Abstract
The mechanism of nitrogenase remains enigmatic, with a major unresolved issue concerning how inhibitors and substrates bind to the active site. We report a crystal structure of carbon monoxide (CO) inhibited nitrogenase MoFe-protein at 1.50 Å resolution, revealing a CO molecule bridging Fe2 and Fe6 of the FeMo-cofactor. The μ2 binding geometry is achieved by replacing a belt-sulfur atom (S2B) and highlights the generation of a reactive iron species uncovered by the displacement of sulfur. The CO inhibition is fully reversible as established by regain of enzyme activity and reappearance of S2B in the 1.43 Å resolution structure of the reactivated enzyme. The substantial and reversible reorganization of the FeMo-cofactor accompanying CO binding was unanticipated and provides insights into a catalytically competent state of nitrogenase.
Biological nitrogen fixation is Nature‘s pathway to convert atmospheric dinitrogen (N2) into a bioavailable form, ammonia (NH3). Nitrogenase, the only known enzyme capable of performing this multi-electron reduction, consists of two component metalloproteins, the Fe- (Av2) and MoFe- (Av1) protein (1–3). The Fe-protein, containing a [4Fe:4S]-cluster, mediates the adenosine triphosphate (ATP) dependent electron transfer to the MoFe-protein to support dinitrogen reduction (4). The MoFe-protein is an α2β2 heterotetramer with one catalytic unit per αβ heterodimer (5). To achieve the elaborate redox properties required for reducing the N-N triple bond, two metal centers are present in the MoFe-protein: the P-cluster and the FeMo-cofactor. The P-cluster, an [8Fe:7S] entity, is the initial acceptor for electrons, donated from the Fe-protein during complex formation between the two proteins (6–8). Electrons are subsequently transferred to the FeMo-cofactor, a [7Fe:9S:C:Mo]-R-homocitrate cluster that constitutes the active site for substrate reduction, and is the most complex metal center known in biological systems (5, 9–12).
Substrates and inhibitors bind only to forms of the MoFe-protein reduced by two to four electrons relative to the resting, "as-isolated" state, which can only be generated in the presence of reduced Fe-protein and ATP (1). Mechanistic studies must take into account the dynamic nature of the nitrogenase system, requiring multiple association and dissociation events between the two component proteins, as well as the ubiquitous presence of protons that are reduced to dihydrogen even in competition with other substrates (1, 13–15). The resulting distribution of intermediates under turnover conditions significantly complicates the structural and spectroscopic investigation of substrate interactions. Hence, even the fundamental question whether molybdenum or iron represents the site for substrate binding at the FeMo-cofactor is still under debate, and as a consequence, a variety of mechanistic pathways have been proposed based on either molybdenum or iron as the catalytic center mainly following Chatt-type chemistry (16).
Inhibitors are potentially powerful tools for the preparation of stably trapped transient states that could provide insight into the multi-electron reduction mechanism. In this regard, carbon monoxide (CO), a non-competitive inhibitor for all substrates except protons (17, 18), has a number of attractive properties; CO is isoelectronic to the physiological substrate, is a reversible inhibitor, and only binds to partially reduced MoFe-protein generated under turnover conditions. While non-competitive inhibitors are traditionally considered to bind at distinct sites from the substrate, for complex enzymes such as nitrogenase with multiple oxidation states and potential substrate binding modes, this distinction is not required (19). More recently, it has also been shown that CO is a substrate, albeit a very poor one, whose reduction includes concomitant C-C bond formation to generate C2 and longer-chain hydrocarbons, in a reaction reminiscent of the Fischer-Tropsch synthesis (20, 21). Therefore, CO binding as inhibitor/substrate must involve important active site properties common to the reduction of the natural substrate dinitrogen. For this reason, CO binding has been investigated by a variety of spectroscopic methods, most notably EPR and IR, and depending on the partial pressure, multiple CO-bound species have been observed; yet, a structurally explicit description of any CO binding site has been elusive (18, 22–27).
