SUMMARY
Pellino proteins are RING E3 ubiquitin ligases involved in signaling events downstream of the Toll and interleukin-1 (IL-1) receptors, key initiators of innate immune and inflammatory responses. Pellino proteins associate with and ubiquitinate proteins in these pathways, including the interleukin-1 receptor associated kinase-1 (IRAK1). We determined the X-ray crystal structure of a Pellino2 fragment lacking only the RING domain. This structure reveals that the IRAK1-binding region of Pellino proteins consists largely of a previously unidentified forkhead-associated (FHA) domain. FHA domains are well-characterized phosphothreonine-binding modules, and this cryptic example in Pellino2 can drive interaction of this protein with phosphorylated IRAK1. The Pellino FHA domain is decorated with an unusual appendage or ‘wing’ composed of two long inserts that lie within the FHA homology region. Delineating how this E3 ligase associates with substrates, and how these interactions are regulated by phosphorylation, is crucial for a complete understanding of Toll/IL-1 receptor signaling.
INTRODUCTION
The first line of defense in the human body against invading microorganisms comprises a set of relatively non-specific innate immune responses. Members of the Toll/Interleukin-1 receptor (TIR) superfamily (Dunne and O’Neill, 2003) play a crucial role in regulating cellular aspects of these innate immune responses. Engagement of TIR receptors by cytokines or pathogen derived molecules leads to the activation of several different key cellular signaling pathways, including the nuclear factor kappaB (NF-κB) and mitogen-activated protein kinase (MAPK) pathways – mediated by transforming growth factor-β-activated kinase 1 (TAK1) – and/or the activation of interferon regulatory factors (Dunne and O’Neill, 2003; Kawai and Akira, 2007). Crosstalk among the different members of the TIR superfamily generates a complex signaling network that can elicit, and then dampen, responses to pathogens. Perturbations in innate immune signaling can result in debilitating illnesses such as septic shock, systemic autoimmune diseases and atherosclerosis (Cook et al., 2004).
Intracellular signal downstream of TIR superfamily receptors is transmitted through kinase cascades that involve an array of adaptor proteins and posttranslational modifications (Kawai and Akira, 2007). The first step in TIR signaling involves receptor homo- or hetero-dimerization and the resulting recruitment of one or more of five adapter proteins (including MyD88 and MAL) through homotypic interactions between so-called TIR domains in both receptor and adapter (O’Neill and Bowie, 2007). The TIR-domain-containing adapter proteins in turn recruit Ser/Thr kinases of the IL-1 receptor associated kinase (IRAK) family, which are regulated by both phosphorylation and ubiquitination. Initial phosphorylation of IRAK1 by IRAK4 leads to full activation of IRAK1 and its subsequent hyper-autophosphorylation (Janssens and Beyaert, 2003). Hyperphosphorylated IRAK1 is released from the TIR complex, allowing it to activate downstream molecules including IRF7, TRAF6 and TAK1. This activated IRAK1 also becomes highly ubiquitinated. Lysine 63 linked poly-ubiquitin (K63-pUb) chains on IRAK1 serve as docking sites for NEMO (the NFκB essential modulator), a regulatory subunit of the IκBα kinase (IKK) complex (Conze et al., 2008; Windheim et al., 2008). By contrast, K48-linked poly-ubiquitination promotes proteasomal degradation of IRAK1 (Yamin and Miller, 1997). The Pellino proteins are primary candidates for E3 ligases responsible for these ubiquitination events. In particular, the Pellino proteins have been shown to interact with IRAKs specifically following receptor stimulation and IRAK1 phosphorylation (Jensen and Whitehead, 2003b; Jiang et al., 2003; Yu et al., 2002), and can K63 poly-ubiquitinate IRAK1 (Butler et al., 2007; Ordureau et al., 2008; Schauvliege et al., 2006; Xiao et al., 2008).
Pellino proteins were originally found to interact with IRAKs in a yeast two-hybrid screen for proteins that interact with the Drosophila IRAK homologue Pelle (Grosshans et al., 1999). In mammals there are four Pellino proteins, with between 418 and 479 amino acids (Fig. 1A): Pellino1, Pellino2, and two splice variants of Pellino3 (Pellino3a and 3b). Pellino1 is involved in TIR signaling to NF-κB (Choi et al., 2006; Jiang et al., 2003), whereas both Pellino3 isoforms have been implicated in the activation of the MAP kinase pathway (Butler et al., 2005; Jensen and Whitehead, 2003b) and Pellino3b is a negative regulator of NF-κB activation (Xiao et al., 2008). Pellino2 is implicated in activation of both NF-κB (Liu et al., 2004; Yu et al., 2002) and MAP kinase (Jensen and Whitehead, 2003a). These differences suggest that each Pellino may ubiquitinate a different set of substrates, or that their activities may be modulated by isoform-specific posttranslational modifications. By analogy with other RING E3 ubiquitin ligases, Pellino proteins are likely to contain a substrate recognition domain (Ardley and Robinson, 2005). We show here that the C-terminal 140 amino acids of Pellino proteins, which contain the RING domain, are not required for interaction with phosphorylated IRAK1. Instead, the N-terminal domain of Pellinos 1, 2, and 3b are sufficient to interact with this ubiquitination substrate. To shed light on how Pellino proteins interact with their substrates, such as IRAK1, we solved the X-ray crystal structure of the Pellino2 N-terminal region (amino acids 15–275). Unexpectedly, this region of Pellino2 consists entirely of an FHA domain that had not been predicted from the primary sequence. The cryptic Pellino2 FHA domain contains an unusual ‘wing-like’ appendage made up from two long sequence insertions in the FHA domain. Key amino acids known to be important in phosphothreonine-binding by canonical FHA domains (Durocher, 2005) are conserved in all Pellino proteins, and mutation of these amino acids in Pellino2 abolishes interaction with IRAK1. Our studies argue that Pellino proteins bind directly to phosphorylated threonines on IRAK1 and other phosphorylated substrates. The identification of an FHA domain in Pellino proteins not only highlights the involvement of this domain in innate immune signaling, but also illustrates a molecular basis for the regulation and substrate specificity of Pellino proteins.
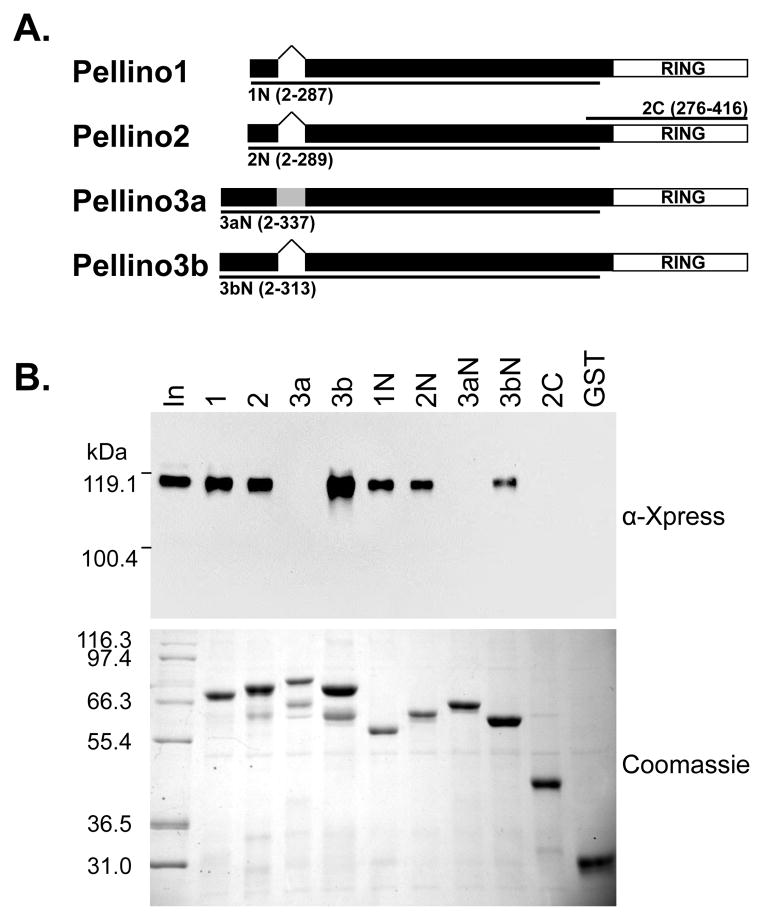
Figure 1. The Pellino C-terminal RING motif is not required for interaction with IRAK1.
