Abstract
The discovery of small RNA silencing pathways has greatly extended our knowledge of gene regulation. Small RNAs have been presumed to play a role in every field of biology because they affect many biological processes via regulation of gene expression and chromatin remodeling. Most well-known examples of affected processes are development, fertility, and maintenance of genome stability. Here we review the role of the three main endogenous small RNA silencing pathways in Caenorhabditis elegans: microRNAs, endogenous small interfering RNAs, and PIWI-interacting RNAs. After providing an entry-level overview on how these pathways function, we discuss research on other nematode species providing insight into the evolution of these small RNA pathways. In understanding the differences between the endogenous small RNA pathways and their evolution, a more comprehensive picture is formed of the functions and effects of small RNAs.
Keywords: nematode, C. elegans, small RNA, Argonaute, miRNA, endo-siRNA, piRNA, RNA silencing, RNA interference
Introduction
Since their discovery 15 y ago, RNA silencing pathways mediated by small RNAs have been presumed to play a role in all fields of biology.1 Small RNAs are key because, as regulators of gene expression, they affect many processes in most organisms. The best-known examples are development, fertility, and maintenance of genome stability.2,3 Additionally, small RNAs are widely used as research tools; for example, to study gene function or genetic variation.4,5 The accumulated knowledge about small RNAs is scattered over numerous research papers. The objective of this review is to provide an overview of the three main endogenous small RNA silencing pathways in nematodes.
In general, small RNAs affect gene expression specifically via a complex formed by the binding of a small, non-coding, single-stranded RNA to an Argonaute protein. This complex can influence gene expression via translation inhibition, mRNA degradation, mRNA storage, or epigenetic changes.3,6 Together, and in addition to the “classic” transcription regulator genes, the RNA silencing pathways form an extra layer of gene regulation. Most of these pathways are conserved throughout the animal kingdom.7,8 Related pathways performing similar functions have also been found in plants, fungi, and bacteria.3,8,9
Nematodes
Nematodes are the most abundant animals on earth and live in a wide range of habitats, including fresh water, salt water, and soil.10 Some are free-living, while others parasitize plants, vertebrates, insects, or other nematodes.10 Studying nematodes provides knowledge that can have practical applications in the fields of agriculture, human, and animal health, as well as fundamental knowledge of evolutionarily conserved processes.
A large part of nematode research has been conducted on the model species Caenorhabditis elegans (Nematoda; Rhabditidae) because of its transparency, ease of cultivation, convenience of manipulation, short life cycle, genetic tractability, and relatively small, fully sequenced, genome.11,12 Most of the knowledge we have on endogenous RNA silencing pathways in nematodes comes from studies in C. elegans.
Molecular phylogenetic research has given insight into nematode evolution.10,13-15 Figure 1 shows an overview of phylogenetic relationships of nematodes that have (part of) their genome published. Comparing different sequenced nematode species with C. elegans provides a great opportunity to study the evolution of small RNAs in evolutionarily near and distant species. Sequencing of several nematode species has revealed that many proteins found to be essential for the small RNA pathways in C. elegans seem not to have orthologs in all species.16 Additionally, there is evidence to support the view that throughout evolution small RNAs provide a way for nematodes to adapt to changes in their environment and life style.17-19
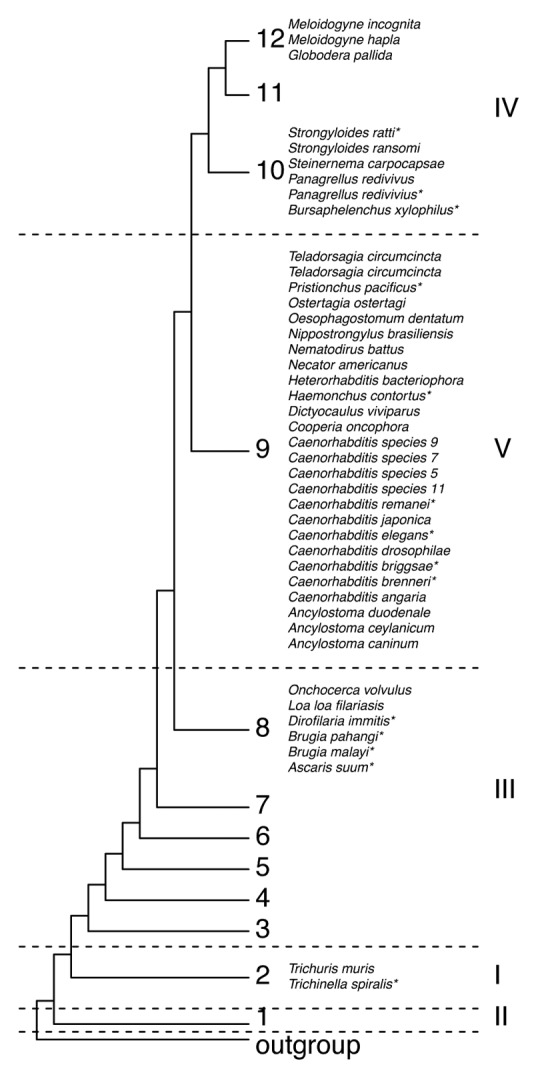
Figure 1. Phylogeny of sequenced nematodes. Phylogenetic relationship of nematodes that have (part of) their DNA sequences published or are being sequenced (as indicated on www.wormbase.org). The clades are shown according to Van Megen et al. in Arabic numbers and to Blaxter et al. in Roman numbers.14,96 In both studies, the phylogenetic relationships were determined by the DNA sequences of the small ribosomal subunits, with 1215 and 53 sequences, respectively. An asterisk indicates that this species has been studied for (some aspects of) the small RNA response.