Building on these observations, we have determined a high-resolution crystal structure of a CO-bound state of the MoFe-protein from Azotobacter vinelandii. This necessitated overcoming several obstacles. First, the experimental setup for all protein handling steps, including crystallization, was deemed to require the continuous presence of CO. Second, because inhibition requires enzyme turnover, a prerequisite was the ability to obtain crystals of the MoFe-protein from activity assay mixtures, rather than from isolated protein (see supplementary material for assay details), conditions that are typically contradictory to crystallization requirements. Finally, rapid MoFe-protein crystallization (≤ 5 hrs) was crucial and was achieved based on previously developed protocols in combination with seeding strategies, while maintaining the presence of CO throughout this process (10).
Crystals of the inhibited MoFe-protein (Av1-CO) yielded structural data at 1.50 Å resolution, which allowed clear identification of a CO ligand (Figure 1A, C, S1A–D). The Av1-CO structure directly demonstrates the binding of one molecule CO per active site in a μ2-bridging mode between Fe2 and Fe6 that form one edge of the trigonal six-iron prism (Fe2-3-4-5-6-7) of the FeMo-cofactor (Fig. 1A, C, D). Remarkably, CO binding is accompanied by a displacement of one of the belt sulfur atoms (S2B) while retaining the essentially tetrahedral coordination spheres for Fe2 and Fe6. As a result, the two Fe are coordinated by two sulfur and two carbon atoms, a geometry, to our knowledge, not previously observed in metallo-clusters (although higher coordination number geometries have been observed in FeFe-hydrogenases (28)). Confirmation of the S2B displacement was provided by anomalous difference Fourier maps calculated with diffraction data measured at 7100 eV (Fig. 1B); this energy is just below the Fe K-edge so that the anomalous scattering from S is significantly enhanced relative to Fe. The carbon atom of CO is located at a distance of 1.86 Å from each of the irons (Fe2 and Fe6), compared to a previous distance of 2.2 Å for S2B (Figure 1C). The altered ligand environment results in a small adjustment of the FeMo-cofactor geometry, with the Fe2–Fe6 distance (2.5 Å) slightly shortened relative to the unchanged Fe4–Fe5 and Fe3–Fe7 distances (2.6 Å) (Figure 1C).
Figure 1. CO-inhibited MoFe-protein (Av1-CO).
Refined structure of the CO-bound FeMo-cofactor at a resolution of 1.50 Å. A) View along the Fe1-C-Mo direction. The electron density (2Fo-Fc) map is contoured at 4.0 σ and represented as blue mesh. The density at the former S2B site is significantly decreased and in excellent agreement with bound CO (see also C)). B) Same orientation as A) superimposed with the anomalous density map calculated at 7100 eV (green) at a resolution of 2.1 Å contoured at 4.0 σ showing the significant reduction of anomalous electron density at the CO site. C) Side view of FeMo-cofactor highlighting the μ2 binding geometry of CO. The electron density (2Fo-Fc) map (blue mesh) surrounding CO-Fe2-Fe6-C is contoured at 1.5 σ. D) Same orientation as C) highlighting the ligand environment of the metal center. The catalytically important side chain residues α-Val70 and α-His195 are in close proximity to the CO-binding site. Iron atoms are shown in orange, sulfur in yellow, molybdenum in turquoise, carbon in grey, nitrogen in blue and oxygen in red.
Given the complete displacement of S2B, we assessed whether the CO-inhibited protein could be reactivated or if it was irreversibly modified. Crystals of CO-inhibited MoFe-protein were active when dissolved in an assay mixture in the absence of CO. Furthermore, when CO-inhibited MoFe-protein from the original inhibition preparations was newly assayed after removal of CO (see supplementary material for assay details), a quantitative recovery (94 ± 4 %) of the initial activity was obtained (Table 1). The reactivated MoFe-protein was subsequently re-isolated from activity assay mixtures and crystallized, yielding a structure at 1.43 Å resolution. The structural data of the protein (Av1-reactivated) revealed that S2B is regained by replacing the previously bound CO-ligand, which results in the recovery of the resting state FeMo-cofactor. The full occupancy of the sulfur at the S2B-site in the reactivated enzyme is evident by inspecting the 2Fo-Fc electron density map as well as the anomalous electron density map verifying the anomalous scattering contribution expected for S2B (Figure 2A, B).
