Abstract
Ubiquilin1 (UBQLN1) is a ubiquitin-like domain and a ubiquitin-associated domain containing protein that has been reported to be involved in shuttling proteins to the proteasome, especially during endoplasmic reticulum-associated protein degradation (ERAD). Thus, UBQLN1 function has been shown to be critical for combating a number of neurological disorders caused by protein aggregation, such as Amyotrophic lateral sclerosis (ALS), Alzheimer’s disease and Huntington’s disease. A role for UBQLN1 in regulating processes involved in tumorigenesis has not been demonstrated. Herein, we show that loss of UBQLN1 causes increased cell migration and invasion, actin cytoskeleton reorganization and induction of epithelial mesenchymal transition (EMT). Loss of UBQLN1 results in a significant decrease in the expression of epithelial markers including E-cadherin and claudin1, whereas expression of mesenchymal markers including Vimentin, Snail and ZEB1 are significantly elevated. Interestingly, we found that ZEB1 is required for induction of mesenchymal-like properties following loss of UBQLN1 and ZEB1 is capable of repressing expression of UBQLN1, suggesting a physiological, reciprocal regulation of EMT by UBQLN1 and ZEB1. Further, we find evidence for a role for UBQLN2 in also regulating EMT and cell migration. These observations have potential clinical relevance because the UBQLN1 gene is lost and under-expressed in a large percentage of human cancer cell lines and primary human lung cancer samples and recurrent mutations in both all five Ubiquilin family members have been identified in human lung cancers. Taken together, our results suggest for the first time a role for Ubiquilin family members in cancer biology.
Keywords: UBQLN1, ERAD, EMT, ZEB1, metastasis
Introduction
Ubiquilin1 (UBQLN1) belongs to a family of proteins that have been suggested to be involved in endoplasmic reticulum-associated protein degradation (ERAD) (1, 2). The UBQLN family consists of five closely related members (UBQLN1, UBQLN2, UBQLN3, UBQLN4, and UBQLNL) that have a high degree of similarity at the level of both amino acid and domain structure. This family of proteins is characterized by the presence of an amino-terminal ubiquitin-like (UBL) domain, a carboxy-terminal ubiquitin-associated (UBA) domain and a series of four chaperonin-like domains within the central portion of the protein. Disruption of UBQLN function has been linked to a variety of human neurodegenerative disorders, including ALS, Alzheimer’s disease and Huntington’s disease (3–6). Previous work from our lab, using a variety of approaches, including immunoaffinity and mass spectrometry identified Ubiquilin1 (UBQLN1) as a regulator of the ubiquitination, stability and the sub-cellular localization of BCL2L10/BCLb (7). In this work we also suggested that UBQLN1 mRNA levels vary widely among primary human lung adenocarcinoma samples (7). Further, work from Chen et. al. reported UBQLN1 as an autoantibody antigen in the serum of patients with lung adenocarcinoma (8). These are the only reports linking UBQLN1 with a potential role in human cancer, however the cellular processes regulated by UBQLN1 during tumorigenesis have never been examined.
Epithelial-to-mesenchymal transition (EMT) is a process during which epithelial cells lose their polarity, cell-to-cell contact and acquire more migratory, and invasive properties (9, 10). Several transcription factors are known to be involved in the regulation of EMT including Snail, Slug, Twist, and ZEB1/2 which simultaneously induce the expression of genes required for mesenchymal phenotype and repress the expression of genes required for maintaining epithelial phenotype such as E-cadherin (11). E-cadherin is responsible for maintaining the cell polarity and loss of E-cadherin is a hallmark of EMT (12). Several oncogenic pathways including TGFß, integrin-linked kinase (ILK), Wnt, and hypoxia-inducing factor-2α (HIF-2α) are involved in cancer progression and have been shown as inducers of EMT (13).
