Abstract
Developmental biologists have been fascinated with the long-standing mystery of how multicellular organisms, such as plants and animals, sense and control their organ size. In plants, leaves are a suitable experimental system for elucidation of the mystery, because they, like animal organs, inherently exhibit a determinate growth pattern, meaning that they possess genetic information for the control of their final size. The cell proliferation and expansion processes are prerequisites for growth, so that the genetic controls should converge on the 2 cellular processes and decide their rate or duration during leaf growth. Plant scientists have found dozens of genes involved in the control of the cellular processes, including the Arabidopsis thaliana GRF-INTERACTING FACTOR (GIF) family. The GIF family consists of 3 members, GIF1 to GIF3, and encodes a class of transcription co-activators. Although the GIF family genes have been shown to play an essential role in the control of cell proliferation of the leaf organ, understanding of the spatio-temporal behaviors of GIF expression, in both aspects of their promoters and proteins, has been limited to GIF1 (also known as ANGUSTIFOLIA3, AN3). Here, we define kinematic growth properties of wild-type and gif leaf organs and present spatio-temporal expression patterns of all GIF genes, thus providing comprehensive insights into biological roles and expression behaviors of the whole GIF family members during leaf growth.
Keywords: leaf growth, organ size, GROWTH-REGULATING FACTOR, GRF-INTERACTING FACTOR, ANGUSTIFOLIA3, cell proliferation, cell expansion
The leaf organ initiates as a small bump of cells from the periphery of the shoot apical meristem and takes a finger-like shape. Afterwards, the leaf primordium develops into a flat lamina structure and starts to enlarge into a mature leaf through a complex coordination between cell division and expansion (Fig. 1).1 The leaf organ inherently exhibits a determinate growth pattern, meaning that it possesses genetic information that controls the rate or duration of cell proliferation and expansion and thus determines its final size. The cell proliferation and expansion processes occur partially overlapping during the conversion of a leaf primordium to a mature leaf.1-3 The early primordium of the wild-type leaf (WT) consists entirely of highly proliferative cells whose expansion occurs limitedly (Fig. 1B-D). As the primordium grows further, however, its distal region cells lose their proliferative activities and start to expand vigorously unto their final volume.1,4 The transition boundary between the proliferative and actively expanding cells is referred to the cell cycle arrest front, which moves from the distal region down to the proximal region in proportion as the leaf organ becomes mature. In consequence, the leaf organ at later stages is mostly composed of cells at the postmitotic expansion phase with no more proliferation (Fig. 1).1,4