(A) Schematic representation of Pellino proteins indicating the location of the RING domain and of the 24 amino acid insert in Pellino3a (grey). Lines indicate the deletion variants. The Pellino amino acids in these GST-fusion proteins are indicated.
(B) The indicated GST-fusion of Pellino proteins or of deletion variants were incubated with HEK293T lysates that contained overexpressed IRAK1. Following washing, bound proteins were eluted by denaturation, analyzed by SDS-PAGE, and visualized by immunoblotting with α-Xpress antibody to detect IRAK1 and Coomassie blue staining to detect the GST-fusion proteins. “In” indicates 0.8 % of lysate. Positions of molecular mass markers are shown.
RESULTS
The C-terminal RING motif of Pellino is not required for interaction with IRAK1
To define which region(s) of Pellino proteins direct(s) their interaction with IRAK1, we used E. coli expressed glutathione S-transferase (GST)-fusions of Pellino proteins to precipitate, from cell lysates, epitope-tagged IRAK1 that was overexpressed (and thus highly phosphorylated) in HEK293T cells. As shown in Fig. 1B, robust interaction of IRAK1 is observed with the purified GST-fusion proteins of full-length Pellino1, 2 and 3b. To investigate whether the C-terminal RING domain of Pellino proteins is required for this interaction, we generated fragments of each Pellino protein corresponding to the N-terminal 287 to 337 amino acids: i.e. lacking only the C-terminal ~120-residue RING domain (Fig. 1A). Removal of the RING domain does not abolish IRAK1 binding (Fig. 1B) in this qualitative assay; fragments 1N, 2N and 3bN all retain substantial affinity. By contrast, removing the N-terminal region from Pellino2, leaving only amino acids 276–416 (the RING domain: 2C) abolishes its ability to pull down IRAK1 from these lysates (Fig. 1B). The Pellino N-terminal region appears to be sufficient for association with IRAK1, although additional stabilization of the interaction by C-terminal regions of the protein cannot be ruled out – particularly for Pellino3b.
Structure determination
The IRAK1-binding N-terminal 287 amino acids of Pellino2 contain no recognizable domains or motifs based on primary sequence analysis. As a first step towards understanding how Pellino employs this region to recognize their ubiquitination targets, we chose to determine its structure. When expressed in E.coli, full-length Pellino proteins are highly sensitive to proteolysis that leads to removal and degradation of the C-terminal RING domain-containing region. The stable proteolytic fragments that remain were used to guide selection of boundaries for expressing variants of the N-terminal IRAK-binding region suitable for crystallization. IRAK1 binding of each variant was confirmed using the GST-pull down assay (Fig. S1A). Optimal crystals were obtained for a selenomethionine-containing protein comprising amino acids 15–275 of Pellino2 (P2(15–275)*) containing two methionine substitutions at positions where methionine is observed in at least one other Pellino protein (V61 and L232: Fig. S2). The structure of P2(15–275)* was solved by the method of multiwavelength anomalous dispersion (MAD) and was refined against data to 1.8Å resolution (Table 1). A different crystal form was also obtained for a slightly longer fragment of Pellino2 (without methionine substitutions) comprising amino acids 7–289 (P2(7–289)). The structure of P2(7–289) was solved to 3.3Å resolution using molecular replacement (MR) methods with P2(15–275)* as the search model. The P2(15–275)* and P2(7–289) structures are almost identical (Fig. S1B). The two methionine substitutions in P2(15–275)* do not perturb the overall structure of the Pellino2 N-terminal domain, nor do they affect the ability of Pellino2 to interact with IRAK1 (Fig. S1A).
TABLE 1.
Data collection and refinement statistics
| P2(7–289) | P2(15–275)* | |||
|---|---|---|---|---|
| Data collectiona | APS 23ID-D | APS 23ID-B | ||
| Space group | P212121 | F222 | ||
| Cell dimensions | ||||
| a, b, c | 43.7 Å, 89.5 Å, 125.6 Å | 83.6 Å, 86.1 Å, 162.8 Å | ||
| α, β, γ | 90°, 90°, 90° | 90°, 90°, 90° | ||
| Native | Peak | Inflection | Remote | |
|
|
||||
| Wavelength | 0.9793 Å | 0.9795 Å | 0.9796 | 0.9495 Å |
| Resolution limit | 3.25 Å | 1.8 Å | 1.8 Å | 1.9 Å |
| Rsymb | 0.16 (0.46) | 0.1 (0.5) | 0.074 (0.43) | 0.08 (0.456) |
| <I/σ> | 19.0 (4.5) | 37 (2.4) | 41.7 (2.6) | 32 (2.9) |
| Completeness (%) | 99.9 (99.0) | 98.5 (86.9) | 98.4 (86.8) | 99.3 (93.7) |
| Redundancy | 8.4 (7.7) | 8.2 (3.9) | 8.4 (4.8) | 8.6 (5.7) |
|
| ||||
| Refinement | ||||
| Resolution limits | 45 – 3.25 Å | 43.1 – 1.8 Å | ||
| No. of reflections | 8,229 | 27,047 | ||
| R factor (Rfree)c | 0.25 (0.32) | 0.21 (0.24) | ||
|
| ||||
| Model | ||||
| Protein | 2 × P2(7–289) aa 15–27, 39–66, 71–120, 132–197, 203–282d |
1 × P2(15–275)* aa 15–27, 38–119, 130–196, 202–258 1 sulfate ione, 130 water molecules |
||
| Total number atoms | 3,221 | 1,836 | ||
| R.M.S deviations | ||||
| bond lengths | 0.010 Å | 0.014 Å | ||
| bond angles | 1.262° | 1.510° | ||
Numbers in parentheses refer to last resolution shell
Rsym = Σ|Ih-<Ih>|/ΣIh, where <Ih> = average intensity over symmetry equivalent measurements
R factor = Σ|Fo-Fc|/ΣFo, where summation is over data used in the refinement; Rfree includes only 5% of the data excluded from the refinement
Numbers in Table are for chain A. For chain B: aa 16–26, 40–64, 73–119, 130–196, 204–269
Sulfate ion is at special position, modeled with half occupancy
The IRAK binding domain of Pellino is a modified FHA domain
The Pellino2 N-terminal region forms a single globular domain containing seventeen β-strands (Fig. 2) that form four distinct β-sheets. Unexpectedly, parts of this domain share remarkable structural similarity with the forkhead-associated (FHA) domain, a well-known, and extensively characterized, phosphothreonine-binding module (Durocher et al., 1999; Sun et al., 1998). The region colored green in Fig. 2 corresponds very closely to an FHA domain, comprising two β-sheets that form a β-sandwich. The remaining two sheets (blue in Fig. 2) are formed by two insertions (of 45 and 26 amino acids respectively) in the FHA module that constitute a significant ‘wing’ or appendage on the FHA domain structure. FHA domains are found in a large number of proteins in eukaryotes and bacteria with a diverse array of functions including DNA repair and cell cycle regulation (Durocher, 2005). Of particular note among the 26 known FHA-containing proteins in humans are the RING E3 ubiquitin ligases Chfr and RNF8 (Huen et al., 2007; Kang et al., 2002) and TIF2A/T2BP, which is involved in innate immune signaling and (like Pellino) has been suggested to bind both IRAK1 and TRAF6 (Takatsuna et al., 2003).