Endogenous Small RNAs and Argonautes
A wide range of small, non-coding RNAs have been found in C. elegans (Fig. 2A). The three main endogenous small RNA pathways that can be distinguished are: microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs), and PIWI (P-element-induced wimpy testis) interacting RNAs (piRNAs). Each of these pathways use RNAs with different characteristics and are active during different processes (Fig. 2B and C). Since the full impact of small RNAs has become apparent in C. elegans, researchers have been trying to find and analyze small RNAs in non-model nematodes, for example, in the animal parasite Ascaris suum.20
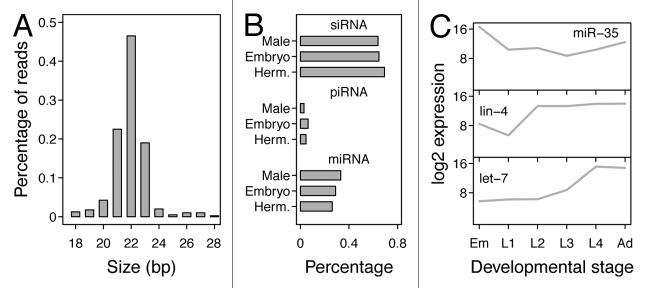
Figure 2. Small RNAs and developmental dynamics. Panel A shows an example of the size distribution of small RNAs in adult hermaphrodites, adapted from reference 42. These can be divided over several classes of small RNAs, which are shown in panel B. In panel B, it can be seen that the relative amounts of small RNAs change throughout development and are different between sexes, adapted from reference 42. This is illustrated in more detail in panel C, where the expression of three different microRNAs is shown throughout the hermaphroditic development, adapted from reference 97.
Argonautes, the effector proteins, fulfill a central role in all small RNA pathways. C. elegans has 25 different Argonautes clustered into three clades: the Argonaute-like proteins, the PIWI-like proteins, and a worm-specific clade of Argonautes, the WAGO (Worm Argonautes) proteins.19,21 Several Argonautes function only in one specific small RNA pathway. However, the exact function, role, and specificity of many Argonautes remains unknown.
miRNAs Enforce Robust Developmental Programs
MicroRNAs (miRNAs) were first discovered in C. elegans. The lin-4 gene, known to control the timing of C. elegans larval development, was found not to encode for a protein, but for a small RNA.22 These small RNAs reduced the amount of LIN-14 protein without noticeably changing the levels of the lin-14 mRNA. The second miRNA discovered in C. elegans, let-7, was found to be present in a wide range of animal species.23,24 Many miRNAs have since been discovered both in plant and animal species.25 It is very clear that the miRNA pathway as a whole has an essential role during development as well as during adulthood (Fig. 2C). C. elegans double mutants of both miRNA-specific Argonautes ALG-1 and ALG-2, show lethality due to a range of severe developmental defects.26,27 Overall, miRNAs help to ensure the robustness of developmental and physiological pathways.2
Primary-miRNAs (pri-miRNAs) are the transcripts of miRNA genes synthesized by RNA polymerase II (Fig. 3).28 These pri-miRNAs are cleaved by the RNase III enzyme Drosha into ~65 nucleotide precursor-miRNAs (pre-miRNAs).29 Pre-miRNAs have a characteristic stem-loop structure and are transported into the cytoplasm.30,31 There, they are processed into mature 22 nucleotide miRNAs by the RNase III enzyme Dicer and one of the miRNA-specific Argonaute proteins ALG-1 or ALG-2 (Table 1).26,32 The mature double stranded miRNA is then bound by ALG-1 or ALG-2.26 The Argonaute protein will release one of the strands and remain bound to the other strand. The complex, consisting of the Argonaute bound to the single stranded miRNA, is called the miRNA-induced silencing complex (miRISC).33,34 The miRISC binds to an mRNA, which is complementary to the seed region (see below) of the single stranded miRNA in the complex and causes silencing of the mRNA. The seed region is approximately six to eight nucleotides near the 5′ end of the miRNA. These nucleotides are thought to be very important for the specificity of targeting mRNAs.35,36 The mechanism by which the bound mRNA is post-transcriptionally silenced is still debated.37,38 One currently popular model is that miRNAs repress translation and promote mRNA de-adenylation. The de-adenylation will cause degradation of the target mRNA.6,37,39 Because the target mRNA is only complementary to the short seed region, accurate in silico prediction of miRNA targets is problematic. Recently, a web accessible database (miRNA_Targets: http://mamsap.it.deakin.edu.au/mirna_targets/) specifically aimed at miRNA target predictions has been created to share results.40 But also at miRBase validated and predicted targets can be found.25,41
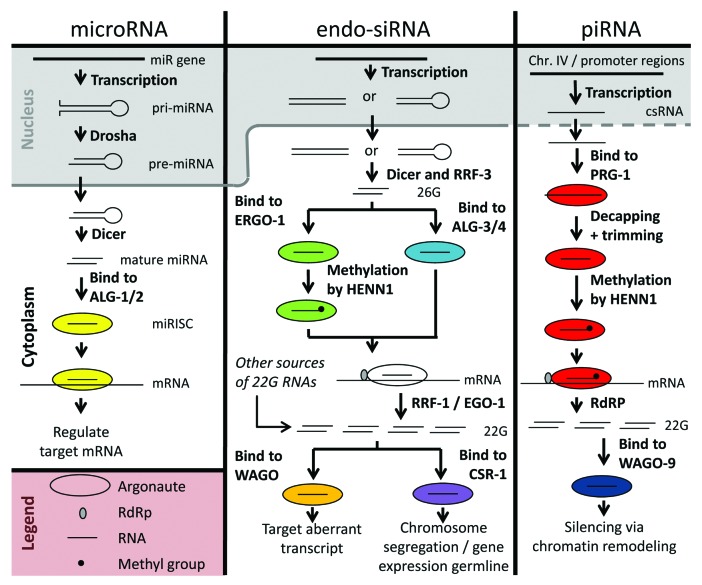
Figure 3. Small RNA pathways in C. elegans. Overview of the discussed endogenous small RNA pathways in C. elegans. The dashed line between nucleus and cytoplasm in the piRNA pathway means that it is not clear when the piRNAs leave the nucleus. For the miRNAs and endo-siRNAs, it is known exactly, indicated by a solid line.