UBQLN1 and its interacting partner Valosin-containing protein (VCP/p97) have been suggested to regulate ER stress, by potentiating ERAD (14). However, in the present study, we found that loss of VCP but not UBQLN1 resulted in ER stress associated unfolded protein response (UPR). Furthermore, cells expressing a TCR-yfp fusion protein, which is well-studied substrate of ERAD, were found to significantly accumulate TCRyfp following loss of VCP, however there was absolutely no change after loss of UBQLN1, indicating that the loss of UBQLN1 does not alter ERAD. In an effort to understand what cellular processes UBQLN1 may be altering, we performed microarray analysis after siRNA-mediated loss of UBQLN1. The most striking signatures that were the result of UBQLN1 loss were processes, such as cell migration, that suggested a role of UBQLN1 in tumorigenesis. Validation experiments clearly demonstrate that loss of UBQLN1 induces cellular migration, invasion and epithelial-to-mesenchymal transition. We suggest that UBQLN1 plays a key and important role in the regulation of EMT in non-small cell lung cancer.
Results
Loss of UBQLN1 does not cause ER stress or alter ERAD
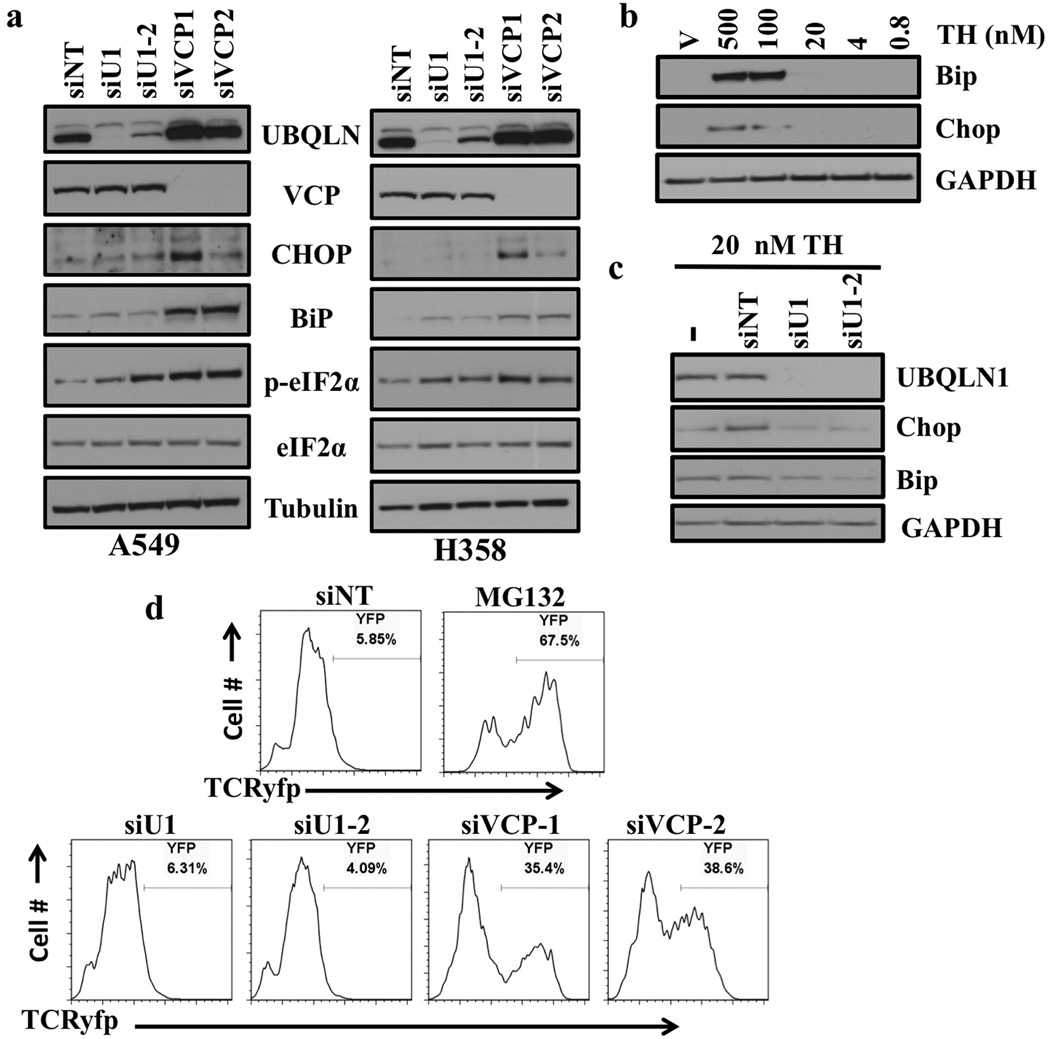
Previously, UBQLN1 and its partner Valosin containing protein (VCP/p97) have been reported as regulators of ER stress and ER-associated protein degradation (ERAD) (15, 1). We therefore wanted to determine if loss of UBQLN1 would induce ER stress and cell death in human lung adenocarcinoma cells. Our data demonstrated that loss of VCP but not UBQLN1 caused ER stress and altered ERAD in non-small cell lung cancer cell lines (A549 and H358) (fig. 1). Western blot analysis of the proteins involved in ER stress including Chop and Bip showed a significant increase in expression following siRNA-mediated knockdown of VCP but not knockdown of UBQLN1 (fig. 1a). In addition, we observed a dramatic increase in phosphorylation of eukaryotic initiation factor (eIF2-α) which has been shown to be phosphorylated following ER stress which ultimately leads to subsequent inhibition of protein translation (16) (fig. 1a).