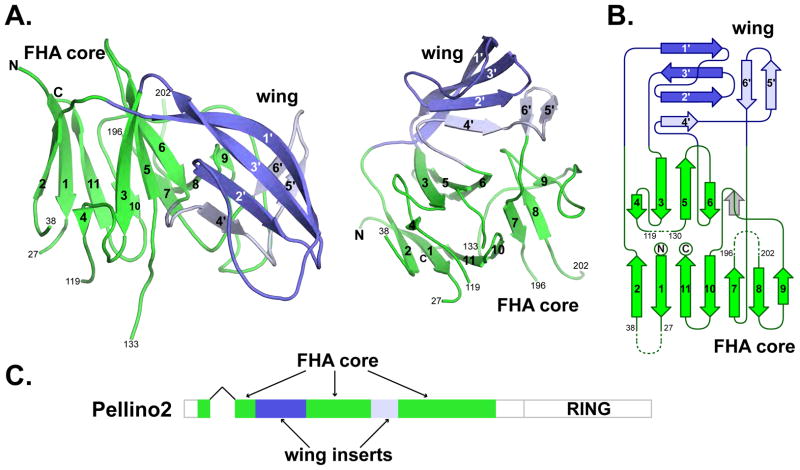
Figure 2. The IRAK1-binding region of Pellino2 is an FHA-like domain.
(A) Cartoon views of the Pellino2 structure with the FHA core in green and the non-canonical wing in blue (amino acids 51–98 and 159–184 in dark and light blue, respectively). β-strands of the FHA core are numbered 1–11, while those of the wing are 1′-6′. The N- and C-termini are labeled, as are the amino acids numbers at the beginning and end of each missing loop.
(B) Topology diagram of Pellino with the strands colored as in A. Missing loops are shown with dashed lines. The position of β9 in canonical FHA domains is shown in grey.
(C) The domain architecture of Pellino2.
The Pellino FHA domain
The 11-stranded Pellino2 β-sandwich, colored green in Fig. 2, is readily recognized as an FHA domain by fold or structure recognition algorithms, such as the DALI server (Holm et al., 1992). This Pellino2 FHA core region is superimposed on the Rad53 N-terminal FHA domain in Fig. 3A. In the structure-based amino acid sequence alignment (Fig. 3B), regions of significant structural overlay of the Pellino2 FHA core with selected FHA domains are underlined. The structural similarities extend beyond the β-strands well into many of the loop regions of the domain. Many of the key conserved amino acids in the β3/β4, β4/β5 and β6/β7 loops of canonical FHA domains (bold in Fig. 3B) are also found in Pellino2. A subset of these conserved amino acids is involved in binding to phosphothreonine-containing peptides.
Figure 3. The Pellino FHA core shares features required for phosphothreonine recognition.
(A) Superposition of the Pellino FHA core (green) with the Rad53-FHA1 (grey, PDB ID 1gxc). The Rad9 peptide bound to the Rad53-FHA1 is shown in cyan with the phosphothreonine in stick representation. The beginning and end of each wing loop are shown in blue. Numbers indicate the chain breaks. β9 of Pellino and Rad53 are labeled 9P and 9R respectively.
(B) Structure-based sequence alignment of the Pellino FHA core with select FHA domains. β-strands, as defined by the program DSSP (Kabsch and Sander, 1983), are highlighted in green. Green arrows indicate Pellino2 β-strands, grey arrow shows β9 of canonical FHA domains. The amino acids underlined in red were used for the structural alignment and superimpose with Pellino to yield pairwise RMSD values for Cα positions of < 3.2Å. Bold text indicates identity/close homology (represented 4 or more times in this alignment). Numbers indicate omitted amino acids. For Pellino2, black numbers indicate structurally disordered amino acids, blue numbers the wing insertions. Conserved amino acids that contact the phosphopeptide are highlighted with triangles. The sequence of the TIFA FHA domain is aligned based on amino acid homology only. FHA/50% is the 50 % consensus for the FHA homology region (Letunic et al., 2006). PDB IDs used to generate this alignment: 2ff4, 2aaf, 1lgp, 1gxc, 1g6g, 1mzk, 1yjm, 2pie and 1j4l.
(C) A detailed view of the superposition of Pellino2 and the Rad53-FHA1/Rad9 complex, colored as in A. Conserved side chains are show, labeled in green for Pellino2 and in black for Rad53. Hydrogen bonds between Rad53 and Rad9 are indicated with dashed lines.
The Pellino FHA domain shares features known to be critical for phosphothreonine-binding
Structures of 8 canonical FHA domains bound to phosphothreonine peptides (Bernstein et al., 2005; Byeon et al., 2005; Byeon et al., 2001; Durocher et al., 2000; Huen et al., 2007; Lee et al., 2008; Li et al., 2002; Yuan et al., 2001), together with mutational analyses (Durocher, 2005; Durocher et al., 1999; Li et al., 1999) have identified highly conserved amino acid side chains that are crucial for binding phosphorylated peptides and proteins. The locations of these amino acids, immediately following β3 and preceding both β5 and β7, are marked with triangles above the sequences in Fig. 3B. These amino acids are also conserved in Pellino proteins (Fig. 3B, S2). Moreover, the conformations of these side-chains, shown interacting with a phosphopeptide bound to Rad53-FHA1 in Fig. 3C, are remarkably well conserved in the Pellino2 FHA core. An invariant arginine (R70 in Rad53-FHA1) immediately follows strand β3 and interacts with the phosphothreonine γ oxygen. A highly conserved serine (S85 in Rad53-FHA1) in the loop preceding β5 interacts with one of the ε oxygens. The amino acid following this serine frequently has a basic side-chain and makes key contacts (although an asparagine [N86] plays this role in Rad53). In the β6/β7 loop a well-conserved asparagine (N107 in Rad53) interacts with the phosphopeptide’s backbone in all known FHA domain/phosphopeptide complex structures, and contributes to the binding pocket for the γ methyl group of the phosphothreonine. The amino acid preceding this position is often a threonine, which makes additional interactions with the phosphate moiety. The Pellino2 FHA core maintains all five of these key residues (R106, S137, R138, T187 and N188), which may contribute directly to phosphothreonine-dependent binding of Pellino2 to IRAK1.
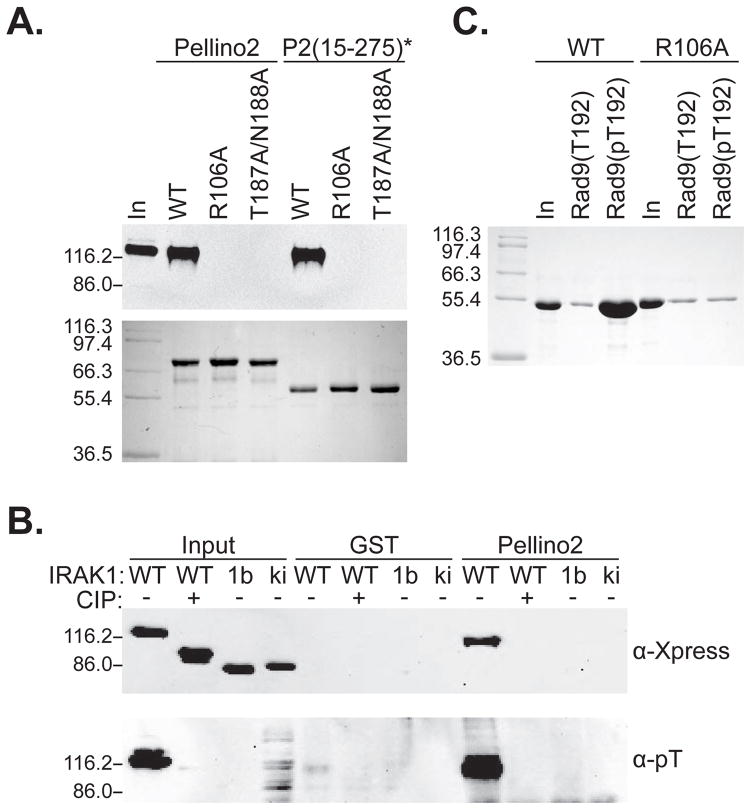
To determine whether these conserved amino acids are indeed important in Pellino2 binding to phosphorylated IRAK1, GST-Pellino variants were generated with either R106 or both T187 and N188 replaced with alanine (R106A and T187A/N188A respectively). Both alterations abolished the ability of GST-fusions of Pellino2 or P2(15–275)* to precipitate IRAK1 from HEK293 lysates, without affecting expression level or stability of the Pellino protein (Fig. 4A). This observation is consistent with the hypothesis that the Pellino2 FHA core binds phosphothreonines in IRAK1 through a mechanism seen for binding of other FHA domains to their phosphorylated targets. In further support of this hypothesis, we showed that enzymatic dephosphorylation of IRAK1 abolishes its ability to interact with GST-Pellino2 fusion proteins (Fig. 4B). Lysates from HEK293T cells transfected with epitope-tagged IRAK1were incubated with calf intestinal alkaline phosphatase (CIP) for six hours at 37°C. The CIP-treated IRAK1 migrates further in SDS-PAGE, and cannot be detected with anti-phosphothreonine or anti-phosphoserine specific antibodies (Fig. 4B and data not shown). Following CIP treatment, IRAK1 is no longer precipitated by GST-Pellino2, arguing that its phosphorylation is critical for interaction with the Pellino2 FHA core. Moreover, two kinase-inactive forms of IRAK1 (IRAK1b and IRAK1-ki) fail to become autophosphorylated and are not precipitated by the Pellino2 GST fusion protein.
