Abstract
CAM7, a member of CaM family in Arabidopsis, acts as a transcriptional regulator and enhances photomorphogenic growth and light regulated gene expression under various light conditions. HY5, a bZIP transcription factor, promotes photomorphogenesis at multiple wavelengths of light including far red, red, and blue light. Very recently, it has been shown that CAM7 and HY5 directly interact with the HY5 promoter to regulate the transcriptional activity of HY5 during Arabidopsis seedling development. In this study, we have investigated the root phenotype of cam7 hy5 double mutants and shown that CAM7 and HY5 genetically interact to control the root growth. We have further shown an interdependent function of HY5 and CAM7 in abscisic acid (ABA) responsiveness.
Keywords: Photomorphogenesis, CAM7, HY5, bZIP, transcription
Light plays a major role for proper growth and development of plants from seed germination till flowering.1 During the transition from dark to light, the seedlings follow photomorphogenesis, which is characterized at the cellular level by suppression of cell elongation and expansion in hypocotyl and cotyledon, and onset of growth by cell division in both shoot and root meristem. On the other hand, phytohormones also play important roles for the regulation of cell expansion and division. Thus, light and hormone signaling pathways are likely to intersect with each other to evoke the desired responses. It has already been shown that various plant hormones are involved in light signaling and influence cell expansion, division, and light-regulated gene expression.2-4
In Arabidopsis thaliana, several transcription factors involved in switching the skotomorphogic to photomorphogenic growth have been identified. These transcription factors are either positive or negative regulators of light signal transduction pathways and are specific to single or multiple wavelengths of light. Among them, LONG HYPOCOTYL 5 (HY5), the basic leucine zipper (bZIP) transcription factor, and CALMODULIN7 (CAM7/ZBF3) are the only known transcription factors acting downstream of multiple wavelengths of light including far red light, red light, and blue light, and positively regulate light signaling pathways.5-8 It has been shown that unlike other calmodulins, CAM7 acts as a unique transcriptional factor that directly binds to promoters of several light-inducible genes.8 The ectopic expression of CAM7 causes hyperphotomorphogenic growth under various light conditions. Whereas cam7 mutants do not exhibit any altered photomorphogenic phenotype, cam7 hy5 double mutant seedlings exhibit a super-tall phenotype.8 Recently it has been revealed that CAM7 and HY5 directly interact with the E- and T/G box of HY5 promoter and positively regulate the expression of HY5 in a concerted manner at various stages of development.9 HY5 is a major and versatile molecule, which plays an important role in light-regulated gene expression. Loss-of-function hy5 mutants exhibit dark-grown characteristics in the light10 with the elongated hypocotyl and severely affected root morphogenesis. The most obvious phenotype of hy5 seedlings is the increased number of lateral roots. These lateral roots along with primary roots grow faster and are negatively gravitropic.10
HY5, the key molecule of light signal transduction pathway, has also been involved in multiple hormone signaling pathways like GA, cytokinin, auxin, and abscisic acid (ABA),11-14 and thus acts as a convergence point of these pathways. Recently, the gene ontology analysis by Zhang et al., 2011, has shown that HY5-dependent developmental programs are mostly controlled by HY5-regulated transcription factors. Chip-on-chip analyses have also shown that genes involved in auxin, ethylene, cytokinin, and jasmonic acid pathways are extremely enriched in HY5-regulated genes. Thus, HY5 acts as a master molecule that coordinates the hormonal responses with the light signaling pathways.15 Although the molecular and genetic interactions between HY5 and CAM7 have been studied,8,9 the cross talk of these two molecules with respect to phytohormones is still to be elucidated. In this study, we have reported that CAM7 and HY5 genetically interact to control the root phenotype. We have also shown the genetic relationship between CAM7 and HY5 in ABA responsiveness.
Additional mutation in CAM7 partly suppresses the root phenotype of hy5 mutants
It has been found that mutation in HY5 causes an increase in the number of lateral roots.10 To determine the genetic relationship between cam7 and hy5 in controlling the lateral root formation, we examined the lateral roots formed in cam7 hy5 double mutant seedlings grown for 10 d in white light (100 μmol/m2/s). While counting the number of lateral roots, the number was found to be significantly higher in hy5 mutants, consistent with the observation made by Oyama et al., 1997. Although cam7 mutants did not show any significant alteration in the number of lateral roots formed, the number of lateral roots were significantly reduced in cam7 hy5 double mutants. These results suggest that additional mutation in CAM7 is able to suppress the defect of lateral root formation in hy5 (Fig. 1).