Table 1. Nitrogenase activity.
The acetylene reduction activity of “as-isolated” Av1, Av1-CO and Av1-reactivated was measured by the quantification of ethylene production. Nitrogenase activity is quantitatively recovered upon reactivation. Errors represent standard deviations of three measurements.
| Sample | Specific reduction activity [nmol(acetylene) min−1 mg(Av1)−1] |
Specific reduction activity [%] |
|---|---|---|
| Av1-as-isolated | 1930 ± 90 | 100 ± 5 |
| Av1-CO | < 2 ± 2 | < 0.1 ± 0.1 |
| Av1-reactivated | 1820 ± 80 | 94 ± 4 |
Figure 2. Reactivated MoFe-protein (Av1-reactivated).
Refined structure of the FeMo-cofactor at a resolution of 1.43 Å. A) View along the Fe1-C-Mo direction. The electron density (2Fo-Fc) map is contoured at 4.0 σ and represented as blue mesh. Electron density at the S2B site is in excellent agreement with a regained sulfur. B) Same orientation as A) superimposed with the anomalous density map (green) at a resolution of 2.15 Å contoured at 4.0 σ showing the presence of anomalous density at the S2B site. Color scheme is according to Figure 1.
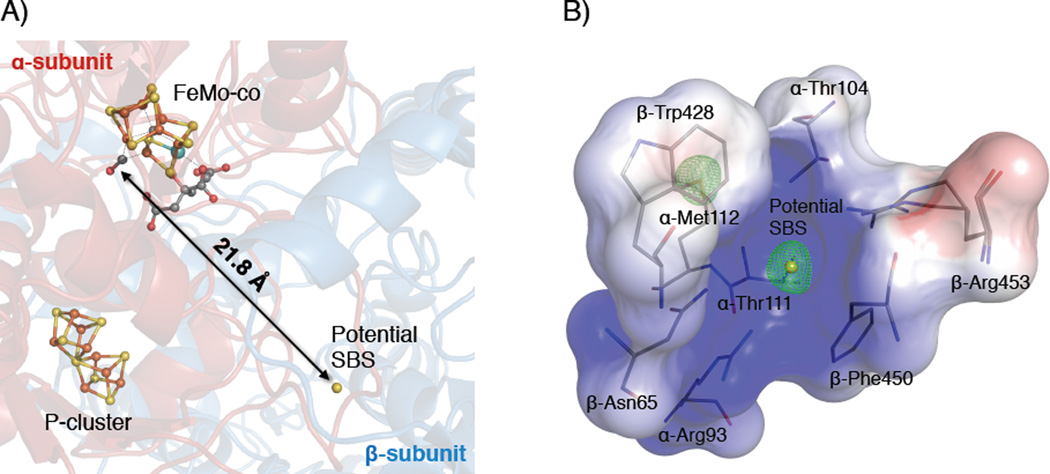
The finding that S2B can be reversibly replaced by CO raises the question of where this atom is located in the CO-inhibited state. If the S2B binding site is ordered, candidate locations should be evident by an inspection of the 7100 eV anomalous difference Fourier map. In this manner, one site per catalytic unit was identified with anomalous density compatible with sulfur. This potential sulfur binding site (SBS) is positioned ~22 Å away from the S2B position in the FeMo-cofactor and consists of a small protein pocket at the interface of the α- and β-subunits, formed by the side chains of residues α-Arg93, α-Thr104, α-Thr111, α-Met112, β-Asn65, β-Trp428, β-Phe450 and β-Arg453 (Figure 3A, B). The positive surface charge of the cavity is suited to accommodate an anionic species such as HS− or S2− (Figure 3B). In previous structures of the resting state enzyme (pdb-IDs: 1M1N and 3U7Q), this site has been assigned as water; intriguingly, the density at this site is also decreased in the structure of reactivated Av1 (Figure S2). The potential SBS is connected to the FeMo-cofactor binding pocket by a non-continuous water channel, and conformational rearrangements would be needed to accommodate the reversible migration of S2B from and to the active site. Although we have observed a correlation between density at the potential SBS and the CO-inhibited state of the MoFe-protein, the identity of this site as the displaced S2B cannot be achieved solely based on crystallographic data. In assessing the relevance of this site, it should be noted that the residues forming the pocket are poorly conserved, the site seems rather remote from the FeMo-cofactor, and since sulfur and chloride have similar anomalous scattering properties at 7100 eV, it is possible that this is a general anion-binding site in Av1.