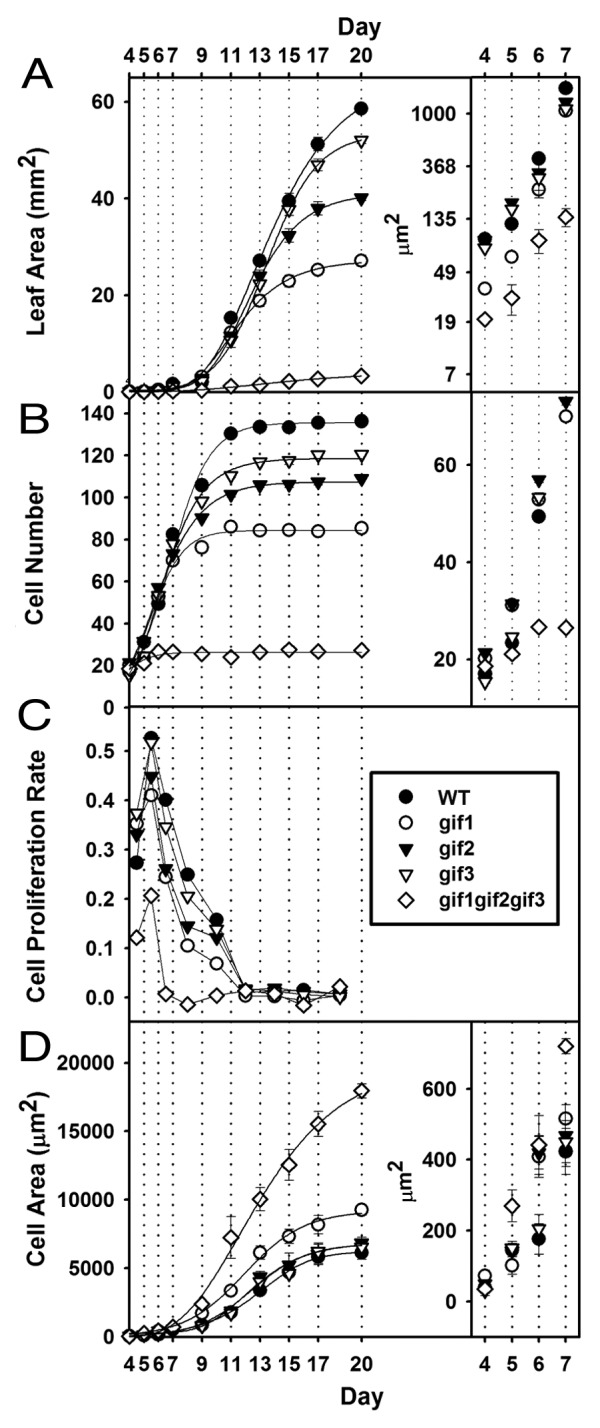
Figure 1. Kinematic features of cell proliferation and expansion of wild-type and gif mutant leaves. Adaxial palisade cells of the first 2 leaves were analyzed over the indicated time-lapses after germination on soil. (A) Leaf blade area. (B) Numbers of cells in a line along the maximum-width region. (C) Relative cell proliferation rate. (D) Cell areas. Insets: magnified versions to show differences in the early stages of leaf growth. Note that the inset of (A) was plotted on the natural log scale. n = 10 for determination of organ-level parameters; n = 200 from 10 different organs for cellular parameters. Results are means ± SE.
The Arabidopsis thaliana GRF-INTERACTING FACTOR (GIF) family genes, GIF1 to GIF3, encode a class of transcription co-activators that form a functional complex with GROWTH-REGULATING FACTOR (GRF) transcription factors, and are required for the cell proliferation activities of lateral organs, including leaves.5-7 Here in this study, we set out to define, in the context of kinematic properties of leaf growth, the spatio-temporal expression pattern of all the GIF family genes by employing the GUS (for β-glucuronidases) reporter. For a solid comparison between the leaf growth and GUS expression patterns, kinematic analyses of leaf growth and the GUS staining assays were performed with plants grown in parallel in the same growth conditions, although a similar kinematic analysis of leaf growth had been presented before.8
The first 2 leaves of the wild type attained almost a mature size at 20 d after germination, and their constituent cells ceased to proliferate at 11 d (Fig. 1A,B). On the other hand, the loss-of-function, single mutant leaf of GIF1, which is also known as ANGUSTIFOLIA3 (AN3), exhibited half the wild-type size and moderate decreases in both the rate and duration of cell proliferation, and the gif2 and gif3 single mutants also displayed substantial reductions in those parameters (Fig. 1A,B).7,8 Moreover, the gif1 gif2 gif3 triple mutation deteriorated, synergistically and extremely, the defects in leaf growth and cell proliferation. In more detail, cell proliferation of the gif triple mutant ceased completely already at 6 d, while that of the wild type and even single mutants was still fairly active (Fig. 1B,C). On the other hand, the triple mutant cells started to expand markedly around the same day (Fig. 1D). These results indicate that GIF1/AN3, GIF2, and GIF3 genes are all positive regulators of cell proliferation and functionally redundant, confirming the previous results.7,8
