United Kingdom) was generated from Bluescript KS (Stratagene, La Jolla, California, USA) by linearizing with NotI and transcribing with T3.
Proliferation assay and TUNEL
Cell proliferation was assessed by pulsed BrdU incorporation. Mice were injected intraperitoneally with 10 ∝l/g body weight BrdU (Zymed, San Francisco, California, USA) and sacrificed after 2 hours. Experiments were in accordance with the Animal Welfare Committee of The University of Helsinki. Explants were cultured for 20 hours, and then BrdU was added to the medium (1:200) for 4 hours. Zymed’s streptavidin-biotin staining system was used according to the manufacturer’s instructions, and sections were counterstained with hematoxylin. The number of BrdU-positive cells was recorded by two individuals, with the mean taken. The total number of cells was not found to differ between genotypes. Thus, the value of BrdU-positive cells reflects the percentage of the total cell population undergoing proliferation and is not a reflection of varying cell density. A total of 11 WT littermates, 6 Fgfr2b–/– mice, and 6 Fgf10–/– mice were used, and 58 WT, 54 Fgfr2b–/–, and 43 Fgf10–/– sections counted. Using a grid, at E12 BrdU-positive cells were counted in a 0.02 mm2 area of mesenchyme in the apex of the palatal projection and a 0.006 mm2 area in the bend region. A 286-∝m length of epithelium was counted in the apex of the palatal projection, and a 143-∝m length in the bend region was counted. At E13 the areas were 0.02 mm2 and 0.007 mm2 for the apex and bend areas, respectively, and 329 ∝m and 164 ∝m for the lengths of epithelium (Figure 3L).
Apoptotic cells were identified on paraffin sections using Promega’s DeadEnd Colorimetric TUNEL System (Promega, Madison, Wisconsin, USA).
Acknowledgments
We thank Stalin Kariyawasam, Merja Mäkinen, Heide Olsen, Ludmila Rasskozova, and Riikka Santalahti for their technical assistance and Bethan L. Thomas for her help with the manuscript. We thank David Ornitz for generously providing the Fgf10–/– mice. This work was supported by grants from the Medical Research Council UK and the Royal Society (to D.P.C. Rice), the Academy of Finland (to I. Thesleff), the Finnish Cultural Foundation (to R. Rice), the Swedish MRC (2789, 14100, to A. Gritli-Linde), and the NIH (NS33462, to A.P. McMahon).
Footnotes
See the related Commentary beginning on page 1676.
Nonstandard abbreviations used: apical ectodermal ridge (AER); bone morphogenetic protein (Bmp); embryonic day (E); FGF receptor 2 (Fgfr2); heterozygote (Het); Keratin 14 (K-14); medial epithelial edge (MEE); newborn (NB); paraformaldehyde (PFA); Patched 1 (Ptc1); Smoothened (Smo); Sonic hedgehog (Shh).Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Gorlin, R.J., Cohen, M.M., Jr., and Hennekam, R.C.M. 2001. Syndromes of the head and neck. 4th edition. Oxford University Press. New York, New York, USA. 850–853.