Table 1. Overview of the endogenous small RNAs in C. elegans that are discussed in the main text (for references, see main text).
| Signaturea | Argonaute | Methylation by Henn-1 | Functions in | |
|---|---|---|---|---|
| miRNA | 22 | ALG1/2 | No | Germline + somatic tissue |
| endo-siRNA | 26G | ERGO-1 | Yes | Oocyte + embryo |
| 26G | ALG3/4 | No | Sperm | |
| 22G | WAGOs | No | Germline + somatic tissue | |
| 22G | CSR-1 | No | Germline | |
| piRNA | 21U | PRG-1 | Yes | Germline |
| 22G | WAGO-9 | No | Germline |
a) Signature = length (no. of nucleotides) of small RNA + nucleotide at its 5′ end.
miRNAs are identified by sequencing small RNAs or by predicting miRNAs by means of bioinformatics analyses. All known miRNAs can be found online in the miRBase database.25,41 To predict miRNA candidates the conservation of the seed region is often used as an indicator. When several mature miRNAs are derived from different genomic loci, but share the same seed sequence, they form a “miR family” or “seed family.”20,42
To study miRNA evolution, de Wit et al.43 sequenced the small RNAs of C. elegans, C. briggsae, C. remanei, and Pristionchus pacificus. They showed that more than half of all sequenced miRNAs were conserved at the seed region, with only a few miRNAs that were species specific within these four nematode species.43 This was supported by deep sequencing of small RNAs from C. elegans, C. briggsae, C. remanei, and C. brenneri, which identified hundreds of new miRNAs and revealed a high conservation of miRNAs across C. briggsae, C. remanei, and C. brenneri. 44 Table 2 gives an overview of studies on microRNAs in non-model nematodes. Although much work still needs to be done to gain a thorough understanding of miRNA evolution some surprising observations have already been made. For instance, Guo et al.45 showed that different miRNAs have different evolutionary patterns. This, at least in part, was explained by Chen and Rajewsky.46 They argued that miRNAs co-evolve with their targets, so when studying miRNA evolution, also the evolution of the targets should be taken in account. Altogether (see papers mentioned in Table 2), closely related nematode species have similar miRNAs while distantly related species have dissimilar miRNAs.
Table 2. Overview of studies on microRNAs (miRNAs) in non-model nematodes.
| Species (clade number)a | Number of loci/ miRNAs/miRNA familiesb | Observations of conservation and uniquenessc | Original paper |
|---|---|---|---|
| Hemonchus contortus (V) | 192 mature miRNAs (S/C) | 54 of the 192 mature miRNAs are conserved * | Winter et al., 201281 |
| Pristionchus pacificus (V) | 124 loci (S) | 30 species-specific loci ** | De Wit et al., 200943 |
| 362 miRNA genes (S) | the majority of miRNA genes of distantly related nematodes are not conserved *** | Ahmed et al., 201398 | |
| Caenorhabditis elegans (V) | 185 loci (S) | 7 loci are species-specific ** | De Wit et al., 200943 |
| 106 miRNA families (C) | > 20% of miRNA families are unique, 54 miRNA families are conserved **** | Shi et al., 201342 | |
| 257 miRNA genes (S) | the majority of miRNA genes of distantly related nematodes are not conserved *** | Ahmed et al., 201398 | |
| Caenorhabditis briggsae (V) | 141 loci (S) | 8 loci are species-specific** | De Wit et al., 200943 |
| 84 miRNA families (C) | > 20% of miRNA families are unique, 54 miRNA families are conserved **** | Shi et al., 201342 | |
| Caenorhabditis remanei (V) | 109 loci (S) | 1 locus is species-specific ** | De Wit et al., 200943 |
| 85 miRNA families (C) | > 20% of miRNA families are unique, 54 miRNA families are conserved **** | Shi et al., 201342 | |
| Caenorhabditis brenneri (V) | 87 miRNA families (C) | > 20% of miRNA families are unique, 54 miRNA families are conserved **** | Shi et al., 201342 |
| Strongyloides ratti (IV) | 106 miRNA genes (S) | the majority of miRNA genes of distantly related nematodes are not conserved *** | Ahmed et al., 201398 |
| Bursaphelenchus xylophilus (IV) | 810 miRNAs (C) of which 57 miRNAs (S) | 10 species-specific miRNAs * | Huang et al., 201099 |
| Brugia pahangi (III) | 104 mature miRNAs (S/C) | 42 of the 104 mature miRNAs are conserved * | Winter et al., 201281 |
| Brugia malayi (III) | 32 miRNAs in 24 miRNA families (S/C) | the miRNA-36-family occurs only in helminthes * | Poole et al., 2010100 |
| Ascaris suum (III) | 97 miRNAs in 59 miRNA seed families (S) | 80% of the seed sequences is conserved * | Wang et al., 201120 |
| Dirofilaria immitis (III) | 1063 conserved miRNA candidates (C) + 13 novel miRNA (S) | 11 of the 13 tested conserved miRNA candidates were verified *, ***** | Fu et al., 2013101 |
a Clade numbers are derived from references 14 and 96. bS, sequenced; C, computer predicted; S/C, sequenced or computer predicted. cObservations compared with miRNAs of the following databases/species: *, all (animal) miRBase entries; **, P. pacificus, C. elegans, C. briggsae, and C. remanei; ***, P. pacificus, C. elegans, and S. ratti; ****, C. elegans, C. briggsae, C. remanei, and C. brenneri; *****, Brugia malayi.
Endo-siRNAs: Flexible and Responsive Pathway with a Wide Range of Functions
Small interfering RNAs (siRNAs) consist of two main classes: exogenous siRNAs and endogenous siRNAs. Exogenous siRNAs (exo-siRNAs) are induced by exogenous long, double stranded RNA precursors originating from cellular transfections, microinjections, feeding bacteria expressing dsRNA, or from virus-derived double stranded RNA (either as genomic RNA or as the replication intermediate). Here, however, we will focus on the endogenous siRNAs (endo-siRNAs) (Fig. 3). Endo-siRNAs do not only influence gene expression via the degradation of a transcript or by translational inhibition, as was previously assumed, but also via modification of chromatin.47,48 The functions that are associated with different kinds of endo-siRNAs are very diverse and are discussed separately per group. Two major classes of endo-siRNAs can be distinguished: 26G RNAs and 22G RNAs (Fig. 3).21 These two classes are distinguished by their length and by which Argonautes they bind to (Table 1).