Figure 3. Overview of the potential sulfur binding site (SBS) in the CO-inhibited MoFe-protein (Av1-CO).
A) Location of the potentially bound sulfur in a protein cavity on the interface between the α- and β-subunit of the α2β2 MoFe-protein. The potential SBS is located 22 Å away from its former position in the FeMo-cofactor (S2B-site). B) Close-up view on the binding cavity. Positive surface charge is represented in blue, negative surface charge in red. The anomalous density map at a resolution of 2.1 Å is represented as green mesh and contoured at 4.0 σ showing the presence of anomalous density at the potential SBS. The side chain sulfur of α-Met112 provides an internal standard for full occupancy. The color scheme is according to Figure 1.
The crystallographic characterization of CO-bound FeMo-cofactor of the MoFe-protein has important implications for the mechanism of substrate reduction by nitrogenase:
The CO binding site is close to the side chains of residues α-His195 (2.8 Å, NE2-OC distance) and α-Val70 (3.4 Å, closest methyl-OC distance). Modifications to both side chains were reported to significantly alter the catalytic properties of the enzyme. An α-His195 to α-Gln195 mutation resulted in the loss of N2 reduction activity while an α-Val70 to α-Ala/Gly70 alteration was reported to enable the reduction of longer carbon-chain substrates such as propyne and 1-butyne, respectively (29–33). In the structure presented here, α-His195 is in hydrogen bonding distance to the oxygen of CO while α-Val70 directly flanks the binding site (Figure 1D).
The displacement of S2B could be facilitated by a proton donation from α-His195 to yield either HS− or H2S, thereby generating a better leaving group than S2−. Although the dissociation of a sulfur may seem surprising, it opens up the ligand binding site, since the large radius of S2− effectively shields the cofactor Fe atoms in the resting state from substrate/inhibitor attack (2). The more general implication that binding of exogenous ligands can be accompanied by the reversible dissociation of at least one belt-sulfur from the metal sites of the FeMo-cofactor, changes the present view of the structural inertness of the [7Fe:9S:C:Mo]-R-homocitrate cluster towards ligand exchange. The relative lack of reactivity of the resting state is a striking property of the FeMo-cofactor, and the requirement for more highly reduced forms to bind substrate and inhibitors may reflect the need to dissociate sulfur ligands from Fe sites.
The displacement of the belt sulfur S2B by carbon monoxide causes the FeMo-cofactor scaffold to lose its intrinsic three-fold symmetry. Additionally, Fe1, the interstitial carbon and molybdenum are no longer aligned, creating an asymmetry in the resulting [7Fe:8S:C:Mo] cluster (Figure 1C). The modest adjustments of the remaining scaffold upon CO binding are suggestive of an important role for the interstitial carbon in stabilizing the cofactor during rearrangements and substitutions to the coordination environment of the irons (34, 35).
The experimental manipulations used to generate the CO-inhibited structure are distinct from those reported in previous spectroscopic studies; hence it is not possible to unambiguously assign the structure to one of the many annotated spectroscopic states. Interestingly, many of the previously identified states undergo dynamic interchanges including photoinduced transitions between states (25). Like the structure presented here, the spectroscopically identified “lo-CO” state has been proposed to involve one molecule of CO bound to the active site in a bridging mode (22, 26). A state with two CO bound to Fe2 and Fe6 could correspond to the “high-CO” form (22, 23) and might represent an intermediate relevant to the C-C coupling reaction.
