Abstract
Longstanding mechanistic questions about the role of protecting osmolyte trimethylamine N-oxide (TMAO) which favors protein folding and the denaturing osmolyte urea are addressed by studying their effects on the folding of uncharged polymer chains. Using atomistic molecular dynamics simulations, we show that 1-M TMAO and 7-M urea solutions act dramatically differently on these model polymer chains. Their behaviors are sensitive to the strength of the attractive dispersion interactions of the chain with its environment: when these dispersion interactions are high enough, TMAO suppresses the formation of extended conformations of the hydrophobic polymer as compared to water, while urea promotes formation of extended conformations. Similar trends are observed experimentally on real protein systems. Quite surprisingly, we find that both protecting and denaturing osmolytes strongly interact with the polymer, seemingly in contrast with existing explanations of the osmolyte effect on proteins. We show that what really matters for a protective osmolyte is its effective depletion as the polymer conformation changes, which leads to a negative change in the preferential binding coefficient. For TMAO, there is a much more favorable free energy of insertion of a single osmolyte near collapsed conformations of the polymer than near extended conformations. By contrast, urea is preferentially stabilized next to the extended conformation and thus has a denaturing effect.
Keywords: Protective Osmolyte, Denaturant, Preferential Binding, Polymer Conformations, FEP, PMF
Introduction
Osmolytes are small cosolutes found endogenously to protect cells against osmotic stress.1 However, they can have profound effects on protein stability.2-8 While some of them are denaturants (e.g. urea), others like trimethylamine N-oxide (TMAO) act as protecting osmolytes in vivo: in denaturing conditions, they bias the protein structure toward the folded conformation.1,9-12 They are thus referred to as chemical chaperones. Hence, TMAO is used by deep-sea organisms to counteract the deleterious effect of pressure and by sharks or skates to compensate for their relatively high concentrations of the denaturing urea.11 Most interestingly, the protein folding propensity of TMAO has been used experimentally to study the mechanisms involved in protein misfolding diseases, including e.g. prion protein,13,14 tau protein15,16 (Alzheimer disease) and alpha-synuclein17 (involved in numerous neurodegenerative diseases); chemical chaperones like TMAO even appear promising as therapeutics,18 even though it was recently found to be related to an increased risk of cardiovascular diseases in humans.19 TMAO is active in endogenous systems for concentrations as low as 200 mM.1,13 Experiments in vitro on alpha-synuclein, an intrinsically disordered protein, have given evidence that this protective effect increases with concentration.17
TMAO is a small amphiphile (chemical formula: (CH3)3NO) consisting of a small hydrophilic group (N+O−) and a bulky hydrophobic part (3 methyl groups). Several mechanisms have been invoked to explain the folding propensity of TMAO. In a first scenario, TMAO would enhance water structure and hydrogen-bond (HB) strength, which would indirectly affect the equilibrium between the folded and the unfolded conformations of a protein.20-22 However, this mechanism has been challenged by several studies, mainly based on molecular dynamics (MD) simulations, where no significant alteration of water structure was found in aqueous solutions of TMAO.23-25 These results may not be surprising since TMAO can only accept 2 to 3 strong HBs at its hydrophilic head, which represents less than 10% of its hydration water HB population.26
Other studies have suggested that direct interactions, or especially the lack thereof, between TMAO and the protein backbone could cause the osmolyte effect. In particular, thermodynamic measurements have highlighted the importance of the interactions between TMAO and the protein backbone and side-chains.9,10 TMAO has favorable interactions with some protein side-chains, especially the positively charged groups that can interact with the O− of TMAO. In contrast, interactions with the protein backbone, in particular with the amide NH, are entropically unfavorable.24 If these unfavorable interactions were to dominate, TMAO would be depleted from the protein surface. It has been suggested that the resulting concentration gradient in the TMAO could lead to an osmotic pressure favoring the folded conformation with respect to the unfolded one.24 Recently, Garcia and co-workers have combined27 computer simulation with the experimental osmotic pressure measurements and have suggested a mechanism where there is preferential exclusion of TMAO from protein surfaces due to repulsive self-interaction in the solvation shell. Others have argued that osmotic pressure itself cannot explain this phenomenon, and that “water mediated interactions” between the osmolyte and the protein could also play a role.25
However, a clear unifying scenario has not yet emerged from the study of the effect of TMAO on proteins. One of the reasons might be the presence of amino acids with different chemical properties which might complicate the role of TMAO as a structure enhancer. Thus instead of struggling with different amino acids, a successful strategy can be to use a simple polymeric chain whose hydrophobicity can be tuned. There have already been studies of the action of TMAO on purely hydrophobic chains,23 but even for such a simple system a consensus has not yet been achieved. A good illustration of the lack of consensus is the opposite conclusions reached in two different investigations. Based on simulations of a small hydrophobic solutes and of hydrophobic chains, one of these studies suggested that TMAO has a negligible effect on the hydrophobic interactions.23 In contrast other studies suggest that TMAO destroys hydrophobic interactions.28
To investigate the molecular mechanism of TMAO’s role as a protective osmolyte, it is interesting to compare it with the effect of urea (chemical formula: (NH2)2CO) solutions that lead to the opposite behavior — unfolding of the protein. For example Pettitt and co-workers29 have recently explored the conformational preferences of decaalanine in TMAO and urea solutions using free energy perturbation techniques. Their analysis, based on the decomposition of the transfer free energy, suggests the differences in the behavior of peptide in the two different solutions arises mainly from differences in the relative importances of van der Waals and electrostatic interactions: urea denaturation is dominated by van der Waals attractions whereas TMAO exerts its effect by causing unfavorable electrostatic interactions. In this contribution we extend our previous work on urea30 by using similar systems and methodologies and apply it to contrast the respective role of a denaturing osmolyte (urea) and a protective osmolyte (TMAO) on uncharged chains in water. We focus on the mechanisms by which these two osmolytes produce opposite actions on the conformations of this Lennard-Jones chain. Following most of the previous simulation studies, we used concentrations higher than that found in vivo to enhance the osmolyte influence on protein stability. Hence we chose a concentration of 7 M for urea (consistent with Ref.30) and a concentration of 1 M for TMAO. Both concentrations are widely used to study the effects of the respective osmolytes in vitro and in silico. Our study shows that while acting on the same chain, TMAO stabilizes the collapsed conformations of the chain while urea destabilizes the collapsed conformations, and the simulations later allow us to offer a molecular explanation to these different behaviors. The paper is organized as follows: the simulation model and methods are described in section II, results are presented in section III, and some conclusions are presented in section IV.