The expression levels of endo-siRNAs are often tissue- or developmental stage-specific. 26G RNAs are 26 nucleotides long and have a 5′ guanine.49 26G RNAs are derived from an initial endogenous double stranded RNA-trigger by Dicer and the RNA-dependent RNA Polymerase (RdRP) RRF-3. They can be divided into two subclasses: ERGO-1-bound-26G RNAs, which function in oocytes and embryos, and ALG3/4-bound-26G RNAs, required for normal sperm development (Table 1).49,50 The ERGO-1 class of 26G-RNAs are methylated by HENN-1 while the ALG-3 and -4 26G-RNAs are not methylated.51-53 This methylation is required to stabilize the ERGO-1 26G-RNAs.51-53 26G RNAs are bound to Argonautes and this complex interacts with perfect complementarity to target transcripts.54 This will lead to a local production of 22G RNAs around the target site by the RdRP RRF-1.55-58 However, not all 22G RNAs are derived from 26G RNAs, some are derived from piRNAs or from exogenous double stranded RNA, and in one case even from a miRNA.59-62
22G RNAs are 22 nucleotides long and have a 5′ guanine.63 They can be divided into two subclasses as well: WAGO-bound-22G RNAs and CSR-1-bound-22G RNAs.63,64 The CSR-1-bound-22G RNAs are essential for proper chromosome segregation by targeting germline-expressed genes and also for protecting gene expression in the germline against piRNA-mediated degradation.64-68 The WAGO-bound-22G RNAs silence transposable elements, aberrant endogenous transcripts, as well as certain genes (Table 1).63 Similarly to 26G RNAs, 22G RNAs interact with target transcripts with perfect complementarity.69 There is evidence that the 22G RNAs that are taken up by the Argonaute CSR-1 are mainly produced by the RdRP EGO-1.70,71 Whereas, the WAGO-bound-22G RNAs are thought to predominantly be produced by the RdRP RRF-1 and to a lesser extent by EGO-1.55,56,58
Until recently, most experiments in non-model nematodes have been focused on exo-siRNAs in hopes of downregulating gene transcription levels to study gene-functions (reviewed in refs. 72 and 73). However, two studies were published in the last few years that shed more light on endo-siRNAs.
Wang et al.20 compared the small RNAs of Ascaris suum to those of C. elegans by sequencing. Several differences could be observed when comparing the endo-siRNAs. For example, no methylation was detected on the 26G RNAs of A. suum. Also, the 22G RNAs mapped more frequently toward the 5′-end of the mRNA, while in C. elegans they map to both ends. Another difference is that the 26G and 22G RNAs in A. suum have a different ratio for spermatogenesis related genes. In C. elegans, the amount of 26G RNAs decreases during spermatogenesis while the amount of 22G RNAs dramatically increases. In contrast, the ratio of 26G to 22G RNAs in A. suum remains the same during spermatogenesis.20 Together, these observations imply that small RNA pathways are flexible and can be adapted throughout evolution.
Shi et al.42 reported on the sequenced small RNAs of C. elegans and three related nematode species: C. briggsae, C. remanei, and C. brenneri. They found that there is almost no conservation of individual endo-siRNAs and that the orthologs of genes targeted by CSR-1 in C. elegans are often also targets of small RNAs in the other species. But genes targeted by WAGO-bound siRNAs in C. elegans are less likely to have complementary siRNAs in other species. Furthermore, many features of the pathway in which the endo–siRNAs operate are conserved. Some conserved features of 26G RNAs (in all four species) are the genomic distribution, low abundance, sex specificity, and the ability to trigger 22G RNA production.42 The conservation of the endo-siRNA pathway, but not the individual sequences, shows the flexibility of the endo-siRNA pathway.16
piRNAs Maintain Genome Stability in the Germline
PIWI-interacting RNAs (piRNAs) are germline-specific small RNAs linked to trans-generational silencing. C. elegans PRG-1 mutants (that completely lack piRNAs) exhibit a broad spectrum of germ line defects.74,75 Knowledge about the targets of piRNAs is now becoming available because the “rules of targeting” have recently been identified more clearly.59,62 It is apparent that the piRNA pathway has an evolutionarily conserved role in maintaining genome stability in the germline of animals by keeping transposons silenced.59,62
C. elegans piRNAs are also known as 21U RNAs because they have a length of 21 nucleotides with a uridine at their 5′ end.74,76 piRNA precursors are thought to be ~26 nucleotide capped small RNAs (csRNAs). csRNAs are transcribed by RNA polymerase II from two large clusters on chromosome IV and from the promoter region of protein coding genes (Fig. 3).76,77 They are bound by a PIWI Argonaute: PRG-1.74 Subsequently, the RNAs are de-capped and two nucleotides are removed from the 5′ end by an unknown mechanism.77 The 3′ ends are trimmed and later methylated by HENN-1 with a 2’-O-methyl group stabilizing the piRNAs over longer time intervals.51-53
The PRG-1-piRNA-complex recognizes transcripts that are partially complementary to the piRNA.59 This leads to a local production of 22G RNAs around the piRNA target site by an RdRP.59,62 The amount of 22G RNAs is dependent on the complementarity of the piRNA to the target: more mismatches lead to less 22G RNAs.59 The 22G RNAs bind to WAGO-9 and cause trans-generational silencing via chromatin remodelling.59,62,78-80 Other Argonautes (WAGO-1 and WAGO-10) have been found to play a role in this process too.80 To summarize, the initiation of silencing depends on PRG-1/piRNAs, but the maintenance of the silencing requires WAGO/RdRP/22G RNAs.62,78,80
The first paper describing 21U RNAs in C. elegans already noted that when a comparison was made with the 21U RNAs of C. briggsae, not a single sequence was shared between these related species.76 De Wit et al.43 found 705 21U RNAs in C. elegans, 250 in C. briggsae, 1314 in C. remanei, and 1123 in P. pacificus. No sequence conservation of 21U RNAs across these four species was found. It was however observed that within the Caenorhabditis clade the genomic regions where 21U RNA precursor clusters are highly conserved, while the 21U RNAs precursor clusters found in P. pacificus are much smaller and more widespread.43
In the earlier mentioned study on the small RNAs of A. suum, the authors also searched for piRNAs.20 Remarkably, it was found that all piRNAs and components of the piRNA pathway were lost in A. suum. Instead, a subset of endo-siRNAs increased and diverged when compared with C. elegans. It was postulated that the extra endo-siRNAs in A. suum may have functions that are regulated by piRNAs in C. elegans.20 The loss of the piRNA pathway is not restricted to A. suum. 21U RNAs are also absent in another clade III parasite Brugia pahangi (for the clades, see Fig. 1).81 Also, no PRG-like Argonautes are present in A. suum, B. pahangi, and other clade III species.19 It is not clear why the piRNA pathway is missing in these nematodes. It is however not linked to parasitism as piRNAs are present in other, non-clade III, parasites.19,81
Recently, it was found that gonochoristic species (which mate every generation) have larger numbers of piRNAs and almost twice as many Argonautes as androdioecious species (which primarily self-fertilize).42 This suggests that species that mate every generation need more piRNAs and Argonautes to deal with the great diversity of paternal transposons mixed in every generation. Overall, the conservation of the piRNA pathway (but not of individual piRNAs) in most nematodes investigated, suggests a conserved role in protecting germline viability.