Figure 4. The Pellino FHA-like domain is a phosphothreonine-binding module.
(A) Substitution of alanine at key conserved amino acids in the Pellino FHA core abolishes interaction with IRAK1. GST-fusions of full-length Pellino2 or of P2(15–275)* with substitutions R106A or T187/N188A fail to pulldown IRAK1 using the assay described in the legend to Fig. 1. Upper panel; α-Xpress immunoblot, lower panel; Coomassie blue staining.
(B) Enzymatic dephosphorylation of IRAK1, by incubation of HEK293 lysates with CIP for 6 hours at 37°C, leads to a further migrating band on SDS-PAGE that is not detected by anti-phosphosthreonine antibody (α-pT), and does not interact with GST-Pellino2. Kinase inactive forms of IRAK1 (an alternative spliceform, 1b, and kinase dead mutant, ki (Jensen and Whitehead, 2001)) are also not detected by anti-phosphothreonine, and do not interact with Pellino2. “In” represents 6% of lysate.
(C) Purified Pellino2 can interact with immobilized, phosphorylated Rad9 derived peptide [Rad9(pT192)] but not with a non-phosphorylated peptide of the same sequence [Rad9(T192)]. Substitution in Pellino2 of R106 with alanine abolishes this interaction. Peptides were amine coupled to agarose beads and incubated with 50 μM purified wild type or R106A Pellino2. Following three washes, bound proteins were eluted by denaturation, analyzed by SDS-PAGE and visualized by Coomassie staining. “In” represents 1 % of the input protein. Positions of molecular mass markers are shown (kDa)
Binding of Pellino2 to phosphothreonine-containing peptides
FHA domains can bind phosphothreonine-containing peptides, and typically show selectivity for phosphothreonine in a particular sequence context (Yaffe and Smerdon, 2004). To determine whether the modified FHA domain of Pellino2 shares this property, we investigated its ability to bind phosphothreonine-containing peptides. We first analyzed binding to a phosphopeptide based on amino acids 188–196 of the Rad9 sequence (where T192 is phosphorylated). This peptide [Rad9(pT192)] has been shown to bind to Rad53-FHA1 (Durocher et al., 2000; Liao et al., 2000) and EmbR-FHA (Alderwick et al., 2006). As shown in Fig. 4C, the Rad9(pT192) peptide can also interact with Pellino2 in the context of a bead pull-down assay. The R106A mutation, which abolished IRAK1 binding by Pellino2, also prevents it from binding the Rad9(pT192) peptide. Moreover, an unphosphorylated control peptide with the same sequence (Rad9(T192)) does not interact with Pellino2. These data strongly suggest that the Pellino2 FHA core resembles other FHA domains in its ability to recognize phosphothreonine-containing sequences, and that this mode of interaction is likely to contribute to recognition of phosphorylated IRAK1 by Pellino proteins. Although Pellino2 binding to the Rad9(pT192) peptide could be detected in the qualitative experiments shown in Fig. 4C, efforts to measure the affinity of this interaction suggest a KD value of greater than 100μM. There are no threonines in IRAK1 that are in a sequence context that resembles Rad9(pT192). We tested binding of Pellino to a series of phosphopeptides based on predicted IRAK1 phosphorylation sites. None of these peptides interacts with Pellino2 in the qualitative bead pull-down assay, suggesting that we have not yet identifying the cognate Pellino binding sequence.
Non-canonical features of the Pellino FHA core
Although many characteristic FHA domain features are conserved in the Pellino FHA core, and the data presented above suggest that the phosphothreonine-binding function is maintained, there are other conserved amino acids in FHA domains that are not found in Pellino. Almost every known FHA domain has a histidine just before β5 and an aspartic acid near the end of β6. Although these do not participate in phosphopeptide binding, it has been proposed that the respective side chains contribute to interactions between the β4/β5 and β6/β7 loops that constitute much of the binding pocket (Durocher et al., 2000). All Pellino proteins have alanine at these two positions (A140 & A157 in Pellino2) so the relationship between the β4/β5 and β6/β7 loops is less constrained than in canonical FHA domains. This difference can be appreciated in Fig. 3C from the altered location of T187 in the Pellino2 β6/β7 loop compared with T106 of Rad53-FHA1. It is also important to note that the β6/β7 loop contains one of the wing inserts, which may participate in Pellino2 binding to phosphorylated substrates (see below).
A striking feature of the Pellino2 FHA core is the increased length of many of the loops separating strands in the β-sandwich (some disordered in the structure; Table 1) compared with the short connecting loops typically seen in canonical FHA domains (Fig. 3B). In the particularly long β1/β2, β4/β5 and β7/β8 loops (Fig. 3B, S2), electron density was poorly defined for 10, 10, and 5 amino acids respectively, suggesting substantial disorder. These three disordered loops all project away from the 7-stranded sheet of the FHA core β-sandwich (Fig. 2A, S3), suggesting that they could act together to form a flexible binding surface for another molecule, or could possibly interact with the C-terminal RING motif region of Pellino itself. These loops are not well conserved between Pellino proteins (Fig. S2). The β1/β2 loop is the location of a long insert found only in Pellino3a, the β4/β5 loop is one of the most divergent regions among human Pellino proteins, and both the β4/β5 and β7/β8 loops are quite divergent in other phyla. The differences in these three loops might dictate binding selectivity (and thus different substrate and signaling specificities) for these proteins.
The topology of the Pellino2 FHA core deviates from that observed in all other FHA domains in the region connecting β8 and β10 (Fig. 2A, 3A). In all canonical FHA domains of known structure the ninth β-strand is antiparallel to strand β6 (grey arrow in Fig. 2B, 3B; 9R in Fig. 3A), completing a five-stranded sheet made up from strands β3, β4, β5, β6 and β9. In the Pellino2 FHA core, the ninth β-strand instead forms an antiparallel interaction with strand β8, so that the larger β-sheet of the Pellino2 FHA core β-sandwich comprises seven strands, rather than the six seen in canonical FHA domains. The loop connecting this alternatively-located β9 strand and β10 is well ordered in both the P2(15–275)* and P2(7–289) structures, stabilized by interactions with β5′ and β6′ from the Pellino2 wing (Fig. 5A), and by a network of hydrogen bonds and salt bridges between the β9/β10 loop and the β7/β8/β10 region of the FHA core. It is interesting that the tip of the β9/β10 loop is close to a region implicated in defining specificity for phosphothreonine sequence context in canonical FHA domains (interacting with the side chain of the pT+3 amino acid (Durocher, 2005)). This region of the Pellino2 FHA core, and the distinct connections of the β9 strand, might therefore play a role in defining the specificity of its phosphopeptide recognition. The β8 to β10 region is highly conserved among all Pellino proteins (Fig. S2), suggesting that this region would not confer isoform specificity.