Simulation model and methods
System and forcefields
Our system consists of a 32-bead polymer solvated in various aqueous solutions. The polymer is uncharged and the beads only interacts with their environment via Lennard-Jones (LJ) potentials. While the bead radius is fixed (σb=0.4 nm) the hydrophobic character of the chain can be tuned by varying the energy parameter εb. Following a previous study,30 four values were employed (εb = 0.4; 0.6; 0.8; 1.0 kJ/mol) even if most of the current work was performed on the εb = 1.0 kJ/mol polymer. Among the chain, 1-4 interactions were removed; parameters for the 1-2 (bonds) and 1-3 (angles) interactions can be found elsewhere.30 For water molecules, we used the SPC/E model,31 while urea interacts through the OPLS/AA forcefield32 and TMAO through the forcefield developed by Kast et al’s.33 The geometric combining rules were used in the cross interactions for ε and arithmetic combination rules were used for σ. Three systems were simulated. The system of pure aquous solution was composed of the polymer solvated by 4092 water molecules. The system of 1 M TMAO solution was composed of the 32-bead polymeric chain, 79 TMAO molecules and 4013 water molecules. On the other hand, the system of 7 M urea solution was composed of the 32-bead polymeric chain, 500 urea molecules and 2727 water molecules. The box size was close to 5 × 5 × 5 nm3 in all cases. We have also repeated our simulation for TMAO at εb = 1.0 kJ/mol using a different forcefield (called herein the “osmotic model”) recently proposed by Garcia and coworkers27 and we found it to follow the same qualitative trends (c.f. the SI Text).
Equilibrium simulations
All simulations were performed using Gromacs 4.5.4 software.34 In a bid to sample different polymer conformations in different osmolyte solutions, unrestrained equilibrium MD simulations of the polymer chain were performed in pure water, 1 M TMAO and 7 M urea solutions. In order to avoid any bias, an extended configuration of the polymeric chain was used as an initial configuration in pure water and in the aqueous solution of 1 M TMAO while a collapsed configuration of the polymer was used as an initial configuration for the simulation in aqueous solution of 7 M urea. The initial extended configuration was an all-trans configuration of the polymer while the collapsed configuration was picked from the simulation of the polymer in water. Each of the systems was first energy-minimized using a steepest-descent algorithm and then subjected to 100 ns of production run in NPT ensemble. The Nose-Hoover thermostat was used for maintaining the average temperature at 300 K and the Parinello-Rahman barostat was used for maintaining the average pressure at 1 bar. For all three aqueous solutions, unrestrained simulations were repeated for the four values of the LJ energy parameter (εb) for the polymer beads.
Potentials of mean-force
We determined the free energy landscape (or potential of mean force [PMF]) of the 32-bead LJ chains along one or several collective reaction coordinates in different solutions by performing umbrella sampling simulations. We chose as reaction coordinate the polymer radius of gyration Rg in pure water, 1 M TMAO and 7 M urea. We employed the PLUMED extension of Gromacs.35 The value of Rg ranged from 0.4 nm to 1.2 nm at a spacing of 0.05 nm between adjacent windows. Restraining harmonic force constants of 7000 kJ/mol/nm2 were used in the umbrella potential in all positions to ensure a Gaussian distribution of the reaction coordinate around each desired value of the reaction coordinate. Finally, we used the Weighted Histogram Analysis Method (WHAM).36,37 to generate unbiased histograms and corresponding free energies. As described later we also compute the joint probability distribiution of Rg and the end-to-end distance and the corresponding potential of mean force as a function of these two variables.
Preferential interaction
We employed two parameters to measure the affinity of the cosolvent (urea or TMAO) for the polymer. First, the local-bulk partition coefficient Kp was calculated, where as7
| (1) |
Here 〈nX〉 is the average number of molecules of type X bound to polymer and is the total number of molecules of type X in the system (where X = s stands for the cosolvent (urea or TMAO) and X = w stands for water). Kp is intensive and reflects the affinity of the cosolvent for the polymer regardless of the exposed surface area of the polymer. The other parameter is the experimentally-relevant preferential binding coefficient,2,3,7,27,38
| (2) |
which is extensive (i.e., it depends on the size of the hydration shell). To determine the dependence on the proximal cut-off distance for the counting of molecules around the polymer, we computed the value of both quantities Kp and Γ as a function of distance from the polymer (i.e., by examining the explicit distance dependance of ns(r) and nw(r)), which is defined as the shortest distance between the central atom of the solvent molecule (O for water, N for TMAO and C for urea) and any polymer bead. Additional simulations were performed as follows. We froze 5 representative configurations of the the polymer either in the collapsed or the extended state for each osmolyte solution, and then propagated each of these simulations for 15 ns (total simulation length of 75 ns for each polymer configuration). In each case, both Kp and Γ were averaged over this ensemble of trajectories, and standard deviations were obtained via block averaging.
Finally, we used the Free Energy Perturbation (FEP) technique to compute the transfer free-energy (chemical potential) for inserting a single TMAO (or urea or water molecule) from bulk solution into the first solvation shell of particular conformations of polymer in 1 M TMAO (or 7 M urea) where the polymer conformation was fixed in either a collapsed or an extended conformation. For these calculations the initial configurations were taken from a representative snapshot of the prior umbrella sampling windows and the TMAO (or urea or water) molecule was grown in presence of other TMAO (or urea or water) molecules in solution. The interactions of the molecule being inserted were slowly turned on in two stages: in the first stage only the van der Waals interactions were turned on, and in the second stage the electrostatic interactions were turned on. Thermodynamic integration gives these two contributions to the transfer free energy. The difference between the free energy for the insertion proximate to the polymer and the insertion in bulk gives the required transfer free energies. Finally, since the choice of the position near the polymer where the osmolyte and solvent molecules is grown is arbitrary, we repeated such calculations at several positions in each case: 5 sites for TMAO and urea and 3 sites for water near each of collapsed and extended conformations and 2 sites of each of them in bulk media. They were finally averaged and the standard deviations were estimated.
Results and discussion
Osmolyte effect on folding equilibria
We first verify that the effects of TMAO and urea on an uncharged Lennard-Jones polymer chain are those observed in protein systems, i.e. that they act respectively as protective and denaturing osmolytes. Towards this end, a reasonable approach is to follow the time-evolution of an order parameter describing the polymer conformation, like its radius of gyration which will allow us to distinguish between the collapsed and extended configurations. Such profiles are shown for a polymer with a bead interaction parameter εb = 1.0 kJ/mol in Figure 1. For these unrestrained simulations, the initial configurations in each system (water, 1 M TMAO and 7 M urea) were chosen anticipating the effect of this aqueous solution on the polymer conformational equilibrium. Thus, simulations were started from an unfolded, state in water and TMAO solutions (Rg = 11.5 Å), and started from a collapsed configuration (Rg = 4.5 Å) in the urea solution. Each of these simulations were then propagated for 100 ns.
Figure 1.