Other Small RNA Pathways
When studying the miRNA, piRNA, and endo-siRNA pathways in C. elegans, it is clear that they follow the same pattern: a single stranded, small RNA binds an Argonaute, interacts with a transcript, and changes gene expression. Next to the Argonautes, also Dicer, RdRPs, HENN-1, and secondary RNAs are present in more than one pathway. Other small RNA pathways exist in C. elegans that contain the same proteins and follow the same pattern. Two of these pathways will be briefly discussed below.
The first of these is the previously mentioned exogenous siRNA (exo-siRNA) pathway.1,82 An exogenous double stranded RNA is cleaved into primary siRNAs (which are 21 nucleotides long) by Dicer, which are bound by the Argonaute RDE-1.32,83 This complex is called the siRNA-induced silencing complex (siRISC). Interaction of this siRISC with the target mRNA leads to RdRP-production of 22-nucleotide, secondary siRNAs.21,57 These secondary siRNAs are bound by secondary-siRNA-defective Argonaute proteins (SAGO proteins, which fall under the WAGO-type Argonautes).
The second small RNA pathway, known as nuclear RNAi, is found in the nucleus instead of in the cytoplasm.84 Certain WAGO-bound 22G RNAs, downstream of ERGO-1-bound 26G RNAs, interact with the Argonaute NRDE-1.85 This association with NRDE-1 causes the small RNAs to be transported to the nucleus where the complex silences target genes.85,86 The transport of 22G RNAs links siRNAs and piRNAs to nuclear processes such as transposon regulation, heterochromatin formation, and genome stability.3,87-89
Argonaute Specificity
With all these very similar small RNAs in the cytoplasm, how do Argonaute proteins distinguish between different kinds of small RNAs? Temporal and spatial distribution of both the small RNAs and Argonaute proteins within an organism and its cells provides an important limiting factor. But there are additional factors that determine the specificity of an Argonaute; for example, the 5′ nucleotide and length of the small RNA and the proteins, which the small RNA is bound to. Another example is the precursor structure; the let-7-miRNA precursor is loaded into RDE-1 (instead of the normal ALG-1) when specific nucleotides are changed to make the stem fully complementary.90 This binding to RDE-1 activates the downstream exo-siRNA pathway, instead of the normal miRNA pathway.90
Some Argonautes however are not specific in binding small RNAs. The Argonaute RDE-1 was found to function as a scavenger protein, taking up small RNAs from many different sources. In this way, many small RNA species can be used in the exo-siRNA pathway.60 Related to this, it remains an open question which factors determine the uptake of small RNAs into particular complexes.
Regulation of Small RNAs
Small RNAs themselves are regulated in different ways. For example, the miRNA, endo-siRNA, and exo-siRNA pathways compete for access to limited resources.91 When C. elegans feeds on E. coli expressing double stranded RNA during controlled experiments to study gene-function, the expression of miRNA-regulated genes increases.91 This indicates that miRNAs are less effective in downregulating target gene expression. Conversely, miRNAs of the “miR-35-41” cluster reduced exo-siRNA sensitivity and enhance endo-siRNA effectiveness, balancing these siRNA pathways.92,93 Next to crosstalk between the pathways there is also feedback between the small RNAs and their targets.94 It has been shown that mRNAs targeted by miRNAs actually protect the miRNA from degradation; so the level of mature miRNAs is modulated by the mRNA target levels.94 This modulation provides a way to maintain a diverse set of miRNAs of which only the “useful” ones are accumulated.
Losing and Gaining Pathway Components
As more genome sequences become available and are studied for components of the small RNA pathways, it becomes apparent that there are many variations in the pathways across the different clades (Fig. 4). All investigated nematode species lack at least one Argonaute or RdRP compared with C. elegans.16 Genes are not only lost, Trichinella spiralis for example was found to have 119 ALG3/4-like genes, a gain of 117 compared with the two C. elegans genes.18,19 The function of these proteins, and which RNAs they bind to, is still unknown.
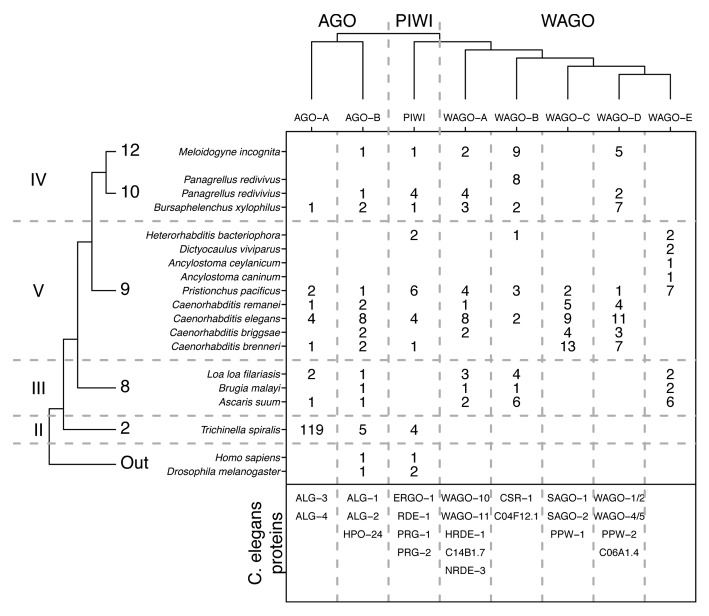
Figure 4. Identified Argonaute orthologs in nematodes. Phylogenetic tree of nematodes researched for Argonaute orthologs and the three groups of Argonautes (Argonaute-like, PIWI, and Worm-specific).19,21 The three major types of Argonaute proteins are sub-divided into eight homologous groups.19 The AGO-A group contains the C. elegans proteins ALG-3 and ALG-4 and the AGO-B group contains the C. elegans proteins ALG-1, ALG-2, and HPO-24. The PIWI-like group contains ERGO-1, RDE-1, PRG-1, and PRG-2. The different WAGO groups contain WAGO-10, WAGO-11, HRDE-1, C14B1.7 and NRDE-3 (WAGO-A); CSR-1 and C04F12.1 (WAGO-B); SAGO-1, SAGO-2 and PPW-1 (WAGO-C); WAGO-1, WAGO-2, WAGO-4, WAGO-5, PPW-2, and C06A1.4 (WAGO-D); no Caenorhabditis WAGO’s (WAGO-E).