Figure 5. The Pellino2 wing packs tightly against the FHA core.
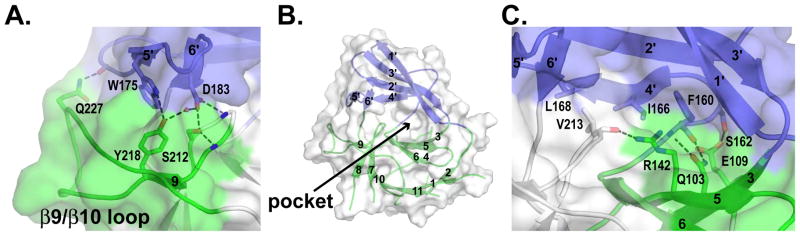
Cartoon representations of Pellino2 FHA looking onto the face of the protein that opposes the phosphopeptide-binding site. Colors are as in Fig. 2. A transparent molecular surface is shown.
(A) The β5′/β6′ hairpin of the wing interacts with the non-canonical β8-β10 region of the FHA core, stabilized by both van der Waals and polar interactions.
(B) A solvent-filled pocket lies at the center of this face of the protein (see also Fig. S4).
(C) The wing region is additionally stabilized against the FHA core by interaction between the β4′ strand (plus the preceding loop) and the β3/β4/β5/β6 sheet of the FHA core.
The Pellino2 FHA domain wing
As mentioned above, the Pellino2 FHA domain has a unique additional ‘wing’ or appendage that has not been seen in any other FHA domain structure, but is well conserved in sequence across Pellino proteins (suggesting functional significance). The wing comprises two large inserts in the β2/β3 and β6/β7 loops of the FHA core, of 45 and 26 amino acids respectively (Fig. 3B, S2). Such long inserts have not previously been seen in an FHA domain, although the Rad53-FHA1 does have a short α-helical elaboration between strands β2 and β3 (Fig. 3A). The presence of these inserts confounded the recognition of the FHA core in Pellino sequences; indeed, with these inserts removed from the sequence, the Pellino2 FHA core can be identified by threading algorithms.
The first of the two inserts (amino acids 51–98, dark blue in Fig. 2) contributes three strands (β1′-β3′) to a four-stranded β-sheet. The second insert (amino acids 159–184, light blue in Fig. 2) contains three shorter strands (β4′-β6′). Strand β4′ is parallel to strand β2′ as part of the larger β-sheet of the wing, whereas β5′ and β6′ form a β-hairpin that lies orthogonal to this larger β-sheet. The wing is tightly packed against the FHA core (Fig. 5) and is highly unlikely to be independently stable. The β5′/β6′ hairpin interacts with the non-canonical β8-β10 region of the Pellino2 FHA core, stabilized by both van der Waal’s interactions and a network of hydrogen bonds (Fig. 5A). Hydrophobic side-chains, largely from strand β4′ and the preceding loop, pack against part of the outer face of the four-stranded sheet (β4/β3/β5/β6) of the FHA core. The β4′ edge of the wing is further stabilized against the FHA core by polar interactions (Fig. 5B). Strands β1′, β2′ and β3′ make few contacts with the FHA core, other than those near the covalent linkages. As a consequence, a deep pocket is created that is also rimmed by the β4/β3/β5/β6 sheet and β9/β10 loop of the FHA core, as well as the β5′/β6′ wing loop (Fig. 5B). This pocket is surrounded by negative charges, filled with solvent, and lies on the opposite face of the Pellino2 FHA core from the likely phosphopeptide binding site. This binding pocket is well conserved among Pellino proteins (Fig. S4A), suggesting that it might provide an important interaction surface for a positively charged binding partner. Alternatively, the pocket might be involved in interactions with the RING module of Pellino2, in a manner reminiscent of the interaction between the RING domain of c-Cbl and its tyrosine kinase binding domain (Zheng et al., 2000).
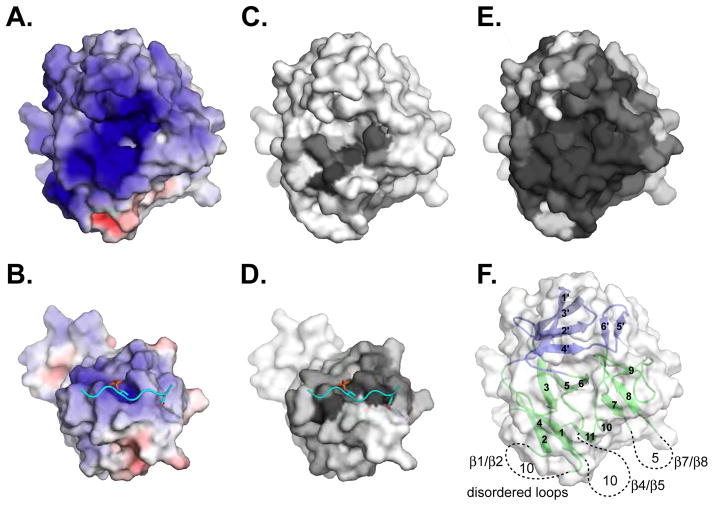
It is highly probable that the wing of the Pellino2 N-terminal domain plays an important role in its interaction with phosphorylated IRAK1 (or other substrates), in addition to the FHA core. Fig. 6A shows a view of the phosphopeptide binding face of Pellino. The apposition of the wing and the FHA core effectively expands the phosphopeptide binding region of this atypical FHA domain, suggesting that the cognate binding target will include elements expected of a canonical FHA domain ligand plus additional features that interact with the wing region. In Fig. 6B the Rad53-FHA1/Rad9 peptide complex is shown superimposed on the Pellino2 FHA-like domain. The Rad9 peptide lies in a groove on the surface of Rad53-FHA1 (Fig. 6C), which is conserved in Pellino 2 (green in Fig. 6A). The wing of the Pellino2 FHA-like domain (top of Fig. 6A, colored blue) extends this binding site substantially, presenting polar residues including R85, K69 and E170, plus hydrophobic side-chains such as F167. The wing of the Pellino2 FHA-like domain may play a role in IRAK1 binding analogous to that of the RT loop of an SH3 domain bound to HIV Nef (Lee et al., 1996). SH3 domain recognition of Nef involves two components: the canonical PXXP sequence recognition and additional interactions of the SH3 domain RT loop with specific features on the Nef surface. Cooperation between these two types of interaction allows the SH3 domain to bind Nef with an affinity that is ~100-fold higher than for the proline-rich peptide mimetic. Similarly, the wing in the Pellino2 FHA-like domain might interact with surface features of IRAK1 that supplement the recognition of adjacent phosphothreonine-containing sequences by the FHA core – allowing increased affinity and specificity. Another possibility, since the Pellino2 binding pocket is very highly positively-charged – substantially more so than other FHA domains (compare Fig. 7A, B) – is that the wing could allow binding to a multiply phosphorylated region. This is not unprecedented for FHA domains (Byeon et al., 2005; Lee et al., 2008). Whereas only the region surrounding the canonical binding site is conserved when all FHA domains are considered (Fig. 7C, D), the entire positively-charged face of the Pellino2 FHA-like domain is highly conserved across all Pellino proteins (Fig. 7E), suggesting that the entire surface is used for key interactions. The less well-conserved loops, such as that between β4 and β5 (Fig. 7F), may project away from this conserved surface, or could possibly participate in binding to phosphorylated substrates, and confer specificity (or regulation) of interactions for different Pellino proteins.