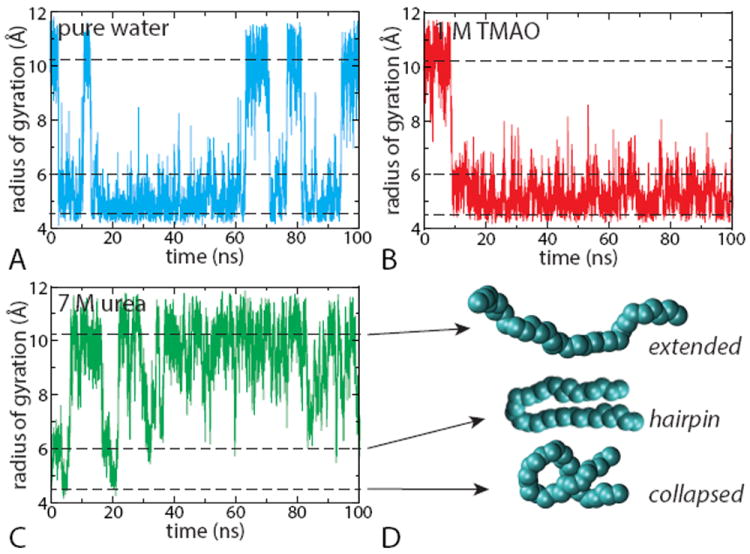
Time profile of polymer’s radius of gyration for εb = 1.0 kJ/mol as obtained from unrestrained simulations in different aqueous solutions: (A) in water, (B) in 1 M TMAO solution and (C) in 7 M urea solution. The horizontal dashed lines corresponding to Rg = 4.5, 6.0 and 10.2 Å represent the most probable values of the radius of gyration for the collapsed (folded), hairpin-like and extended (unfolded) conformations respectively, depicted in (D).
Figure 1 shows that the polymeric chain behaves very differently in the three environments. In water (Figure 1A), the initially-extended polymer quickly collapses and then fluctuates between the collapsed and the extended configurations. In the TMAO solution (Figure 1B), the polymer collapses and remains compact for the whole 100-ns timescale: the extended configuration sampled in pure water is not observed in TMAO on the time scale of the simulation. In contrast, the polymer in urea unfolds, (Figure 1C) but very occasionally revisits more compact states like the hairpin at Rg ≈ 0.6 nm, and very rarely visits the most compact states seen in water.
We thus see that for εb=1.0 kJ/mol, TMAO and urea act respectively as protective and a denaturing osmolytes with respect to the hydrophobic chain. This was already observed for urea30 albeit for a different water model (TIP4P39). It is remarkable that these osmolytes have similar effects on the hydrophobic chain as they do on real proteins. In the following, we aim to better understand this interesting behavior.
The semi-compact hairpin configuration of the chain (Rg ≈ 0.6 nm) observed in the urea solution30 (see Figure 1C) and to some extent in water and TMAO solution (Figure 1A and Figure 1B) can be better understood by considering a two dimensional collective coordinate consisting of the radius of gyration and the end-to-end distance of the chain. Figure 2 shows the joint probability P(L, Rg) of finding a polymer (εb=1.0 kJ/mol) with end-to-end distance L and radius of gyration Rg, for each of the systems shown in Figure 1. To avoid possible biases due to limited sampling in unperturbed simulations, the probability distribution of Rg is first recovered from the PMF obtained via umbrella-sampling simulations. In each window, we later estimate the conditional probability P(L∣Rg) of finding L given Rg. The joint probability is finally recovered using the relation
| (3) |
The distributions shown in Figure 2 confirm our previous findings: in urea the extended configurations are significantly more populated than in water whereas in TMAO they are essentially absent. Moreover, the hairpin state observed at Rg ≈ 0.6 nm and small L in the 2D plots, although more prominent in urea solution, also appear in TMAO solution and in pure water. In contrast, the collapsed state around Rg ≈ 0.45 nm corresponds to higher L, showing that the two order parameters are largely decoupled in this region of the distribution. Unfolded configurations correspond to high values of both L and Rg and give rise to distributions elongated along the diagonal in the 2D plots and only urea has a strong peak there.
Figure 2.
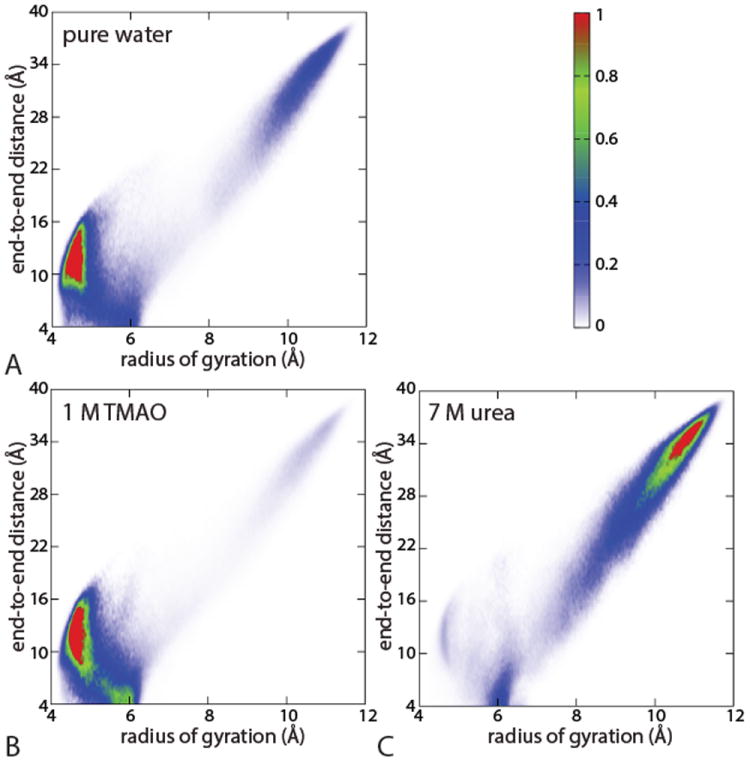
Comparison of joint-probability distribution of polymer chain (with εb = 1 kJ/mol) along radius of gyration and end-to-end distance (A) in water, (B) in 1 M TMAO and (C) in 7 M urea, obtained from umbrella sampling simulations.
The potentials of mean force W (Rg) as a function of the polymer radius of gyration Rg, obtained via umbrella sampling, corresponding probability distributions exp[−βW(Rg)] (for εb=1.0 kJ/mol) and for the three aqueous solutions are shown in Figure 3A and B respectively (and in the Figure S1 for other values of εb). These correspond to the projections of the joint-probability distribution onto the radius of gyration axis. These provide a more reliable and quantitative description of the polymer conformational equilibrium than simulations based on unrestrained MD trajectories (Figure 1) because those would require much longer runs (as later illustrated by the free-energy barriers of 2 to 4 kcal/mol between states).
Figure 3.