In which part of the leaf organ, in regard to the cell cycle arrest front of leaf growth, are GIF genes expressed? How could the expression patterns of GIF genes be associated with their manifestation of positive effects on cell proliferation? The answer to these questions came from an examination of the GIF1/AN3 promoter activity.7,9 The GIF1/AN3 promoter activity was detected only in the proliferative region of the leaf organ and, as the leaf primordium develops further, became constrained to the proximal region below the arrest front.9 In the long run, the promoter activity disappeared completely in the leaf lamina, in which cell proliferation ceases and cell expansion occurs vigorously. Recently, it was discovered that the GIF1/AN3 gene is transcribed and translated only in the mesophyll, but not in the epidermis, and that the resulting GIF1/AN3 proteins have the capacity to move from the former tissue to the latter, coordinating cell proliferation of these clonally distinct tissues.10 These results indicate that the GIF1/AN3 gene is required for leaf cells, both cell-autonomously and non-cell-autonomously, to promote cell proliferation. In contrast to such advances in understanding of GIF1/AN3, similar approaches to aim GIF2 and GIF3 has been not taken, in spite of their crucial impacts on leaf growth (Fig. 1), in consequence hampering understanding of the GIF family genes as a whole. Moreover, GIF2 and GIF3 genes have unusually short promoters, since their genomic loci closely abut neighboring genes in a head-to-head manner (Fig. 2A).11 This, unavoidably, summons the doubt whether or not the promoters of GIF2 and GIF3 are able to trigger their own transcription.
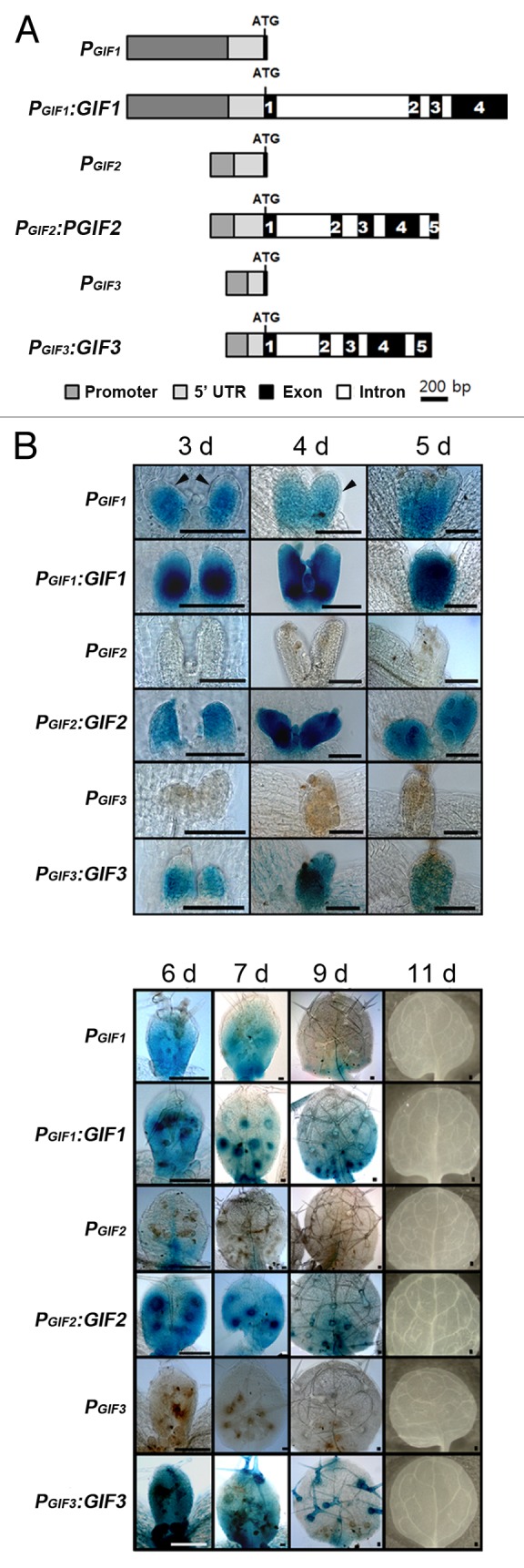
Figure 2. Kinematic GUS assays for promoter activities and protein distribution of the GIF family genes. (A) Schematic representation of GIFs and GUS fusion constructs in a single scale. All GIF DNA fragments were inserted, in in-frame, into the GUS open reading frame of pBI101 to produce PGIF:GUS and PGIF:GIF:GUS (briefly PGIF and PGIF:GIF). ATG denotes the translational start codon; the numbers in black boxes the exon number. (B) Staining patterns of each fusion construct during leaf growth. The GUS staining assays were performed in a single staining batch, except that the GIF1 and GIF2 fusion transgenics were incubated for 1h with X-gluc and the GIF3 transgenics overnight. The numbers on each column indicate days after germination. Scale bars = 100 μm.