Research in A. suum, B. pahangi, and other clade III species indicates that even complete pathways can be lost. The loss of the piRNA pathway in A. suum seems to be compensated for by extending the endo-siRNA pathway.20 These findings could suggest that variations in these pathways are linked to ecology and natural history. Devaney et al.17 for instance, discusses that it is very likely that drug resistance in parasitic nematodes is at least in part mediated by miRNAs, much like the well-studied drug resistance in tumor cells.
The loss of the piRNA pathway in clade III species provides a great opportunity to study how the loss of a complete pathway is dealt with. If this loss is an adaptation to a parasitic life cycle, the question can be asked why other parasitic nematodes, like the clade V nematodes Hemonchus contortus and Heligmosomoides polygyrus, do not show this loss. Studying the small RNA pathways of these and other parasitic species is likely to give more insight into these questions.19,20,81 The before mentioned expansion of ALG3/4-like genes in Trichinella spiralis might also be an adaption associated with parasitism. It was argued that Argonaute diversity in parasitic nematode species could be part of regulatory or sensing adaptations associated with parasitism.19
Although not an endo-siRNA-mediated pathway, exo-RNA-mediated interference studies in nematode species other than C. elegans can also provide some insights into pathway functionality. An example is a recent study on the effectiveness of exo-siRNA/RNAi in non-model grassland inhabiting nematodes, Oscheius sp FVV-2., Rhabditis sp, Mesorhabditis sp., and Acrobeloides sp. Here it was shown that both feeding bacteria expressing dsRNA and injection of dsRNA into the gonad did not result in the expected knockdown phenotypes.95 For plant parasitic nematodes, treatments with double stranded RNA have shown the expected knockdown phenotype (indicating a functional exo-siRNA pathway) for many species (for a detailed overview, see the review by Maul et al.).73 In the future, functional studies need to be coupled to genome analysis to find the cause of this variation.
Concluding Remarks
In this review we have provided an introduction into the endogenous small RNA pathways and the evolutionary conservation of these regulatory pathways in nematodes. We discussed that endogenous small RNAs show similar patterns in their pathways and in some cases appear in different pathways across different species. Yet, “new” small RNAs that bind to other Argonautes might still be discovered. All animal cells contain a flexible, constantly changing reservoir of small RNAs of different sizes and sources. We have just started to scratch the surface when it comes to studying the impact of small RNAs and their interactions. It is expected that, with the increasing sequencing capacity and more powerful bioinformatics, our understanding of small RNAs in nematodes and more animal species will rapidly increase.
For Further Reading
Nematode evolution:
Sommer RJ, Streit A. Comparative genetics and genomics of nematodes: genome structure, development, and lifestyle. Annual review of genetics 2011; 45:1–20.
The many effects of RNA on the genome:
Sabin LR, Delás MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Molecular cell. 2013; 49(5):783–94.
Small RNAs in C. elegans:
Azimzadeh Jamalkandi S, Masoudi-Nejad A. RNAi pathway integration in Caenorhabditis elegans development. Functional and integrative genomics. 2011; 11(3):389–405.
piRNAs in animals:
Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nature Reviews Genetics 2013; 14(8):523–34.
siRNAs, piRNAs and nuclear RNAi in animals, plants and yeast:
Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nature Reviews Genetics 2013; 14(2):100–12.
miRNA evolution in animals:
Berezikov E. Evolution of microRNA diversity and regulation in animals. Nature Reviews Genetics 2011; 12(12):846–60.
Exogenous siRNA in parasitic nematodes:
Maule AG, McVeigh P, Dalzell JJ, Atkinson L, Mousley A, Marks NJ. An eye on RNAi in nematode parasites. Trends in Parasitology 2011; 27(11):505–13.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Régis Lopes-Corrêa, Josien van Wolfswinkel, and René Ketting for critically reading the manuscript and Hans Helder for help with the phylogeny of the nematode species.
Funding
Kammenga JE was funded by the European Union Seventh Framework Programme (FP7/2007-2013) project PANACEA, contract nr. 222936, Snoek LB was funded by the ERASysbio-plus ZonMW project GRAPPLE (90201066), Volkers RJM was funded by the NWO-ALW (project 855.01.151), Sterken MG was funded by Graduate School PE&RC.