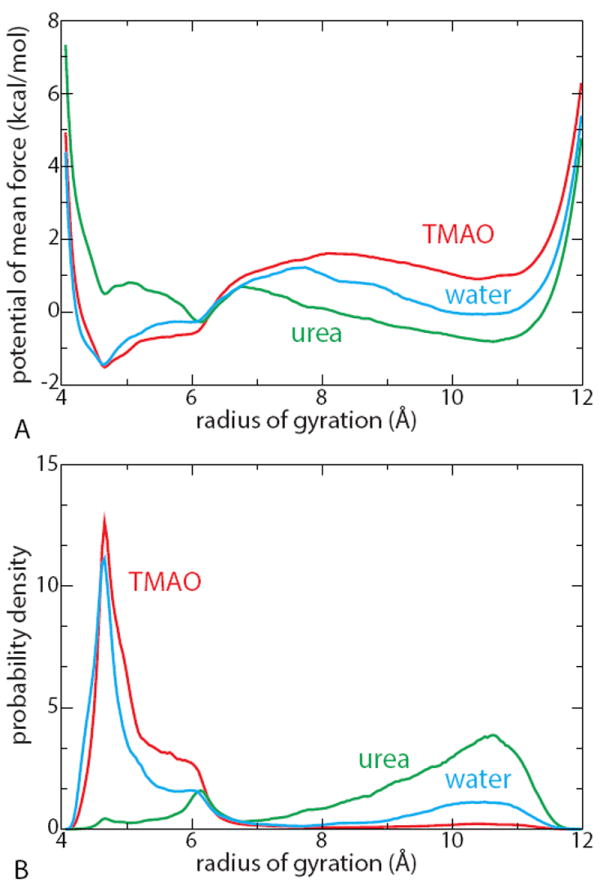
Potential of mean force along the radius of gyration (A) and the corresponding probability distributions (B) in the three aqueous solutions (water, blue; TMAO, red and urea, green) for polymer chains with εb = 1.0 kJ/mol. The PMFs W (Rg) are normalized so that .
As can be seen in Figure 3, the unfolded state in TMAO solution gets destabilized with respect to pure water, whereas the collapsed state is not dramatically affected. Quite remarkably, the unfolded state is almost totally suppressed for TMAO and its collapsed state is more compact for this case of εb = 1.0 kJ/mol.
In previous molecular dynamics simulations of a similar system,23 it was found that that TMAO has little affect on the conformational equilibrium of a hydrophobic polymer chain. In that study a lower polymer bead parameter of εb = 0.6 kJ/mol was used than in our simulations. It is therefore of importance to determine the effect of smaller εb on the behavior of TMAO, as was done earlier for urea solutions.30 In the SI Text and Figure S1, we show that as the hydrophobicity of the chain is increased or equivalently as εb is decreased the chain responds differently to TMAO. The protecting effect of 1 M TMAO solution is thus very sensitive to the value of εb. We find that the protecting role of TMAO is very weak when the chain is strongly hydrophobic (low values of εb = 0.4 and 0.6 kJ/mol), in agreement with the conclusions of the previous study.23 However TMAO’s protecting role becomes much more prominent for larger values εb = 0.8 and 1.0 kJ/mol.
As previously suggested using a different water model,30 the response of the polymer to urea on decreasing εb is radically different. The trend is clearly opposite to that found in water or in TMAO solution: the unfolded state gets stabilized while the collapsed state population progressively disappears, and a significant fraction of the population is found in the hairpin state. Urea therefore exhibits a typical denaturing effect. In strong contrast with TMAO and in agreement with a previous study,30 we show in the Figure S1 that urea readily denatures hydrophobic polymers (e.g. εb = 0.4 kJ/mol) yet its denaturing effect becomes more prominent as εb increases.
To assess the robustness of our results, we also repeated our simulations εb = 1.0 kJ/mol using a different water model39 and found very similar results (see SI Text and Figure S3). Although the force-field we have employed for TMAO has been widely employed in the past and clearly behaves as a protective osmolyte,24,25,29 it has recently been criticized because it underestimates osmotic pressure at high solute concentrations.27 We have repeated our simulations at 1 M using Garcia et al’s modified (“osmotic”) version of this force-field.27 As shown in Figure S2, its effect on the polymer chain nonetheless differs very little from what we observed using the aforementioned forcefield.
Our simulations show that TMAO acts as a protective osmolyte and urea as a denaturant on the polymer chain for εb = 1.0 kJ/mol. TMAO thus acts on this chain similarly to the way it acts on many proteins as found experimentally,6 showing that its effect extends to purely uncharged polymer chains of moderate hydrophobicity. TMAO’s ability to act as a protecting osmolyte depends on the properties of the polymer it is acting on as is shown by its sensitivity to the value of the polymer-bead εb: TMAO seems to have little effect on strongly hydrophobic chains. Urea, on the other hand still denatures them. In the following, we aim to better understand the effect of both osmolytes on the polymer chain with εb = 1.0 kJ/mol.
Molecular Mechanism of Osmolyte-induced (un)folding
Interpretation based on preferential binding and chemical potential of osmolytes
It has been suggested in the literature that denaturants exhibit preferential interaction with protein surfaces while protective osmolytes are preferentially excluded from the surface4-8 because of unfavorable interactions.6,8 Therefore such behavior should be observed in both the local-bulk partition coefficient Kp (Eq. 1) and the preferential binding coefficient Γ (Eq. 2).
In Figure 4A-B we compare the dependence of Kp on polymer conformation as a function of distance from polymer in both TMAO and urea solutions. If the local domain is defined as the polymer’s first solvation shell, then the representatives values for Kp (and later Γ) should be taken near ≈ 6 Å (dashed line), which corresponds to the first minima of the radial distribution functions between the polymer and cosolvent molecules (urea or TMAO). As expected, urea molecules accumulate next to both collapsed and extended states of the polymer, leading to Kp > 1 (Figure 4B). Kp is little sensitive to the polymer conformation but it is always slightly higher in the extended state. The local-bulk partition coefficient of TMAO is also greater than 1 (Figure 4A), implying that TMAO also binds to the polymer surface. This result contradicts the popular view that protective osmolytes are believed to be preferentially excluded from the protein surface.4-8 However, this should not be surprising since TMAO is mostly hydrophobic and thus might be better accommodated in the polymer hydration shell than in bulk solution. But a key observation is that Kp displays significant conformation-dependence: it is higher near a collapsed configuration than near an extended configuration. Therefore, TMAO strongly interacts with the polymer (Kp > 1) but there is an effective depletion next to extended conformations of polymer relative to collapsed conformations.
Figure 4.