In order to clarify these obscurities, we set out to examine the promoter activities and protein distribution patterns of all GIF family members. To generate reporter lines, GUS cDNA was fused to GIF promoters, including the 5′ untranslated region (UTR) and the first couple of codons, or to the whole genomic fragment of each GIF gene (PGIF:GUS or PGIF:GIF:GUS, respectively); we took maximum lengths of GIF2 and GIF3 promoters, permitting only a minimum overlap with a 5′ part of the promoters of the head-to-head neighboring genes (Fig. 2A).
The GIF1/AN3 promoter activities appeared throughout almost the entire leaf primordium at 3 to 5 d, after which the staining pattern faded out from the distal end, gradually being restricted only to the proximal region (6 to 9 d), and, finally, disappeared completely at 11 d (Fig. 2B). This distribution patterns of the GIF1/AN3 promoter activity are similar to previous results.7,9 We also analyzed distributive behaviors of GIF1/AN3 proteins during leaf growth, which had not been examined before, although their local movement between the mesophyll and epidermis was well documented.10 We found that the kinematic distribution patterns of the GIF1/AN3 protein in developing leaves were also, in the main, similar to those of the GIF1/AN3 promoter activity. It should be noted, however, that GIF1/AN3 promoter activities were detected only in the mesophyll, but not in the epidermis, whereas GIF1/AN3 proteins were present in both tissues (Fig. 2B, compare the regions indicated by arrowheads). These results are well in agreement with the previous result in that the GIF1/AN3 promoter activities are absent in the epidermis and its proteins move to the epidermis from mesophyll cells in which the proteins are synthesized.9,10 It should be also noted that the GIF1/AN3 protein signals are much stronger than those of its promoter activities, raising the possibility that the GIF1/AN3 coding sequence, probably introns, may contain additional regulatory cis-elements for transcription enhancement.
Next, we found that GIF2 and GIF3 promoters exhibited almost no activities throughout leaf development, indicating that their short promoters are not enough to activate their own transcription. In contrast, the whole genomic fragments of GIF2 and GIF3 were able to produce staining signals, indicating that GIF2 and GIF3 genes were effectively transcribed and translated into proteins, and thereby suggesting that their coding sequences should contain cis-elements for transcription. Distribution patterns of GIF2 and GIF3 proteins resemble that of GIF1/AN3 in regard to the arrest front, and the signals were detected in both the mesophyll and epidermis, as in case of GIF1/AN3 proteins. In addition, all GIF proteins, but not their promoter activities, were detected in a concentric ring of trichome support cells, and GIF3 proteins even in the trichome cell itself (Fig. 2B, arrows).