The generation and successful stabilization of CO-bound MoFe protein under turnover conditions has culminated in a crystal structure that provides a detailed view of a ligand bound to the nitrogenase active site. The observations that CO is isoelectric to N2, is a potent yet reversible inhibitor of substrate reduction without impeding proton reduction to dihydrogen, and is bound in close proximity to previously determined catalytically important residues emphasizes the relevance of the CO-bound structure towards understanding dinitrogen binding and reduction. This sheds light on N2 activation based on a di-iron edge of the FeMo-cofactor and in this respect resembles the Haber-Bosch catalyst that also uses an iron surface to break the N-N triple bond. The demonstrated structural accessibility of CO-bound MoFe-protein opens the door for comparable studies on a variety of inhibitors and substrates, with the goal of understanding the detailed molecular mechanism of dinitrogen reduction by nitrogenase.
Supplementary Material
Acknowledgments
We thank J. Peters, L. Zhang, J. Rittle, C. Morrison, B. Wenke and K. Dörner for informative discussions. This work was supported by NIH grant GM45162 (D.C.R.), Deutsche Forschungsgemeinschaft grants EI-520/7 and RTG 1976 and the European Research Council N-ABLE project (O.E.). We gratefully acknowledge the Gordon and Betty Moore Foundation, the Beckman Institute, and the Sanofi–Aventis Bioengineering Research Program at Caltech for their generous support of the Molecular Observatory at Caltech, and the staff at Beamline 12–2, Stanford Synchrotron Radiation Lightsource (SSRL) for their assistance with data collection. SSRL is operated for the DOE and supported by its OBER and by the NIH, NIGMS (P41GM103393) and the NCRR (P41RR001209). We thank the Center for Environmental Microbial Interactions for their support of microbiology research at Caltech. Coordinates and structure factors have been deposited in the Protein Data Bank of the Research Collaboratory for Structural Bioinformatics, with IDs 4TKV (Av1-CO) and 4TKU (Av1-reactivated).
Footnotes
“This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
Supplementary Materials:
Materials and Methods
Figures S1&2
Tables S1&2
References 36–44
References and Notes
- 1.Burgess BK, Lowe DJ. Chem. Rev. 1996;96:2983–3011. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- 2.Howard JB, Rees DC. Proc. Natl. Acad. Sci. USA. 2006;103:17088–17093. doi: 10.1073/pnas.0603978103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seefeldt LC, Hoffman BM, Dean DR. Ann. Rev. Biochem. 2009;78:701–722. doi: 10.1146/annurev.biochem.78.070907.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgiadis MM, Komiya H, Chakrabarti P, Woo D, Kornuc JJ, Rees DC. Science. 1992;257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 5.Kim JS, Rees DC. Nature. 1992;360:553–560. doi: 10.1038/360553a0. [DOI] [PubMed] [Google Scholar]
- 6.Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Nature. 1997;387:370–376. doi: 10.1038/387370a0. [DOI] [PubMed] [Google Scholar]
- 7.Peters JW, Fisher K, Newton WE, Dean DR. J. Biol. Chem. 1995;270:27007–27013. doi: 10.1074/jbc.270.45.27007. [DOI] [PubMed] [Google Scholar]