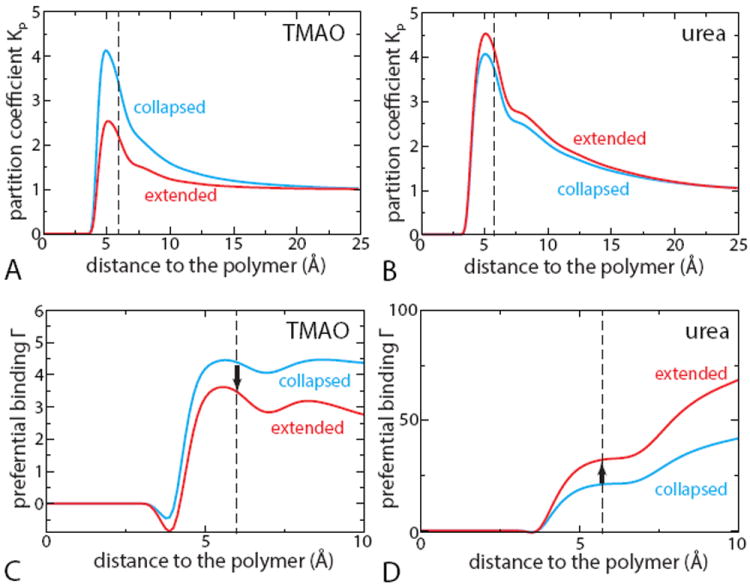
Preferential binding constants of TMAO in 1 M TMAO (red) and urea in 7 M urea (green) as a function to the distance to the polymer (for chains with εb = 1.0 kJ/mol), frozen in an collapsed (black) or extended (red) configurations extended configuration. Vertical dashed lines indicate the position of the polymer first solvation shell in each case (6 Å for TMAO and 5.7 Å for urea), and the black arrows in C and D represent the relevant value of ΔΓ.
Although the local-bulk partition coefficient provides a better description of the preferential interaction because of its intensive nature, it is not directly connected to experimental observables. In contrast, the preferential binding coefficient Γ can be measured experimentally, e.g. using the vapor-pressure osmometry technique.7 The effect of preferential binding on a conformational equilibrium between the folded and the unfolded configurations F ⇌ U (with an equilibrium constant K) is usually understood in terms of the thermodynamic calculation first introduced by Wyman and Tanford,2,3 which leads to
| (4) |
where as is the activity of the cosolvent in the binary solution. According to Eq. 4, an increase in the concentration of the cosolvent would lead to the biomolecule unfolding if ΔΓ > 0, and in contrast would favor the folded state over the unfolded one if ΔΓ < 0.
In Figure 4C-D we show the average preferential binding coefficients G for both collapsed and extended conformations in TMAO and urea solutions. As already discussed above, one should consider the Γ values at the distance corresponding to the polymer first solvation shell. Not surprisingly, in all cases Γ is positive, which is equivalent to Kp > 1 (Figure 4A-B). Similarly, the trends in the difference between the extended and collapsed configurations ΔΓ follow that of ΔKp: ΔΓ is negative for TMAO, which stabilizes the folded state over the unfolded one, while the positive ΔΓ for urea clearly corresponds to its denaturing effect. Our results are therefore in agreement with the current consensus on the osmolyte effect4-8 summarized by Eq.4.
The main difference between our work and previous studies on proteins is that the sign of ΔΓ is different from that of G for TMAO (they have the same sign for urea). This surprising observation can be understood if we consider the relationship between Kp and Γ. Indeed combining Eqs. 1 and 2 with the hypothesis that the bulk domain is large with respect to the local domain (i.e., ), leads to
| (5) |
Therefore ΔΓ will depend on both Δ〈ns〉 and Δ(1/Kp) (note that these two terms are not independent of each other). In experimental studies of proteins, it was found that Γ is proportional to the solvent surface accessible area S.7 Since 〈ns〉 is also proportional to S, Kp is expected to be the same whether the protein is folded or not. This may not be surprising given that the nature of the exposed groups is the same in the folded and the unfolded state. However for our polymer, Kp is very much conformation-dependent, while ns is only marginally higher in the extended state. This therefore provides an explanation for why ΔΓ is negative while TMAO always accumulates in the hydration shell (Γ > 0). Finally, it is interesting to note that simulations of decaalanine have found G to be positive for TMAO29 even though it was observed to behave as a protective osmolyte.
Conformation-dependence of the osmolyte chemical potentials in the polymer first solvation shell
To obtain a better understanding of how TMAO can preferentially bind to the polymer surface, yet still behave as a protective osmolyte favoring the polymer collapsed state, we investigate the free-energy changes (chemical potential) associated with the insertion of a single osmolyte or water molecule next to different conformations (both collapsed and extended) of the polymer. These chemical potentials were determined from thermodynamic integration (see Methods, SI Text and Figures S4 and S5). Insertion of a TMAO and a water molecule in the 1 M TMAO solution (or urea and water in the 7 M urea solution) was considered in three different cases: in the bulk, i.e. far from the polymer; in the first hydration shell of the polymer frozen in a collapsed configuration; and in the first hydration shell of polymer frozen in an extended configuration.
Table 1 lists the results of thermodynamic integration; namely, the van der Waals and electrostatic contributions to the chemical potentials of urea and TMAO in the different cases. In all cases insertion of a osmolyte molecule is more favorable next to the polymer than it is in the bulk: this is in agreement with the preferential binding values discussed above. However, the chemical potentials of TMAO and urea, relative to bulk values follow opposite trends as far as the conformation-dependence is concerned. An inserted single TMAO molecule is more stable (has lower free energy) next to the collapsed conformation of polymer than next to the extended. Its free-energy is lower by 0.8 kcal/mol per TMAO molecule. This is mainly due to the more favorable free energy contribution from the van der Waals interaction, which overcomes the slight destabilization in the electrostatic contribution in the collapsed conformation due to less exposure to water or other TMAO molecules in the collapsed state than in the extended state. Given the importance of the van der Waals term for this (mostly) hydrophobic molecule, the total free-energy change is dominated by this contribution. In urea, however, the van der Waals term is small and does not totally compensate the electrostatic contribution. Therefore the insertion of a urea molecule is more favorable next to the extended state.
Table 1.
Different free-energy contributions (in units of kcal/mol) for inserting single osmolyte molecules in the first solvation shell of the polymer (with εb = 1.0 kJ/mol) in 1 M TMAO and 7 M urea. Δμbulk represents the difference with respect to the bulk solution, and the average number of molecules in first solvation shell of the polymer Nk is also given. Standard deviations are given within parentheses.