Figure 6. The Pellino2 wing extends the substrate-binding surface.
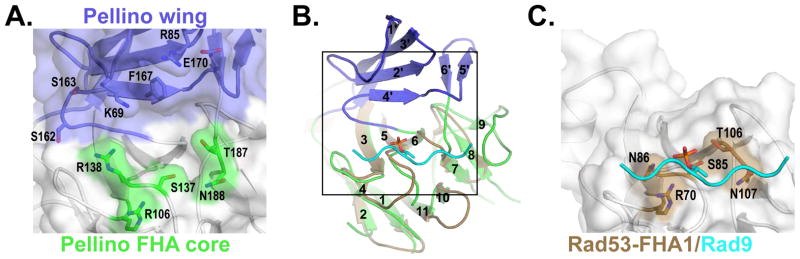
The Pellino2 FHA-like domain and the Rad53 FHA1/Rad9 complex are shown looking onto the phosphothreonine binding site, a 180° rotation about a vertical axis with respect to Fig. 5.
(A) The apposition of the Pellino2 wing and FHA core result in a binding pocket that is lined with hydrophobic amino acids and rimmed on one side by amino acids (green) implicated in direct contact to the phosphothreonine, and on the other side by charged and polar side chains from the wing loops (blue).
(B) The Rad53-FHA1 (beige) is show superimposed on the Pellino FHA core (green). The phosphopeptide bound to Rad53-FHA1 is shown in cyan. The box indicates the area of detail shown in A and C.
(C) The phosphopeptide binds to a groove on the surface of Rad53-FHA1 analogous to the lower rim of the pocket highlighted in A.
Figure 7. Features of the Pellino2 substrate-binding surface.
Surface representations are shown of the Pellino2 FHA-like domain and of the Rad53 FHA1/Rad9 complex in the same orientation as Fig. 6.
(A) The substrate-binding pocket on Pellino2 is highly electropositive, substantially more than the equivalent region of the Rad53 FHA-1 domain (B). The electrostatic potential at the solvent accessible surface, calculated using APBS (Baker et al., 2001), is projected onto a molecular surface, and colored in a gradient (red-white-blue) from −6kT to +6kT.
(C–E) Amino acid conservation is projected onto the same molecular surfaces using the program CONSURF (Landau et al., 2005). A gradient from grey to white represents most conserved to most divergent positions. Conservation taken from: (C) a structure based alignment of Pellino2 to all FHA domains of known structure, (D, E) pfam alignments of (D) all FHA domains or (E) all Pellino proteins.
(F) In view of the Pellino2 FHA domain the location and length of the disordered loops are represented with dashed lines.
DISCUSSION
It has previously been shown that the interaction of Pellino proteins with IRAK1 is dependent on activation and phosphorylation of the kinase, whether it be by agonist stimulation or through autoactivation by overexpression (Jensen and Whitehead, 2003b; Jiang et al., 2003; Strelow et al., 2003; Yu et al., 2002). Further, kinase-inactive IRAK1 dose not interact with Pellino proteins (Grosshans et al., 1999; Jensen and Whitehead, 2003b; Schauvliege et al., 2006). In this work we describe the molecular basis for these observations. Pellino proteins contain an unexpected FHA-like domain that interacts with IRAK1, and likely with other substrates, through a binding event that requires at least one phosphorylated threonine. We describe an unanticipated high degree of structural homology between the core β-sandwich of Pellino2 and FHA domains, particularly in the phosphopeptide binding region. We show that mutation of R106 and other key conserved amino acids in this region of Pellino2 leads to complete loss of its interaction with IRAK1. We also show that the Pellino2 FHA-like domain can bind – albeit with modest affinity – to phosphothreonine-containing peptides. However, the structure of the complete FHA-like domain suggests that key additional binding interactions that increase affinity and specificity are contributed by the unusual wing-like appendage that is intimately associated with the Pellino FHA core. If each Pellino protein has a different binding specificity, this is most likely conferred by the loops that are disordered in this structure (Fig. 7F), or, for Pellino3a, by the isoform specific insertion in the β1/β2 loop.
It is possible that simultaneous recognition of multiple phosphorylated threonines (and/or serines) is required to support high affinity interaction of Pellino proteins with IRAK1. This is suggested by the high positive potential on the large conserved binding surface of the Pellino2 FHA-like domain (Fig. 7A), and there is precedent for related modes of binding for other FHA domains. For example, the FHA domain from the Dun1 kinase, which is involved in the yeast DNA-damage response and is structurally and functionally similar to Rad53, specifically recognizes a double threonine-phosphorylated motif, whereas the Rad53 FHA domain can bind the same motif when it is only singly phosphorylated (Lee et al., 2008). The interaction of the human nucleolar protein hNIFK with the FHA domain from the cell proliferation regulator Ki67 also requires sequential phosphorylation on three amino acids of hNIFK (Byeon et al., 2005). Structural studies of a triply phosphorylated fragment of hNIFK show that interaction with the Ki67 FHA domain involves both the phosphorylated and non-phosphorylated regions (Byeon et al., 2005). In an interesting parallel, activation of IRAK1 is known to require initial sequential phosphorylation steps (Kollewe et al., 2004), which are followed by multiple phosphorylation events at Ser/Thr rich regions of the protein (Janssens and Beyaert, 2003; Kollewe et al., 2004). It is not yet clear which of these events are required for interaction with Pellino proteins.
Finally, additional regulation at the level of phosphoamino acid recognition may result from phosphorylation of Pellino proteins themselves. Pellino proteins are phosphorylated in a manner that depends on IRAK1 kinase activity, and in vitro by recombinant IRAK1 and 4 (Ordureau et al., 2008; Strelow et al., 2003). Our data suggest that phosphorylation of Pellino3a may be required for its interaction with IRAK1. We are unable to detect interaction of Pellino3a with IRAK1 in the GST-pulldown assay (Fig. 1B), although interaction of this isoform of Pellino3 with IRAK1 is seen when both proteins are expressed in mammalian cells (Butler et al., 2005; Jensen and Whitehead, 2003b). In further support of the importance of phosphorylation in the regulation of Pellino proteins, phosphorylation of Pellino1 and 3b has been shown to enhance their ubiquitin ligase activity (Ordureau et al., 2008). The identification of a phosphothreonine-binding module in Pellino proteins provides a valuable molecular framework to investigate the function and regulation of Pellino proteins.
EXPERIMENTAL PROCEDURES
Molecular biology methods
cDNA clones for mouse Pellino1 (MGC-29041) and human Pellino2 (MGC-15066) were obtained from ATCC. Pellino3 clones were as described (Jensen and Whitehead, 2003b). The appropriate coding regions for full length or truncated Pellino proteins were amplified using PCR and inserted into pGEX-4T1 (GE Healthcare; GST-Pellino1N, GST-Pellino3aN and GST-Pellino3bN), or into one of two pET28 derived vectors that direct expression of tobacco etch virus (TEV) protease cleavable N-terminal His6- or GST-fusion proteins (HTUA and pGV67 plasmids generously provided by Dr. Gregory Van Duyne, UPenn). The linker in pGV67 contributes an additional 15 amino acids to the fusion proteins compared to the proteins expressed in pGEX-4T1. Site-specific alterations in Pellino2 were created using Quickchange site-directed mutagenesis system (Stratagene). All PCR-generated DNAs were sequence verified. Human IRAK1, IRAK1b and a kinase inactive mutant of IRAK1 (IRAK1ki), in pcDNA4/HisMax vector (Invitrogen), have been described (Jensen and Whitehead, 2001).