- 2.Jones MC. Etiology of facial clefts: prospective evaluation of 428 patients. Cleft Palate J. 1988;25:16–20. [PubMed] [Google Scholar]
- 3.Murray J. Gene/environment causes of cleft lip and/or palate. Clin. Genet. 2002;61:248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- 4.Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat. Rev. Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- 5.Rice DP, et al. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- 6.Wilkie AO, Oldridge M, Tang Z, Maxson RE., Jr Craniosynostosis and related limb anomalies. Novartis. Found. Symp. 2001;232:122–133; discussion 133–143. doi: 10.1002/0470846658.ch9. [DOI] [PubMed] [Google Scholar]
- 7.Kreiborg S, Cohen MM., Jr The oral manifestations of Apert syndrome. J. Craniofac. Genet. Dev. Biol. 1992;12:41–48. [PubMed] [Google Scholar]
- 8.Ferguson MW. Palate development. Development. 1988;103(Suppl.):41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- 9.Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- 10.Tyler MS, Koch WE. In vitro development of palatal tissues from embryonic mice. III. Interactions between palatal epithelium and heterotypic oral mesenchyme. J. Embryol. Exp. Morphol. 1977;38:37–48. [PubMed] [Google Scholar]
- 11.Ferguson MWJ, Honig LS. Epithelial-mesenchymal interactions during vertebrate palatogenesis. Curr. Top. Dev. Biol. 1984;19:137–164. doi: 10.1016/s0070-2153(08)60398-1. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, et al. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
- 13.Francis-West PH, Robson L, Evans DJ. Craniofacial development: the tissue and molecular interactions that control development of the head. Adv. Anat. Embryol. Cell Biol. 2003;169:III–VI, 1–138. doi: 10.1007/978-3-642-55570-1. [DOI] [PubMed] [Google Scholar]
- 14.Igarashi M, Finch PW, Aaronson SA. Characterization of recombinant human fibroblast growth factor (FGF)-10 reveals functional similarities with keratinocyte growth factor (FGF-7) J. Biol. Chem. 1998;273:13230–13235. doi: 10.1074/jbc.273.21.13230. [DOI] [PubMed] [Google Scholar]
- 15.Beer HD, et al. Fibroblast growth factor (FGF) receptor 1-IIIb is a naturally occurring functional receptor for FGFs that is preferentially expressed in the skin and the brain. J. Biol. Chem. 2000;275:16091–16097. doi: 10.1074/jbc.275.21.16091. [DOI] [PubMed] [Google Scholar]
- 16.De Moerlooze L, et al. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 17.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 18.Martin GR. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- 19.Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 20.Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- 21.Gritli-Linde A, et al. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323–5337. doi: 10.1242/dev.00100. [DOI] [PubMed] [Google Scholar]
- 22.Byrne C, Tainsky M, Fuchs E. Programming gene expression in developing epidermis. Development. 1994;120:2369–2383. doi: 10.1242/dev.120.9.2369. [DOI] [PubMed] [Google Scholar]
- 23.Sekine K, et al. Fgf10 is essential for limb and lung formation. Nat. Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 24.Revest JM, et al. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev. Biol. 2001;231:47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- 25.Revest JM, Suniara RK, Kerr K, Owen JJ, Dickson C. Development of the thymus requires signaling through the fibroblast growth factor receptor R2-IIIb. J. Immunol. 2001;167:1954–1961. doi: 10.4049/jimmunol.167.4.1954. [DOI] [PubMed] [Google Scholar]
- 26.Kettunen P, et al. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev. Dyn. 2000;219:322–332. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1062>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 27.Tyler MS, Pratt RM. Effect of epidermal growth factor on secondary palatal epithelium in vitro: tissue isolation and recombination studies. J. Embryol. Exp. Morphol. 1980;58:93–106. [PubMed] [Google Scholar]
- 28.Miettinen PJ, et al. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat. Genet. 1999;22:69–73. doi: 10.1038/8773. [DOI] [PubMed] [Google Scholar]