| System | Gvdw | Gcoulomb | Gtotal | Δμbulk | < Nk > |
|---|---|---|---|---|---|
| TMAO | |||||
| bulk | +1.99(0.02) | −13.97(0.02) | −11.97(0.01) | 0 | − |
| collapsed | −0.52(0.36) | −13.08(0.22) | −13.60(0.27) | −1.63 | 6.2(0.2) |
| extended | +1.04(0.45) | −13.80(0.08) | −12.76(0.38) | −0.79 | 6.3(0.6) |
|
| |||||
| urea | |||||
| bulk | −0.17(0.04) | −13.27(0.05) | −13.44(0.09) | 0 | − |
| collapsed | −1.33(0.71) | −12.45(0.53) | −13.78(0.20) | −0.33 | 28.6(0.3) |
| extended | −0.99(0.29) | −12.91(0.23) | −13.90(0.15) | −0.46 | 41.9(0.8) |
The observed differences and respective contributions of van der Waals and electrostatic free-energy, which drive the preferential interaction with one state or another, suggest a mostly enthalpic origin to this behavior. This is confirmed by considering the distributions of both van der Waals and electrostatic energy distributions for single osmolyte molecule in the hydration shell of the polymer, either in a collapsed or in an extended conformation (see SI Text and Table S1). The observed trends are similar to that found from the FEP study — TMAO interacts preferentially with the polymer collapsed state because of the van der Waals contribution, leading to a difference of ≈ 0.2 kcal/mol as compared to the extended state and ≈ 0.5 kcal/mol with respect to the bulk phase. In contrast, a slight stabilization of urea in the extended polymer hydration shell with respect to the collapsed state is found (≈ 0.2 kcal/mol) and appears to be driven by the electrostatic contribution. In both cases, a significant stabilization is found as compared to the bulk phase (≈ 1.5 kcal/mol).
A free-energy based model for action of protecting and denaturing osmolyte
The discussion in the previous paragraphs have focussed on only the contributions of the chemical potential of osmolyte (TMAO or urea) to the free-energy difference between the polymer collapsed and the extended states. We now try to build a free energy-based thermodynamic model based on the above FEP data, with the goal of validating it against the net PMF profiles of the polymer in the respective solutions (as previously discussed in Figure 3). The net free-energy change for going from a collapsed to an extended configuration can be expressed as
| (6) |
where (respectively ) is the average number of solvent molecules of type k (water w or osmolyte s) in the first hydration shell of the polymer in a collapsed (respect. extended) configuration, and (respect. ) their associated chemical potentials. We assume here that the chemical potentials of molecules beyond the first hydration shell are similar for both polymer configurations.
To evaluate Eq. 6, we must separately determine three individual contributions:
The first term of the equation corresponds to the free-energy difference between the collapsed and extended states of the polymer itself in gas phase. It was obtained by repeating our simulations in the gas phase and by performing umbrella sampling calculations to estimate the free-energy difference between the collapsed and the extended state in the absence of solvent, which was found to be kcal/mol in favor of the collapsed conformation.
To calculate the second term, which describes water’s contribution to the total free energy difference, we repeated our FEP calculations for water as well using the same method as detailed above for the osmolyte molecules. Results are reported in Table 2. In all cases (bulk, TMAO or urea solutions), the difference in chemical potential between inserting a water molecule next to the collapsed or next to the extended state is very small, and slightly negative with respect to bulk. At the same time, the hydration number is significantly increasing because of a larger surface area in the extended state. Both the changes in chemical potential and that of the number of molecules contribute to the total free-energy difference between the collapsed and the extended state due to water molecules. A detailed analysis discussed in the SI Text and Table S2 shows that in all cases, the dominant contributions logically arise from δNw.
The third term describes contribution of osmolyte molecules (TMAO or urea) to the net free energy difference. The chemical potentials in each configuration were already discussed and are given in Table 1, which also contains the total number of osmolyte molecules in the polymer first hydration shell. Similarly to water, we can define and . For urea, both the δ μs and δNs bring a negative contribution to the solvent-induced free-energy difference, which therefore favors the extended state. For TMAO, the two terms bring opposite contributions (SI Text and Table S2). While δNs > 0 favors the extended conformation, it is the dominant contribution of the large δμs that leads to a positive , stabilizing the collapsed state relative to the extended one.
Table 2.
Total free-energy cost Δμbulk (with respect to the bulk solution reference) for inserting single water molecules in the first solvation shell of the polymer (with εb = 1.0 kJ/mol) in pure water, 1 M TMAO and 7 M urea (in units of kcal/mol), and average number of water molecules in the polymer first hydration shell Nk. Standard deviations are given within parentheses.
| System | Δμbulk | < Nk > | |
|---|---|---|---|
| pure water | |||
| collapsed | −0.17(0.04) | 73.8 | |
| extended | −0.15(0.02) | 111.3 | |
|
| |||
| TMAO | |||
| collapsed | −0.08(0.05) | 66.9 | |
| extended | −0.11(0.04) | 106.6 | |
|
| |||
| urea | |||
| collapsed | −0.13(0.14) | 33.0 | |
| extended | −0.17(0.31) | 43.5 | |
Figure 5 represents these three different individual contributions schematically. In pure water, the water contribution favors the extended state, as shown above. However, it does not totally compensate the polymer contribution (this will depend on the value of ε), so that the folded state is still the most stable in this case, as calculated independently with our umbrella sampling simulations (Figure 3). In the TMAO solution, the water contribution is only slightly perturbed with respect to the pure water case, but TMAO molecules will bring a contribution favoring the collapsed state, in agreement with our earlier estimation based on PMF calculations (Figure 3). Finally in the urea solution, the urea contribution stabilizes the extended conformation greatly so that the overall free-energy difference becomes favorable towards extended conformations, as we have observed.
Figure 5.
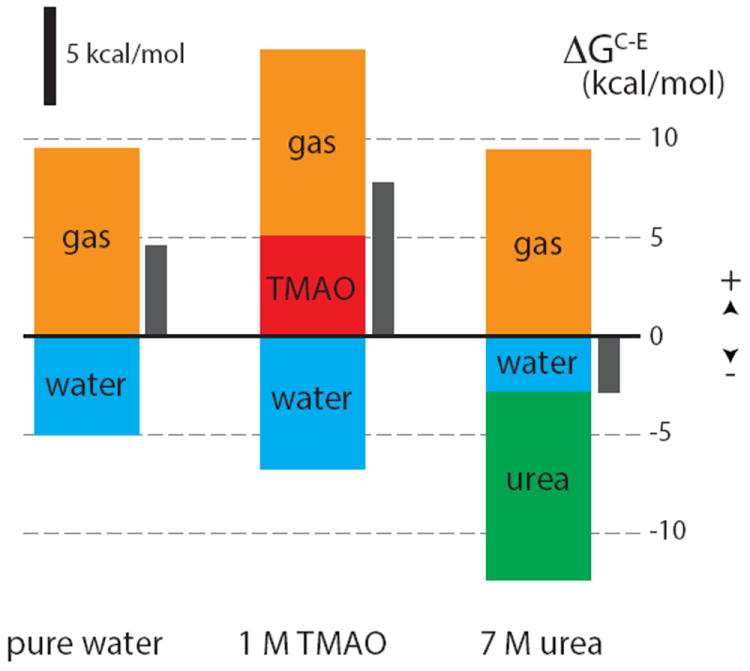
Histograms showing the different contributions to the free-energy cost of inserting water or osmolyte molecules in the polymer hydration shell.