- 8.Lowe DJ, Fisher K, Thorneley RNF. Biochem. J. 1993;292:93–98. doi: 10.1042/bj2920093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Einsle O, Tezcan FA, Andrade SLA, Schmid B, Yoshida M, Howard JB, Rees DC. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 10.Spatzal T, Aksoyoglu M, Zhang L, Andrade SLA, Schleicher E, Weber S, Rees DC, Einsle O. Science. 2011;334:940. doi: 10.1126/science.1214025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lancaster KM, Roemelt M, Ettenhuber P, Hu YL, Ribbe MW, Neese F, Bergmann U, DeBeer S. Science. 2011;334:974–977. doi: 10.1126/science.1206445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu YL, Ribbe MW. Coord. Chem. Rev. 2011;255:1218–1224. doi: 10.1016/j.ccr.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rees DC, Tezcan FA, Haynes CA, Walton MY, Andrade S, Einsle O, Howard JB. Philos. Trans. Roy. Soc. A. 2005;363:971–984. doi: 10.1098/rsta.2004.1539. [DOI] [PubMed] [Google Scholar]
- 14.Lowe DJ, Thorneley RNF. Biochem. J. 1984;224:895–901. doi: 10.1042/bj2240895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorneley RNF, Lowe DJ. Biochem. J. 1983;215:393–403. doi: 10.1042/bj2150393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatt J, Dilworth JR, Richards RL. Chem. Rev. 1978;78:589–625. [Google Scholar]
- 17.Hwang JC, Chen CH, Burris RH. Biochim. Biophys. Acta. 1973;292 doi: 10.1016/0005-2728(73)90270-3. [DOI] [PubMed] [Google Scholar]
- 18.Cameron LM, Hales BJ. Biochemistry. 1998;37:9449–9456. doi: 10.1021/bi972667c. [DOI] [PubMed] [Google Scholar]
- 19.Davis LC, Henzl MT, Burris RH, Orme-Johnson WH. Biochemistry. 1979;18:4860–4869. doi: 10.1021/bi00589a014. [DOI] [PubMed] [Google Scholar]
- 20.Lee CC, Hu YL, Ribbe MW. Science. 2010;329:642–642. doi: 10.1126/science.1191455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu YL, Lee CC, Ribbe MW. Science. 2011;333:753–755. doi: 10.1126/science.1206883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H-I, Cameron LM, Hales BJ, Hoffman BJ. J. Am. Chem. Soc. 1997;119:10121–10126. [Google Scholar]
- 23.Yan LF, Pelmenschikov V, Dapper CH, Scott AD, Newton WE, Cramer SP. Chemistry-a Eur. J. 2012;18:16349–16357. doi: 10.1002/chem.201202072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George SJ, Ashby GA, Wharton CW, Thorneley RNF. J. Am. Chem. Soc. 1997;119:6450–6451. [Google Scholar]
- 25.Maskos Z, Hales BJ. J. Inorg. Biochem. 2003;93:11–17. doi: 10.1016/s0162-0134(02)00480-4. [DOI] [PubMed] [Google Scholar]
- 26.Pollock RC, Lee HI, Camereon LM, DeRose VJ, Hales BJ, Orme-Johnson WH, Hoffman BM. J. Am. Chem. Soc. 1995;117:8686–8687. [Google Scholar]
- 27.Davis LC, Shah VK, Brill WJ, Orme-Johnson WH. Biochim. Biophys. Acta. 1972;256:511–523. doi: 10.1016/0005-2728(72)90079-5. [DOI] [PubMed] [Google Scholar]
- 28.Mulder DW, Shepard EM, Meuser JE, Joshi N, King PW, Posewitz MC, Broderick JB, Peters JW. Structure. 2011;19:1038–1052. doi: 10.1016/j.str.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Benton PMC, Laryukhin M, Mayer SM, Hoffman BM, Dean DR, Seefeldt LC. Biochemistry. 2003;42:9102–9109. doi: 10.1021/bi034595x. [DOI] [PubMed] [Google Scholar]
- 30.Kim CH, Newton WE, Dean DR. Biochemistry. 1995;34:2798–2808. doi: 10.1021/bi00009a008. [DOI] [PubMed] [Google Scholar]
- 31.Dos Santos PC, Mayer SM, Barney BM, Seefeldt LC, Dean DR. J. Inorg. Biochem. 2007;101:1642–1648. doi: 10.1016/j.jinorgbio.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott DJ, May HD, Newton WE, Brigle KE, Dean DR. Nature. 1990;343:188–190. doi: 10.1038/343188a0. [DOI] [PubMed] [Google Scholar]
- 33.Christiansen J, Cash VL, Seefeldt LC, Dean DR. J. Biol. Chem. 2000;275:11459–11464. doi: 10.1074/jbc.275.15.11459. [DOI] [PubMed] [Google Scholar]
- 34.Rittle J, Peters JC. Proc. Natl. Acad. Sci. USA. 2013;110:15898–15903. doi: 10.1073/pnas.1310153110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dance I. Chem. Asian. J. 2007;2:936–946. doi: 10.1002/asia.200700131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.