Protein expression and purification
GST- and His6-Pellino proteins were expressed in E. coli BL21 (DE3) by induction with isoproryl β-D-thiogalactoside (IPTG) at 25°C for 4 hours. Selenomethionine-derivatized (SeMet) P2(15–275)* was expressed in B834 (DE3) E.coli (Novagen). Overnight Luria broth (LB) cultures were pelleted, washed twice with water, re-suspended in SelenoMet™ medium (Molecular Dimensions) and protein expression induced with IPTG at 25°C for 4 hours. His6-Pellino proteins were purified on Ni-NTA agarose and eluted with an increasing imidazole step gradient. TEV protease was added to protein containing fractions, which were dialyzed overnight at 4°C against 100mM Tris-HCl, 70mM NaCl, 0.5mM EDTA, and 1mM DTT, pH 8.0. Following buffer exchanged into 50mM MES, 80mM NaCl, 1mM DTT, pH 6.0, protein was purified by cationic exchange (Source S, GE Healthcare) and size exclusion chromatography (Superose 6, GE Healthcare). SeMet incorporation was confirmed by MALDI (matrix-assisted laser desorption ionization) mass spectrometry. GST-Pellino proteins were incubated with glutathione agarose beads (Sigma) for 90 minutes at 4°C. Beads were washed twice with 50mM Tris-HCl, 150mM NaCl, 0.5mM EDTA, 10% glycerol, 1mM DTT, 1mM PMSF, pH 8.0 and analyzed by SDS-PAGE for normalization of bound protein.
Protein crystallization and structure determination
Pellino proteins in 15mM Hepes, 50mM NaCl, 5mM TCEP, pH 7.0 were crystallized by the hanging-drop vapor diffusion method. Equal volumes of P2(7–289) at 10mg/ml were mixed with a reservoir solution of 50mM sodium citrate, 23% PEG 3350, 1M NaCl, pH 5.5, and equilibrated over this reservoir solution at 22°C. Larger single crystals were obtained using micro-seeding. Crystals of SeMet P2(15–275)* grew spontaneously, at 22°C, from drops of equal volumes 5mg/ml protein and a reservoir solution of 100mM sodium acetate, 26.4% w/v PEG 2000 MME, 200mM (NH4)2SO4, pH 5.5. Both crystals were frozen directly from the drop. Data were collected at the Advanced Photon Source (APS) beamline 23ID and processed with HKL-2000 (Otwinowski et al., 1997).
The structure of SeMet P2(15–275)* was determine using MAD methods implemented in the program SHELX C/D/E (Schneider and Sheldrick, 2002; Sheldrick, 2002). The auto-trace algorithm in RESOLVE (Terwilliger, 2003) was used to obtain an initial model that comprised 70% of the mainchain with the correct sequence placed for one third of the model. This model was improved with cycles of manual building in COOT (Emsley and Cowtan, 2004) and refinement (against the peak dataset) in REFMAC (CCP4, 1994)(Table 1).
The structure of P2(7–289) was solved using MR methods, implemented in the program PHASER (McCoy et al., 2007), using the refined P2(15–275)* as the search model. The model of P2(7–289) was manually rebuilt in COOT and refined using REFMAC. Coordinates and structure factors for the P2(5–275)* and P2(7–289) have been deposited to the PDB with accession codes 3ega and 3egb respectively.
GST-pulldown assay
Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 100U/ml penicillin and 100μg/ml streptomycin. Transfections were performed using FuGENE 6 (Roche) according to the manufacturer’s instructions. Appropriately transfected HEK293T cells were lysed by scraping cell from the tissue culture dish in a 50mM HEPES, 150mM NaCl, pH 7.5 supplemented with 20mM β-glycerophosphate, 1mM EDTA, 1mM benzamidine, 50mM NaF, 1mM Na3VO4, 2mM DTT, 10% Protease Inhibitor Cocktail (Sigma), 0.1% NP-40 (HEK293T lysis buffer). For dephosphorylation, the lysis buffer was 50mM Tris, 100mM NaCl, 10mM MgCl2, 1mM DTT, 1mM PMSF, pH 8.0. Lysates were incubated with calf intestinal alkaline phosphatase (CIP, New England Biolabs; 0.15 units/ml) for six hours at 37°C.
Glutathione agarose beads (50μl), containing equal amounts of immobilized GST or GST-Pellino protein, were incubated overnight with the clarified HEK293T lysates. Beads were washed three times with HEK293T lysis buffer and re-suspended in SDS-PAGE loading buffer for Western blot analysis. Proteins were detected with monoclonal Xpress (Invitrogen) or a polyclonal phosphothreonine antibody (Zymed). Parallel gels were stained with Coomassie to verify normalization of the GST-fusion proteins on the beads.
Peptide binding assay
Peptides were synthesized using standard FMOC chemistry with free N-termini and purified by reverse-phase HPLC. The purity and molecular mass were confirmed by MALDI time-of-flight mass spectrometry. The following peptides were made. IRAK1-derived peptides: WVRDQpTELRL; SGQRpTASVLW; WSPGTpTAPRP; WISRGpTHNFSE; TVRGpTLAY. Rad9 derived peptides: WSLEVpTEADT [Rad9 (pT192)]; WSLEVTEADT [Rad9 (T192)] and YSLEVpTEADT. Lyophilized HPLC-purified peptides were resuspended in DMSO and amine coupled to Affi-Gel 15 agarose beads (Bio-Rad) according to the manufacturer’s protocol. Peptide-coupled beads were equilibrated with binding buffer (25mM HEPES, 300mM NaCl, 1mM DTT, pH 7.0). For each pull-down reaction, 100μl of Pellino2 or of R106A Pellino2 variant, at 50 μM, in binding buffer, were incubated with 20μl of peptide-coupled beads for 1 hour at 4°C. The beads were washed 3 times with binding buffer supplemented with 0.1% Triton X100. Bound protein was eluted by 40μl 3X Laemmli buffer, analyzed by SDS-PAGE, and detected with Coomassie staining.
Supplementary Material
Acknowledgments
We thank Mark Lemmon, Michael May, and members of the Ferguson laboratory for critical comments on the manuscript; Jeannine Mendrola invaluable assistance with cell culture; Bruce Lichtenstein and the Dutton laboratory for assistance with peptide synthesis and purification; and Thanuja Gangi Setty, Yenyi Ho and Eleni Argyropoulou for technical assistance. K.M.F. is supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, and is the Dennis and Marsha Dammerman Scholar of the Damon Runyon Cancer Research Foundation (DRS-52-06). C.-C.L. and K.R.S. were supported in part by the NIH through the UPenn Structural Biology Training Program (GM08275). K.R.S. is also supported in part by a Predoctoral Fellowship (BC051591) from the U.S. Army Breast Cancer Research Program. This work is based upon research conducted at the GM/CA CAT at the Advanced Photon Source (APS) that has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and National Institute of General Medical Science (Y1-GM-1104). Use of APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38.