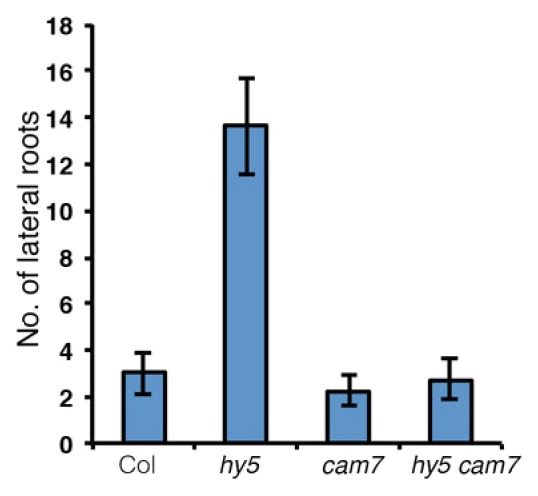
Figure 1. Formation of lateral roots in cam7 hy5 double mutants. Quantification of lateral roots formed in wild type and various mutants of 10-d-old seedlings.
CAM7 and HY5 work in an antagonistic manner in ABA-mediated inhibition of seed germination
Recently, it has been shown that hy5 mutant is tolerant to the inhibitory effect of ABA in seed germination.14 We investigated whether CAM7 mutation alters ABA responsiveness of hy5 mutant. For this study, freshly harvested seeds of wild type, single, and double mutant plants were inoculated on MS media without or with ABA. The rate of germination in wild type and various mutant seeds was found to be similar in the absence of ABA. On the other hand, ABA reduced the germination rate of wild type seeds (~45%). The rate of seed germination was found to be further diminished in cam7 mutants as compared with wild type (~35%), suggesting that cam7 mutants are more susceptible to ABA-mediated inhibition of seed germination. The hy5 mutants showed increased rate of seed germination (~72%) in the presence of ABA (Fig. 2A). However, the rate of germination of hy5 seeds was reduced with the additional mutation in CAM7. The cam7 hy5 double mutant seeds showed ~60% germination in the presence of ABA (Fig. 2A). These results suggest that CAM7 and HY5 work in an antagonistic manner in ABA-mediated inhibition of seed germination, and the additional mutation in CAM7 is able to partly suppress the rate of seed germination in hy5 mutants in the presence of ABA.
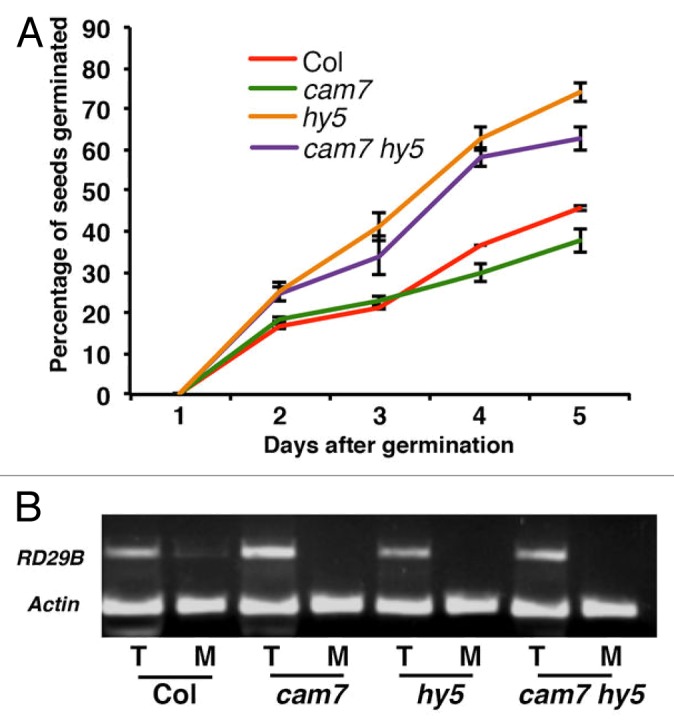
Figure 2. Response of cam7 hy5 double mutant to ABA. (A) Quantification of percentage of seeds germinated in presence of ABA at various days in constant WL (30 µmol/m2/s). (B) Expression of ABA pathway marker gene (RD29B) in the wild type and various mutants from 6-d-old seedlings grown in constant WL (30 µmol/m2/s) mock treated (MS solution) or treated with ABA (5µM). M stands for mock control, and T stands for treated with ABA.