- 29.Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- 30.Chuang PT, Kawcak T, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 2003;17:342–347. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohuchi H, et al. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development. 1997;124:2235–2244. doi: 10.1242/dev.124.11.2235. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Xu J, Colvin JS, Ornitz DM. Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes Dev. 2002;16:859–869. doi: 10.1101/gad.965602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohbayashi N, et al. FGF18 is required for normal cell proliferation and differentiation during osteogenesis and chondrogenesis. Genes Dev. 2002;16:870–879. doi: 10.1101/gad.965702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richman JM, Herbert M, Matovinovic E, Walin J. Effect of fibroblast growth factors on outgrowth of facial mesenchyme. Dev. Biol. 1997;189:135–147. doi: 10.1006/dbio.1997.8656. [DOI] [PubMed] [Google Scholar]
- 35.Richman JM, Crosby Z. Differential growth of facial primordia in chick embryos: responses of facial mesenchyme to basic fibroblast growth factor (bFGF) and serum in micromass culture. Development. 1990;109:341–348. doi: 10.1242/dev.109.2.341. [DOI] [PubMed] [Google Scholar]
- 36.Matovinovic E, Richman JM. Epithelium is required for maintaining FGFR-2 expression levels in facial mesenchyme of the developing chick embryo. Dev. Dyn. 1997;210:407–416. doi: 10.1002/(SICI)1097-0177(199712)210:4<407::AID-AJA5>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 37.Richman JM, Tickle C. Epithelia are interchangeable between facial primordia of chick embryos and morphogenesis is controlled by the mesenchyme. Dev. Biol. 1989;136:201–210. doi: 10.1016/0012-1606(89)90142-5. [DOI] [PubMed] [Google Scholar]
- 38.Yonei-Tamura S, et al. FGF7 and FGF10 directly induce the apical ectodermal ridge in chick embryos. Dev. Biol. 1999;211:133–143. doi: 10.1006/dbio.1999.9290. [DOI] [PubMed] [Google Scholar]
- 39.Guo L, Degenstein L, Fuchs E. Keratinocyte growth factor is required for hair development but not for wound healing. Genes Dev. 1996;10:165–175. doi: 10.1101/gad.10.2.165. [DOI] [PubMed] [Google Scholar]
- 40.Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Boogaard MJ, Dorland M, Beemer FA, van Amstel HK. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat. Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- 42.Schuffenhauer S, et al. De novo deletion (14)(q11.2q13) including PAX9: clinical and molecular findings. J. Med. Genet. 1999;36:233–236. [PMC free article] [PubMed] [Google Scholar]
- 43.Kaartinen V, Cui XM, Heisterkamp N, Groffen J, Shuler CF. Transforming growth factor-beta3 regulates transdifferentiation of medial edge epithelium during palatal fusion and associated degradation of the basement membrane. Dev. Dyn. 1997;209:255–260. doi: 10.1002/(SICI)1097-0177(199707)209:3<255::AID-AJA1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 44.Taya Y, O’Kane S, Ferguson MW. Pathogenesis of cleft palate in TGF-beta3 knockout mice. Development. 1999;126:3869–3879. doi: 10.1242/dev.126.17.3869. [DOI] [PubMed] [Google Scholar]
- 45.Dudas M, Nagy A, Laping NJ, Moustakas A, Kaartinen V. Tgf-beta3-induced palatal fusion is mediated by Alk-5/Smad pathway. Dev. Biol. 2004;266:96–108. doi: 10.1016/j.ydbio.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Min H, et al. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes. Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HJ, Rice DP, Kettunen PJ, Thesleff I. FGF-, BMP- and Shh-mediated signalling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development. 1998;125:1241–1251. doi: 10.1242/dev.125.7.1241. [DOI] [PubMed] [Google Scholar]
- 48.Kettunen P, Karavanova I, Thesleff I. Responsiveness of developing dental tissues to fibroblast growth factors: expression of splicing alternatives of FGFR1, -2, -3, and of FGFR4; and stimulation of cell proliferation by FGF-2, -4, -8, and -9. Dev. Genet. 1998;22:374–385. doi: 10.1002/(SICI)1520-6408(1998)22:4<374::AID-DVG7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 49.Gritli-Linde A, Lewis P, McMahon AP, Linde A. The whereabouts of a morphogen: direct evidence for short- and graded long-range activity of hedgehog signaling peptides. Dev. Biol. 2001;236:364–386. doi: 10.1006/dbio.2001.0336. [DOI] [PubMed] [Google Scholar]
- 50.Vaahtokari A, Aberg T, Thesleff I. Apoptosis in the developing tooth: association with an embryonic signaling center and suppression by EGF and FGF-4. Development. 1996;122:121–129. doi: 10.1242/dev.122.1.121. [DOI] [PubMed] [Google Scholar]