The semi-quantitative agreement that is observed in all three cases with the free-energy differences calculated from umbrella sampling simulations further validates the current approach. Note however that for TMAO and urea solutions, we systematically overestimate the respective stabilization. A possible explanation for this discrepancy is that we consider only a few sites for insertion of a osmolyte or a water molecule in the hydration shell; another source of discrepancy could arise from neglecting the contribution of molecules in the second solvation shell. However, the fact that the trend is correctly and self-consistently predicted and also that the energy distributions exhibit exactly the same behavior makes us confident in this approach. Finally, the decomposition presented in the SI Text in terms of the respective contributions of δμs and δNs also suggests that an osmolyte’s behavior cannot be predicted from the sign of δμs alone, as detailed in the SI Text.1
Conclusions
In this paper, we considered the effect of osmolytes (TMAO and urea) on a simple polymer, consisting of a short Lennard-Jones chain similar to an alkane or lipid chain in aqueous solutions of the two osmolytes. This model is reminiscent of the model used in our previous paper30 directed at understanding urea denaturation of hydrophobic collapse.
Here we determined the free energy landscapes of the hydrophobic polymer as a function of its radius of gyration. We show that 1 M TMAO and 7 M urea act dramatically differently on model polymer chains and their behaviors are sensitive to the strength of the attractive dispersion interactions of the chain with its environment: when these dispersion interactions are high enough, TMAO suppresses the formation of extended conformations of the hydrophobic polymer as compared to water, while urea promotes formation of extended conformations. Quite surprisingly, we find that both protecting and denaturing osmolytes strongly interact with the polymer (with both having a preferential binding constant greater than zero), in contrast with existing explanations of the osmolyte effect. An extensive free energy analysis suggests that protective osmolytes are not necessarily excluded from the polymer surface. What really matters is the effective depletion of the osmolyte as the polymer conformation changes, in agreement with the current consensus on the osmolyte effect.4-8 Indeed for TMAO, it is the much more favorable free energy of insertion of a single osmolyte near the collapsed configurations of the polymer than near the extended configurations that dictates its propensity to drive the system towards the collapsed conformation, and therefore to lead to its protective effect. This appears to be driven by van der Waals interactions. In contrast, urea is preferentially stabilized next to the extended conformation because the smaller van der Waals contributions do not compensate the electrostatic contribution, suggesting this as an explanation of its denaturing effect.
A thermodynamic model taking into account the different contributions (gas-phase, water and osmolyte) to the polymer conformational equilibrium was developed. In the aqueous solution of urea, the free energy contribution coming from urea and water easily cooperates to shift the polymer towards extended conformations of the polymer. In the aqueous solution of TMAO, TMAO’s free energy contribution favors the collapsed conformation and it is able to overcome the water’s free energy contribution which favors the extended conformation: overall, the equilibrium is shifted towards the collapsed conformation.
We believe that this simple thermodynamic model provides an interesting perspective for explaining the role of protecting and denaturing osmolytes on simple macromolecules. Although the model is very simple, it provides fresh insights on the action of various osmolytes. Manipulation of simple polymers at the single molecule level has been recently achieved40 and we believe that the effect of osmolytes such as urea or TMAO on the polymer conformational equilibrium could be probed by such techniques. From a simulation perspective, it will be interesting in the future to extend our free-energy based approach to systems of increasing complexity like charged polymers, real peptides or proteins to shed light on the role of osmolytes on macromolecular conformations.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health [NIH-GM4330 (to B.J.B.)] and by the National Science Foundation through [via Grant No. NSF-CHE-0910943]. We gratefully acknowledge the computational support of the Computational Center for Nanotechnology Innovations (CCNI) at Rensselaer Polytechnic Institute (RPI). This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number OCI-1053575.
Footnotes
Indeed the main δNs contribution (which is equal to ) can overcome that of δμs if the relative variations of the hydration number when going from collapsed to extended conformations are larger than that of the associated chemical potentials, even if the osmolyte molecules are more stable around the collapsed conformations.
Supporting Information Available
Effect of the bead interaction parameter εb on the polymer conformational equilibrium in pure water and in aqueous solutions of TMAO and urea, results for an alternative TMAO forcefield, results for an alternative water model, details about the thermodynamics integrations, energy distribution for solute molecules in the polymer first hydration shell, decomposition of the solvent contribution to the free-energy between the polymer collapsed and the extended states.
This material is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with Water Stress - Evolution of Osmolyte Systems. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 2.Wyman J. Linked Functions and Reciprocal Effects in Hemoglobin: a Second Look. Adv Protein Chem. 1964;19:223–286. doi: 10.1016/s0065-3233(08)60190-4. [DOI] [PubMed] [Google Scholar]
- 3.Tanford C. Extension of the Theory of Linked Functions to Incorporate the Effects of Protein Hydration. J Mol Biol. 1969;39:539–544. doi: 10.1016/0022-2836(69)90143-0. [DOI] [PubMed] [Google Scholar]
- 4.Timasheff SN. Protein-Solvent Preferential Interactions, Protein Hydration, and the Modulation of Biochemical Reactions by Solvent Components. Proc Natl Acad Sci USA. 2002;99:9721–9726. doi: 10.1073/pnas.122225399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsegian VA, Rand RP, Rau DC. Osmotic Stress, Crowding, Preferential Hydration, and Binding: A Comparison of Perspectives. Proc Natl Acad Sci USA. 2000;97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolen D, Rose G. Structure and Energetics of the Hydrogen-Bonded Backbone in Protein Folding. Annu Rev Biochem. 2008;77:339–362. doi: 10.1146/annurev.biochem.77.061306.131357. [DOI] [PubMed] [Google Scholar]