RD29B is one of the marker genes involved in ABA signaling pathways, and its expression is induced by ABA treatment.14 We monitored the expression of RD29B in cam7 hy5 double mutants. RT-PCR analyses without or with ABA revealed that RD29B expression was slightly reduced in hy5 mutants, whereas it was strongly increased in cam7 mutant background. The level of expression of RD29B was found to be similar to wild type in cam7 hy5 double mutants. These results suggest that additional mutation in HY5 is able to suppress the higher level of induction of RD29B in cam7 mutant background (Fig. 2B).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work is financially supported by Department of Science and Technology (JC Bose National Fellowship Grant, SR/S2/JCB-85/2010), Government of India to S.C. N.A. is a recipient of DBT, Government of India fellowship.
References
- 1.Deng XW, Quail PH. Signalling in light-controlled development. Semin Cell Dev Biol. 1999;10:121–9. doi: 10.1006/scdb.1999.0287. [DOI] [PubMed] [Google Scholar]
- 2.Chory J, Reinecke D, Sim S, Washburn T, Brenner M. A role for cytokinins in de-etiolation in Arabidopsis (det mutants have an altered response to cytokinins) Plant Physiol. 1994;104:339–47. doi: 10.1104/pp.104.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neff MM, Fankhauser C, Chory J. Light: an indicator of time and place. Genes Dev. 2000;14:257–71. doi: 10.1101/gad.14.3.257. [DOI] [PubMed] [Google Scholar]
- 4.Vandenbussche F, Vriezen WH, Smalle J, Laarhoven LJ, Harren FJ, Van Der Straeten D. Ethylene and auxin control the Arabidopsis response to decreased light intensity. Plant Physiol. 2003;133:517–27. doi: 10.1104/pp.103.022665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ang LH, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng XW. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–22. doi: 10.1016/S1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 6.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–6. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 7.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–30. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 8.Kushwaha R, Singh A, Chattopadhyay S. Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell. 2008;20:1747–59. doi: 10.1105/tpc.107.057612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbas N, Maurya JP, Senapati D, Gangappa SN, Chattopadhyay S. Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell. 2014;26:1036–52. doi: 10.1105/tpc.113.122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oyama T, Shimura Y, Okada K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–95. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cluis CP, Mouchel CF, Hardtke CS. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004;38:332–47. doi: 10.1111/j.1365-313X.2004.02052.x. [DOI] [PubMed] [Google Scholar]
- 12.Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008;53:324–35. doi: 10.1111/j.1365-313X.2007.03346.x. [DOI] [PubMed] [Google Scholar]
- 13.Vandenbussche F, Habricot Y, Condiff AS, Maldiney R, Van der Straeten D, Ahmad M. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 2007;49:428–41. doi: 10.1111/j.1365-313X.2006.02973.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Zhang J, Neff MM, Hong SW, Zhang H, Deng XW, Xiong L. Integration of light and abscisic acid signaling during seed germination and early seedling development. Proc Natl Acad Sci U S A. 2008;105:4495–500. doi: 10.1073/pnas.0710778105. [DOI] [PMC free article] [PubMed] [Google Scholar]; 10.1073pnas.0710778105
- 15.Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 2011;65:346–58. doi: 10.1111/j.1365-313X.2010.04426.x. [DOI] [PubMed] [Google Scholar]
- 16.Boter M, Ruíz-Rivero O, Abdeen A, Prat S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004;18:1577–91. doi: 10.1101/gad.297704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhai Q, Li CB, Zheng W, Wu X, Zhao J, Zhou G, Jiang H, Sun J, Lou Y, Li C. Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of jasmonate-responsive genes in Arabidopsis. Plant Cell Physiol. 2007;48:1061–71. doi: 10.1093/pcp/pcm076. [DOI] [PubMed] [Google Scholar]