References
- Alderwick LJ, Molle V, Kremer L, Cozzone AJ, Dafforn TR, Besra GS, Futterer K. Molecular structure of EmbR, a response element of Ser/Thr kinase signaling in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:2558–2663. doi: 10.1073/pnas.0507766103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Karimi-Busheri F, Mani RS, Galicia S, Koch CA, Cass CE, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Butler MP, Hanly JA, Moynagh PN. Pellino3 is a novel upstream regulator of p38 MAPK and activates CREB in a p38-dependent manner. J Biol Chem. 2005;280:27759–27768. doi: 10.1074/jbc.M500756200. [DOI] [PubMed] [Google Scholar]
- Butler MP, Hanly JA, Moynagh PN. Kinase-active interleukin-1 receptor-associated kinases promote polyubiquitination and degradation of the Pellino family: direct evidence for PELLINO proteins being ubiquitin-protein isopeptide ligases. J Biol Chem. 2007;282:29729–29737. doi: 10.1074/jbc.M704558200. [DOI] [PubMed] [Google Scholar]
- Byeon IJ, Li H, Song H, Gronenborn AM, Tsai MD. Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67. Nat Struct Mol Biol. 2005;12:987–993. doi: 10.1038/nsmb1008. [DOI] [PubMed] [Google Scholar]
- Byeon IJ, Yongkiettrakul S, Tsai MD. Solution structure of the yeast Rad53 FHA2 complexed with a phosphothreonine peptide pTXXL: comparison with the structures of FHA2-pYXL and FHA1-pTXXD complexes. J Mol Biol. 2001;314:577–588. doi: 10.1006/jmbi.2001.5141. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Choi KC, Lee YS, Lim S, Choi HK, Lee CH, Lee EK, Hong S, Kim IH, Kim SJ, Park SH. Smad6 negatively regulates interleukin 1-receptor-Toll-like receptor signaling through direct interaction with the adaptor Pellino-1. Nat Immunol. 2006;7:1057–1065. doi: 10.1038/ni1383. [DOI] [PubMed] [Google Scholar]
- Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-kappaB activation. Mol Cell Biol. 2008;28:3538–3547. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- Dunne A, O’Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE. 2003:re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- Durocher D. The FHA Domain. In: Cesareni G, Gimona M, Sudo M, Yaffe MB, editors. Modular Protein Domains. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2005. pp. 143–162. [Google Scholar]
- Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Grosshans J, Schnorrer F, Nusslein-Volhard C. Oligomerisation of Tube and Pelle leads to nuclear localisation of dorsal. Mech Dev. 1999;81:127–138. doi: 10.1016/s0925-4773(98)00236-6. [DOI] [PubMed] [Google Scholar]
- Holm L, Ouzounis C, Sander C, Tuparev G, Vriend G. A database of protein structure families with common folding motifs. Prot Sci. 1992;1:1691–1698. doi: 10.1002/pro.5560011217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Beyaert R. Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol Cell. 2003;11:293–302. doi: 10.1016/s1097-2765(03)00053-4. [DOI] [PubMed] [Google Scholar]
- Jensen LE, Whitehead AS. IRAK1b, a novel alternative splice variant of interleukin-1 receptor-associated kinase (IRAK), mediates interleukin-1 signaling and has prolonged stability. J Biol Chem. 2001;276:29037–29044. doi: 10.1074/jbc.M103815200. [DOI] [PubMed] [Google Scholar]
- Jensen LE, Whitehead AS. Pellino2 activates the mitogen activated protein kinase pathway. FEBS Lett. 2003a;545:199–202. doi: 10.1016/s0014-5793(03)00533-7. [DOI] [PubMed] [Google Scholar]
- Jensen LE, Whitehead AS. Pellino3, a novel member of the Pellino protein family, promotes activation of c-Jun and Elk-1 and may act as a scaffolding protein. J Immunol. 2003b;171:1500–1506. doi: 10.4049/jimmunol.171.3.1500. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Johnson HJ, Nie H, Qin J, Bird TA, Li X. Pellino 1 is required for interleukin-1 (IL-1)-mediated signaling through its interaction with the IL-1 receptor-associated kinase 4 (IRAK4)-IRAK-tumor necrosis factor receptor-associated factor 6 (TRAF6) complex. J Biol Chem. 2003;278:10952–10956. doi: 10.1074/jbc.M212112200. [DOI] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kang D, Chen J, Wong J, Fang G. The checkpoint protein Chfr is a ligase that ubiquitinates Plk1 and inhibits Cdc2 at the G2 to M transition. J Cell Biol. 2002;156:249–259. doi: 10.1083/jcb.200108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Kollewe C, Mackensen AC, Neumann D, Knop J, Cao P, Li S, Wesche H, Martin MU. Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J Biol Chem. 2004;279:5227–5236. doi: 10.1074/jbc.M309251200. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- Lee H, Yuan C, Hammet A, Mahajan A, Chen ES, Wu MR, Su MI, Heierhorst J, Tsai MD. Diphosphothreonine-specific interaction between an SQ/TQ cluster and an FHA domain in the Rad53-Dun1 kinase cascade. Mol Cell. 2008;30:767–778. doi: 10.1016/j.molcel.2008.05.013. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Smith GP, Walker JC. Kinase interaction domain of kinase-associated protein phosphatase, a phosphoprotein-binding domain. Proc Natl Acad Sci USA. 1999;96:7821–7826. doi: 10.1073/pnas.96.14.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell. 2002;9:1045–1054. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- Liao H, Yuan C, Su MI, Yongkiettrakul S, Qin D, Li H, Byeon IJ, Pei D, Tsai MD. Structure of the FHA1 domain of yeast Rad53 and identification of binding sites for both FHA1 and its target protein Rad9. J Mol Biol. 2000;304:941–951. doi: 10.1006/jmbi.2000.4291. [DOI] [PubMed] [Google Scholar]
- Liu Y, Dong W, Chen L, Xiang R, Xiao H, De G, Wang Z, Qi Y. BCL10 mediates lipopolysaccharide/toll-like receptor-4 signaling through interaction with Pellino2. J Biol Chem. 2004;279:37436–37444. doi: 10.1074/jbc.M400241200. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:7. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- Ordureau A, Smith H, Windheim M, Peggie M, Carrick E, Morrice N, Cohen P. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys(63)-linked polyubiquitination of IRAK1. Biochem J. 2008;409:43–52. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, Carter Charles W., Jr . Methods in Enzymol. Academic Press; 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Schauvliege R, Janssens S, Beyaert R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: a role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006;580:4697–4702. doi: 10.1016/j.febslet.2006.07.046. [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. Macromolecular phasing with SHELXE. Z Kristallogr. 2002;217:644–650. [Google Scholar]
- Strelow A, Kollewe C, Wesche H. Characterization of Pellino2, a substrate of IRAK1 and IRAK4. FEBS Lett. 2003;547:157–161. doi: 10.1016/s0014-5793(03)00697-5. [DOI] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- Takatsuna H, Kato H, Gohda J, Akiyama T, Moriya A, Okamoto Y, Yamagata Y, Otsuka M, Umezawa K, Semba K, et al. Identification of TIFA as an adapter protein that links tumor necrosis factor receptor-associated factor 6 (TRAF6) to interleukin-1 (IL-1) receptor-associated kinase-1 (IRAK-1) in IL-1 receptor signaling. J Biol Chem. 2003;278:12144–12150. doi: 10.1074/jbc.M300720200. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. 2003;D59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windheim M, Stafford M, Peggie M, Cohen P. Interleukin-1 (IL-1) induces the Lys63-linked polyubiquitination of IL-1 receptor-associated kinase 1 to facilitate NEMO binding and the activation of IkappaBalpha kinase. Mol Cell Biol. 2008;28:1783–1791. doi: 10.1128/MCB.02380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Qian W, Staschke K, Qian Y, Cui G, Deng L, Ehsani M, Wang X, Qian YW, Chen ZJ, et al. Pellino 3b Negatively Regulates Interleukin-1-induced TAK1-dependent NFκB Activation. J Biol Chem. 2008;283:14654–14664. doi: 10.1074/jbc.M706931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Smerdon SJ. The use of in vitro peptide-library screens in the analysis of phosphoserine/threonine-binding domain structure and function. Annu Rev Biophys Biomol Struct. 2004;33:225–244. doi: 10.1146/annurev.biophys.33.110502.133346. [DOI] [PubMed] [Google Scholar]
- Yamin TT, Miller DK. The interleukin-1 receptor-associated kinase is degraded by proteasomes following its phosphorylation. J Biol Chem. 1997;272:21540–21547. doi: 10.1074/jbc.272.34.21540. [DOI] [PubMed] [Google Scholar]
- Yu KY, Kwon HJ, Norman DA, Vig E, Goebl MG, Harrington MA. Cutting edge: mouse pellino-2 modulates IL-1 and lipopolysaccharide signaling. J Immunol. 2002;169:4075–4078. doi: 10.4049/jimmunol.169.8.4075. [DOI] [PubMed] [Google Scholar]
- Yuan C, Yongkiettrakul S, Byeon IJ, Zhou S, Tsai MD. Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53. J Mol Biol. 2001;314:563–575. doi: 10.1006/jmbi.2001.5140. [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.