- 7.Courtenay ES, Capp MW, Anderson CF, Record MT., Jr Vapor Pressure Osmometry Studies of Osmolyte-Protein Interactions: Implications for the Action of Osmoprotectants in Vivo and for the Interpretation of “Osmotic Stress” Experiments in Vitro. Biochemistry. 2000;39:4455–4471. doi: 10.1021/bi992887l. [DOI] [PubMed] [Google Scholar]
- 8.Canchi DR, García AE. Cosolvent Effects on Protein Stability. Annu Rev Phys Chem. 2013;64:273–293. doi: 10.1146/annurev-physchem-040412-110156. [DOI] [PubMed] [Google Scholar]
- 9.Street TO, Bolen DW, Rose GD. A Molecular Mechanism for Osmolyte-Induced Protein Stability. Proc Natl Acad Sci USA. 2006;103:13997–14002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang AJ, Bolen DW. A Naturally Occurring Protective System in Urea-Rich Cells: Mechanism of Osmolyte Protection of Proteins against Urea Denaturation. Biochemistry. 1997;36:9101–9108. doi: 10.1021/bi970247h. [DOI] [PubMed] [Google Scholar]
- 11.Hochachka PW, Somero GN. Biochemical Adaptation. Mechanism and Process in Physiological Evolution. Oxford University Press; 2002. [Google Scholar]
- 12.Lin TY, Timasheff SN. Why Do Some Organisms Use a Urea-Methylamine Mixture as Osmolyte - Thermodynamic Compensation of Urea and Trimethylamine N-oxide Interactions with Proteins. Biochemistry. 1994;33:12695–12701. doi: 10.1021/bi00208a021. [DOI] [PubMed] [Google Scholar]
- 13.Tatzelt J, Prusiner S, Welch W. Chemical Chaperones Interfere with the Formation of Scrapie Prion Protein. EMBO J. 1996;15:6363–6373. [PMC free article] [PubMed] [Google Scholar]
- 14.Nandi PK, Bera A, Sizaret P-Y. Osmolyte TrimethylamineN-oxide Converts Recombinant Alpha-Helical Prion Protein to its Soluble Beta-Structured Form at High Temperature. J Mol Biol. 2006;362:810–820. doi: 10.1016/j.jmb.2006.07.060. [DOI] [PubMed] [Google Scholar]
- 15.Tseng HC, Graves DJ. Natural Methylamine Osmolytes, Trimethylamine N-oxide and Betaine, Increase Tau-Induced Polymerization of Microtubules. Biochem Biophys Res Comm. 1998;250:726–730. doi: 10.1006/bbrc.1998.9382. [DOI] [PubMed] [Google Scholar]
- 16.Scaramozzino F, Peterson DW, Farmer P, Gerig JT, Graves DJ, Lew J. TMAO promotes Fibrillization and Microtubule Assembly Activity in the C-terminal Repeat Region of Tau. Biochemistry. 2006;45:3684–3691. doi: 10.1021/bi052167g. [DOI] [PubMed] [Google Scholar]
- 17.Uversky VN, Li J, Fink AL. TrimethylamineN-oxide-Induced Folding of Alpha-Synuclein. FEBS Lett. 2001;509:31–35. doi: 10.1016/s0014-5793(01)03121-0. [DOI] [PubMed] [Google Scholar]
- 18.Morello JP, Petaja-Repo UE, Bichet DG, Bouvier M. Pharmacological Chaperones: A New Twist on Receptor Folding. Trends Pharmacol Sci. 2000;21:466–469. doi: 10.1016/s0165-6147(00)01575-3. [DOI] [PubMed] [Google Scholar]
- 19.Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennion BJ, Daggett V. Counteraction of Urea-Induced Protein Denaturation by Trimethylamine N-oxide: A Chemical Chaperone at Atomic Resolution. Proc Natl Acad Sci USA. 2004;101:6433–6438. doi: 10.1073/pnas.0308633101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp KA, Madan B, Manas E, Vanderkooi JM. Water Structure Changes Induced by Hydrophobic and Polar Solutes Revealed by Simulations and Infrared Spectroscopy. J Chem Phys. 2001;114:1791–1796. [Google Scholar]
- 22.Hunger J, Tielrooij K-J, Buchner R, Bonn M, Bakker HJ. Complex Formation in Aqueous Trimethylamine N-oxide (TMAO) Solutions. J Phys Chem B. 2012;116:4783–4795. doi: 10.1021/jp212542q. [DOI] [PubMed] [Google Scholar]
- 23.Athawale MV, Dordick JS, Garde S. Osmolyte TrimethylamineN-oxide Does Not Affect the Strength of Hydrophobic Interactions: Origin of Osmolyte Compatibility. Biophys J. 2005;89:858–866. doi: 10.1529/biophysj.104.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho SS, Reddy G, Straub JE, Thirumalai D. Entropic Stabilization of Proteins by TMAO. J Phys Chem B. 2011;115:13401–13407. doi: 10.1021/jp207289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu CY, Lynch GC, Kokubo H, Pettitt BM. Trimethylamine N-oxide Influence on the Backbone of Proteins: An Oligoglycine Model. Proteins. 2010;78:695–704. doi: 10.1002/prot.22598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stirnemann G, Hynes JT, Laage D. Water Hydrogen Bond Dynamics in Aqueous Solutions of Amphiphiles. J Phys Chem B. 2010;114:3052–3059. doi: 10.1021/jp9116886. [DOI] [PubMed] [Google Scholar]
- 27.Canchi DR, Jayasimha P, Rau DC, Makhatadze GI, Garcia AE. Molecular Mechanism for the Preferential Exclusion of TMAO from Protein Surfaces. J Phys Chem B. 2012;116:12095–12104. doi: 10.1021/jp304298c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paul S, Patey GN. The Influence of Urea and Trimethylamine N-oxide on Hydrophobic Interactions. J Phys Chem B. 2007;111:7932–7933. doi: 10.1021/jp0733668. [DOI] [PubMed] [Google Scholar]
- 29.Kokubo H, Hu C, Pettitt BM. Peptide Conformational Preferences in Osmolyte Solutions: Transfer Free Energies of Decaalanine Folding. J Am Chem Soc. 2011;133:1849–1858. doi: 10.1021/ja1078128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zangi R, Zhou R, Berne BJ. Urea’s Action on Hydrophobic Interactions. J Am Chem Soc. 2009;131:1535–1541. doi: 10.1021/ja807887g. [DOI] [PubMed] [Google Scholar]
- 31.Berendsen HJC, Grigera JR, Straatsma TP. The Missing Term in Effective Pair Potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 32.Duffy EM, Severance D, Jorgensen WL. Urea: Potential Functions, log P, and Free Energy of Hydration. lsr J Chem. 1993;33:323–330. [Google Scholar]
- 33.Kast KM, Brickman J, Kast SM, Berry RS. Binary Phases of AliphaticN-Oxides and Water: Force Field Development and Molecular Dynamics Simulation. J Phys Chem A. 2003;107:5342–5351. [Google Scholar]
- 34.Hess B, Kutzner C, Van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 35.Bonomi M, Branduardi D, Bussi G, Camilloni C, Parrinello M. PLUMED: A Portable Plugin for Free-Energy Calculations with Molecular Dynamics. Comp Phys Comm. 2009;180:1961–1972. [Google Scholar]
- 36.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. THE Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules I. The Method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 37.Grossfield A. WHAM: The Weighted Histogram Analysis Method, version 2.0. http://membrane.urmc.rochester.edu/content/wham.
- 38.Shukla D, Shinde C, Trout BL. Molecular Computations of Preferential Interaction Co-efficients of Proteins. J Phys Chem B. 2009;113:12546–12554. doi: 10.1021/jp810949t. [DOI] [PubMed] [Google Scholar]
- 39.Jorgensen W, Chandrasekhar J, Madura J, Impey R, Klein M. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 40.Li ITS, Walker GC. Signature of Hydrophobic Hydration in a Single Polymer. Proc Natl Acad Sci USA. 2011;108:16527–16532. doi: 10.1073/pnas.1105450108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