Conclusion and Perspective
In the present study, we demonstrate that all GIF family proteins are present in almost the entire region of the leaf primordium at the very early stages, after which the protein signals begin to fade away from the distal region and disappear completely at 11 d (Fig. 2B). This distribution pattern of GIF proteins is fairly consistent with the changes in the cell proliferation rate and the commencement of the postmitotic expansion that occur during the wild-type leaf growth (Fig. 1). These results suggest that the whole members of the GIF family are responsible for the behavior of the cell cycle arrest front during leaf growth, and explain how and why the gif triple mutation manifests such severe proliferative defects in a functionally redundant manner. How, then, do the GIF family proteins render leaf cells proliferative? We have recently demonstrated that the GIF family plays an essential role in maintaining the meristematic specification state of proliferative cells, such as the carpel margin meristem cells and archesporial lineage cells.11 Therefore it is a tempting speculation that the GIF family proteins may also enable leaf cells, as far as they express GIFs, to maintain their meristematic specification state. In this regard, a recent result that GIF1/AN3 binds to SWI/SNF chromatin remodeling complexes and, thereby, regulates transcription during leaf development provides an important insight.12
We have observed that GIF1/AN3 proteins, but not its promoter activities, are present in trichome support cells (Fig. 2B). Since GIF1/AN3 proteins translocate from the mesophyll to the epidermis (where there is no synthesis) and since trichome support cells are specialized epidermal cells,10,13 the presence of GIF1/AN3 proteins in trichome support cells may result from the translocation capacity of GIF1/AN3 proteins. This may hold true for GIF2 and GIF3 proteins likewise, since they are also present in both the mesophyll and the epidermis as well as trichome support cells and even in the trichome. The notion, however, remains to be validated by testing whether or not the transcriptional activities of GIF2 and GIF3 are confined to the mesophyll. For that goal, it is a prerequisite to define and verify the possible cis-element for GIF2 and GIF3 transcription through more detailed dissections in the future.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was supported by the National Research Foundation Grants funded by the Korean Government (NRF-2006–331-C00264 and NRF-2009–0076517) and partially by Kyungpook National University Research Fund, 2012.
References
- 1.Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol. 1999;215:407–19. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez N, Vanhaeren H, Inzé D. Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci. 2012;17:332–40. doi: 10.1016/j.tplants.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Powell AE, Lenhard M. Control of organ size in plants. Curr Biol. 2012;22:R360–7. doi: 10.1016/j.cub.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Kazama T, Ichihashi Y, Murata S, Tsukaya H. The mechanism of cell cycle arrest front progression explained by a KLUH/CYP78A5-dependent mobile growth factor in developing leaves of Arabidopsis thaliana. Plant Cell Physiol. 2010;51:1046–54. doi: 10.1093/pcp/pcq051. [DOI] [PubMed] [Google Scholar]
- 5.Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36:94–104. doi: 10.1046/j.1365-313X.2003.01862.x. [DOI] [PubMed] [Google Scholar]
- 6.Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:13374–9. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horiguchi G, Kim GT, Tsukaya H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43:68–78. doi: 10.1111/j.1365-313X.2005.02429.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee BH, Ko J-H, Lee S, Lee Y, Pak J-H, Kim JH. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 2009;151:655–68. doi: 10.1104/pp.109.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ichihashi Y, Kawade K, Usami T, Horiguchi G, Takahashi T, Tsukaya H. Key proliferative activity in the junction between the leaf blade and leaf petiole of Arabidopsis. Plant Physiol. 2011;157:1151–62. doi: 10.1104/pp.111.185066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawade K, Horiguchi G, Usami T, Hirai MY, Tsukaya H. ANGUSTIFOLIA3 signaling coordinates proliferation between clonally distinct cells in leaves. Curr Biol. 2013;23:788–92. doi: 10.1016/j.cub.2013.03.044. [DOI] [PubMed] [Google Scholar]
- 11.Lee BH, Wynn AN, Franks RG, Hwang YS, Lim J, Kim JH. The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev Biol. 2014;386:12–24. doi: 10.1016/j.ydbio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 12.Vercruyssen L, Verkest A, Gonzalez N, Heyndrickx KS, Eeckhout D, Han S-K, Jégu T, Archacki R, Van Leene J, Andriankaja M, et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell. 2014;26:210–29. doi: 10.1105/tpc.113.115907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hülskamp M, Misŕa S, Jürgens G. Genetic dissection of trichome cell development in Arabidopsis. Cell. 1994;76:555–66. doi: 10.1016/0092-8674(94)90118-X. [DOI] [PubMed] [Google Scholar]