Glossary
Abbreviations:
- 21U RNA
21 nucleotides-long RNA with uridine at 5′end
- 22G RNA
22 nucleotides-long RNA with guanine at 5′end
- 26G RNA
26 nucleotides-long RNA with guanine at 5′end
- csRNA
capped small RNA
- Dicer
an endoribonuclease that cleaves dsRNA and pre-microRNA
- endo-siRNA
endogenous small interfering RNA
- exo-siRNA
exogenous small interfering RNA
- miR family
group of microRNAs that share the same seed region
- miRISC
microRNA induced silencing complex
- miRNA
microRNA
- piRNA
PIWI interacting RNA
- PIWI
P-element induced wimpy testis
- pre-miRNA
precursor miRNA
- pri-miRNA
primary miRNA
- RdRP
RNA dependent RNA polymerase
- SAGO
secondary small interfering RNA defective Argonaute protein
- siRISC
small interfering RNA induced silencing complex
- siRNA
small interfering RNA
- WAGO
worm Argonaute
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Abbott AL. Uncovering new functions for microRNAs in Caenorhabditis elegans. Curr Biol. 2011;21:R668–71. doi: 10.1016/j.cub.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–12. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohr SE, Perrimon N. RNAi screening: new approaches, understandings, and organisms. Wiley Interdiscip Rev RNA. 2012;3:145–58. doi: 10.1002/wrna.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elvin M, Snoek LB, Frejno M, Klemstein U, Kammenga JE, Poulin GB. A fitness assay for comparing RNAi effects across multiple C. elegans genotypes. BMC Genomics. 2011;12:510. doi: 10.1186/1471-2164-12-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–40. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy D, Dancis B, Brown JR. The evolution of core proteins involved in microRNA biogenesis. BMC Evol Biol. 2008;8:92. doi: 10.1186/1471-2148-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabach Y, Billi AC, Hayes GD, Newman MA, Zuk O, Gabel H, Kamath R, Yacoby K, Chapman B, Garcia SM, et al. Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence. Nature. 2013;493:694–8. doi: 10.1038/nature11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter C, Chang JT, Fineran PC. Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems. Viruses. 2012;4:2291–311. doi: 10.3390/v4102291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sommer RJ, Streit A. Comparative genetics and genomics of nematodes: genome structure, development, and lifestyle. Annu Rev Genet. 2011;45:1–20. doi: 10.1146/annurev-genet-110410-132417. [DOI] [PubMed] [Google Scholar]
- 11.C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–8. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 12.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bert W, Karssen G, Helder J. Phylogeny and evolution of nematodes. In: Jones J, Gheysen G, Fenoll C, eds. Genomics and molecular genetics of plant-nematode interactions: Springer Netherlands, 2011. [Google Scholar]
- 14.Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–5. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 15.Kiontke KC, Félix MA, Ailion M, Rockman MV, Braendle C, Pénigault JB, Fitch DH. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011;11:339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalzell JJ, McVeigh P, Warnock ND, Mitreva M, Bird DM, Abad P, Fleming CC, Day TA, Mousley A, Marks NJ, et al. RNAi effector diversity in nematodes. PLoS Negl Trop Dis. 2011;5:e1176. doi: 10.1371/journal.pntd.0001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devaney E, Winter AD, Britton C. microRNAs: a role in drug resistance in parasitic nematodes? Trends Parasitol. 2010;26:428–33. doi: 10.1016/j.pt.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, Martin J, Taylor CM, Yin Y, Fulton L, Minx P, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011;43:228–35. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buck AH, Blaxter M. Functional diversification of Argonautes in nematodes: an expanding universe. Biochem Soc Trans. 2013;41:881–6. doi: 10.1042/BST20130086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon GJ, Davis RE. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21:1462–77. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–57. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 22.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 23.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 24.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 25.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouasker S, Simard MJ. The slicing activity of miRNA-specific Argonautes is essential for the miRNA pathway in C. elegans. Nucleic Acids Res. 2012;40:10452–62. doi: 10.1093/nar/gks748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/S0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 30.Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 31.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 34.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/S0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 35.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 37.Izaurralde E. Elucidating the temporal order of silencing. EMBO Rep. 2012;13:662–3. doi: 10.1038/embor.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–7. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar A, Wong AK, Tizard ML, Moore RJ, Lefèvre C. miRNA_Targets: a database for miRNA target predictions in coding and non-coding regions of mRNAs. Genomics. 2012;100:352–6. doi: 10.1016/j.ygeno.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Z, Montgomery TA, Qi Y, Ruvkun G. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 2013;23:497–508. doi: 10.1101/gr.149112.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Wit E, Linsen SE, Cuppen E, Berezikov E. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 2009;19:2064–74. doi: 10.1101/gr.093781.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Z, Montgomery TA, Qi Y, Ruvkun G. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 2013;23:497–508. doi: 10.1101/gr.149112.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo L, Sun B, Sang F, Wang W, Lu Z. Haplotype distribution and evolutionary pattern of miR-17 and miR-124 families based on population analysis. PLoS One. 2009;4:e7944. doi: 10.1371/journal.pone.0007944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3’UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol. 2006;71:149–56. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- 47.Claycomb JM. Caenorhabditis elegans small RNA pathways make their mark on chromatin. DNA Cell Biol. 2012;31(Suppl 1):S17–33. doi: 10.1089/dna.2012.1611. [DOI] [PubMed] [Google Scholar]
- 48.van Wolfswinkel JC, Ketting RF. The role of small non-coding RNAs in genome stability and chromatin organization. J Cell Sci. 2010;123:1825–39. doi: 10.1242/jcs.061713. [DOI] [PubMed] [Google Scholar]
- 49.Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–9. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2010;107:3588–93. doi: 10.1073/pnas.0911685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Billi AC, Alessi AF, Khivansara V, Han T, Freeberg M, Mitani S, Kim JK. The Caenorhabditis elegans HEN1 ortholog, HENN-1, methylates and stabilizes select subclasses of germline small RNAs. PLoS Genet. 2012;8:e1002617. doi: 10.1371/journal.pgen.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamminga LM, van Wolfswinkel JC, Luteijn MJ, Kaaij LJ, Bagijn MP, Sapetschnig A, Miska EA, Berezikov E, Ketting RF. Differential impact of the HEN1 homolog HENN-1 on 21U and 26G RNAs in the germline of Caenorhabditis elegans. PLoS Genet. 2012;8:e1002702. doi: 10.1371/journal.pgen.1002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montgomery TA, Rim YS, Zhang C, Dowen RH, Phillips CM, Fischer SE, Ruvkun G. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS Genet. 2012;8:e1002616. doi: 10.1371/journal.pgen.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, Thierry-Mieg D, Thierry-Mieg J, Kim JK. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–9. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pak J, Maniar JM, Mello CC, Fire A. Protection from feed-forward amplification in an amplified RNAi mechanism. Cell. 2012;151:885–99. doi: 10.1016/j.cell.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–76. doi: 10.1016/S0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 57.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–7. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 58.Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D., Jr. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A. 2010;107:3582–7. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bagijn MP, Goldstein LD, Sapetschnig A, Weick EM, Bouasker S, Lehrbach NJ, Simard MJ, Miska EA. Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science. 2012;337:574–8. doi: 10.1126/science.1220952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corrêa RL, Steiner FA, Berezikov E, Ketting RF. MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet. 2010;6:e1000903. doi: 10.1371/journal.pgen.1000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, Bochud M, Rice KM, Henneman P, Smith AV, et al. LifeLines Cohort Study. EchoGen consortium. AortaGen Consortium. CHARGE Consortium Heart Failure Working Group. KidneyGen consortium. CKDGen consortium. Cardiogenics consortium. CardioGram Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–11. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee HC, Gu W, Shirayama M, Youngman E, Conte D, Jr., Mello CC. C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell. 2012;150:78–87. doi: 10.1016/j.cell.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu W, Shirayama M, Conte D, Jr., Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–44. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell. 2009;139:123–34. doi: 10.1016/j.cell.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wedeles CJ, Wu MZ, Claycomb JM. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev Cell. 2013;27:664–71. doi: 10.1016/j.devcel.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 66.Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D, Jr., Yates JR, 3rd, Mello CC. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell. 2013;155:1532–44. doi: 10.1016/j.cell.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seth M, Shirayama M, Gu W, Ishidate T, Conte D, Jr., Mello CC. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell. 2013;27:656–63. doi: 10.1016/j.devcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–48. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 69.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–53. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Avgousti DC, Palani S, Sherman Y, Grishok A. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J. 2012;31:3821–32. doi: 10.1038/emboj.2012.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maniar JM, Fire AZ. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr Biol. 2011;21:449–59. doi: 10.1016/j.cub.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lilley CJ, Davies LJ, Urwin PE. RNA interference in plant parasitic nematodes: a summary of the current status. Parasitology. 2012;139:630–40. doi: 10.1017/S0031182011002071. [DOI] [PubMed] [Google Scholar]
- 73.Maule AG, McVeigh P, Dalzell JJ, Atkinson L, Mousley A, Marks NJ. An eye on RNAi in nematode parasites. Trends Parasitol. 2011;27:505–13. doi: 10.1016/j.pt.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 74.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–7. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 77.Gu W, Lee HC, Chaves D, Youngman EM, Pazour GJ, Conte D, Jr., Mello CC. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell. 2012;151:1488–500. doi: 10.1016/j.cell.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luteijn MJ, van Bergeijk P, Kaaij LJ, Almeida MV, Roovers EF, Berezikov E, Ketting RF. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012;31:3422–30. doi: 10.1038/emboj.2012.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Jr., Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winter AD, Weir W, Hunt M, Berriman M, Gilleard JS, Devaney E, Britton C. Diversity in parasitic nematode genomes: the microRNAs of Brugia pahangi and Haemonchus contortus are largely novel. BMC Genomics. 2012;13:4. doi: 10.1186/1471-2164-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 83.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–32. doi: 10.1016/S0092-8674(00)81644-X. [DOI] [PubMed] [Google Scholar]
- 84.Bosher JM, Dufourcq P, Sookhareea S, Labouesse M. RNA interference can target pre-mRNA: consequences for gene expression in a Caenorhabditis elegans operon. Genetics. 1999;153:1245–56. doi: 10.1093/genetics/153.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–41. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang C, Montgomery TA, Fischer SE, Garcia SM, Riedel CG, Fahlgren N, Sullivan CM, Carrington JC, Ruvkun G. The Caenorhabditis elegans RDE-10/RDE-11 complex regulates RNAi by promoting secondary siRNA amplification. Curr Biol. 2012;22:881–90. doi: 10.1016/j.cub.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–51. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 2011;7:e1002249. doi: 10.1371/journal.pgen.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2011;108:19683–8. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Steiner FA, Hoogstrate SW, Okihara KL, Thijssen KL, Ketting RF, Plasterk RH, Sijen T. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat Struct Mol Biol. 2007;14:927–33. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- 91.Zhuang JJ, Hunter CP. The influence of competition among C. elegans small RNA pathways on development. Genes (Basel) 2012;3:671. doi: 10.3390/genes3040671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Massirer KB, Perez SG, Mondol V, Pasquinelli AE. The miR-35-41 family of microRNAs regulates RNAi sensitivity in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002536. doi: 10.1371/journal.pgen.1002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Massirer KB, Pasquinelli AE. MicroRNAs that interfere with RNAi. Worm. 2013;2:e21835. doi: 10.4161/worm.21835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chatterjee S, Fasler M, Büssing I, Grosshans H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell. 2011;20:388–96. doi: 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 95.Wheeler D, Darby BJ, Todd TC, Herman MA. Several Grassland Soil Nematode Species Are Insensitive to RNA-Mediated Interference. J Nematol. 2012;44:92–101. [PMC free article] [PubMed] [Google Scholar]
- 96.van Megen H, van den Elsen S, Holterman M, Karssen G, Mooyman P, Bongers T, et al. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology. 2009;11:927–950. doi: 10.1163/156854109X456862. [DOI] [Google Scholar]
- 97.Kato M, de Lencastre A, Pincus Z, Slack FJ. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahmed R, Chang Z, Younis AE, Langnick C, Li N, Chen W, Brattig N, Dieterich C. Conserved miRNAs are candidate post-transcriptional regulators of developmental arrest in free-living and parasitic nematodes. Genome Biol Evol. 2013;5:1246–60. doi: 10.1093/gbe/evt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang QX, Cheng XY, Mao ZC, Wang YS, Zhao LL, Yan X, Ferris VR, Xu RM, Xie BY. MicroRNA discovery and analysis of pinewood nematode Bursaphelenchus xylophilus by deep sequencing. PLoS One. 2010;5:e13271. doi: 10.1371/journal.pone.0013271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Poole CB, Davis PJ, Jin J, McReynolds LA. Cloning and bioinformatic identification of small RNAs in the filarial nematode, Brugia malayi. Mol Biochem Parasitol. 2010;169:87–94. doi: 10.1016/j.molbiopara.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 101.Fu Y, Lan J, Wu X, Yang D, Zhang Z, Nie H, Hou R, Zhang R, Zheng W, Xie Y, et al. Identification of Dirofilaria immitis miRNA using illumina deep sequencing. Vet Res. 2013;44:3. doi: 10.1186/1297-9716-44-3. [DOI] [PMC free article] [PubMed] [Google Scholar]