Figure 1. Loss of UBQLN1 does not induce ER stress or alter ERAD.
(a) Western blot analysis of proteins involved in unfolded protein response and ER stress response. Cells were transfected with either non targeting (siNT) siRNA or two different siRNAs targeting UBQLN1 (siU1 and siU2) and two different siRNAs targeting VCP (siVCP1 and siVCP2). After 72 hrs of transfection, cells were harvested and analyzed for protein expression. (b) Western Blot analysis of proteins involved in ER stress response. A549 cells were treated with vehicle or with indicated concentrations of Thapsigargin (TH). (c) Western Blot analysis of proteins involved in ER stress response. A549 cells were transfected with either non targeting (siNT) siRNA or two different siRNAs targeting UBQLN1 (siU1 and siU2). 48 hrs post siRNA transfections cells were treated with vehicle or with 20nM of Thapsigargin (TH) for 30 min and harvested 24 hrs later. (d) UBQLN1 loss does not alter ERAD. 293T cells stably expressing TCRyfp, a well-known ERAD substrate, were untreated (NT), treated with MG132, or transfected with the indicated siRNA. Cells were subjected to flow cytometry 24 hrs after MG132 treatment or 72 hrs post siRNA transfection to determine the intensity of yfp signal. MG132 treatment is used as a positive control, as ERAD is known to rely on the proteasome for destruction of substrates.
We posited that loss of UBQLN1 may not be sufficient to cause robust ER stress, but perhaps if we combined loss of UBQLN1 with a known ER stressor, such as thapsigargin (TH) we may observe the reported role of UBQLN1 in maintaining ER homeostasis. In the present study, to determine the maximum concentration of TH that did not induce ER stress, we treated A549 cells with different concentrations of TH. We then combined that concentration of TH (20 nM) with loss of UBQLN1 to determine if loss of UBQLN1 and minimal levels of ER stress would manifest as detectable activation of UPR. Interestingly, we observed 20 nM of TH is insufficient to cause ER stress (fig. 1b) and loss of UBQLN1 combined with 20 nM of TH was still not sufficient to induce ER stress (fig. 1c).
To examine if UBQLN1 is able to alter the process of ERAD we used 293T cells expressing T-cell receptor alpha fused to yellow fluorescent protein (TCRyfp), a well-known ERAD substrate that has been extensively studied in the context of ERAD associated pathways (17, 18). TCRyfp cells were untreated (NT), treated with MG132, or transfected with the siRNAs targeting UBQLN1 (siU1 and siU1-2) or VCP (siVCP1 and siVCP1-2). Cells were subjected to flow cytometry 24 hrs after MG132 treatment or 72 hrs post siRNA transfection to determine the intensity of yfp signal. As expected, cells treated with MG132 showed an accumulation of TCRyfp, as ERAD is known to rely on the proteasome for destruction of substrates. Loss of VCP also caused a dramatic accumulation of TCRyfp, consistent with its reported role in potentiating ERAD. However, in contrast to previous reports loss of UBQLN1 did not alter ERAD, as demonstrated by no increase in the TCRyfp signal (fig. 1d and S1). Furthermore, over-expression of UBQLN1 or a truncation mutant of UBQLN1 lacking the UBA-domain did not decrease the TCRyfp signal in these cells (fig. S1). These studies indicate that UBQLN1 does not directly suppress ER stress via potentiating ERAD in these models.
UBQLN1 loss induces cell migration and invasion
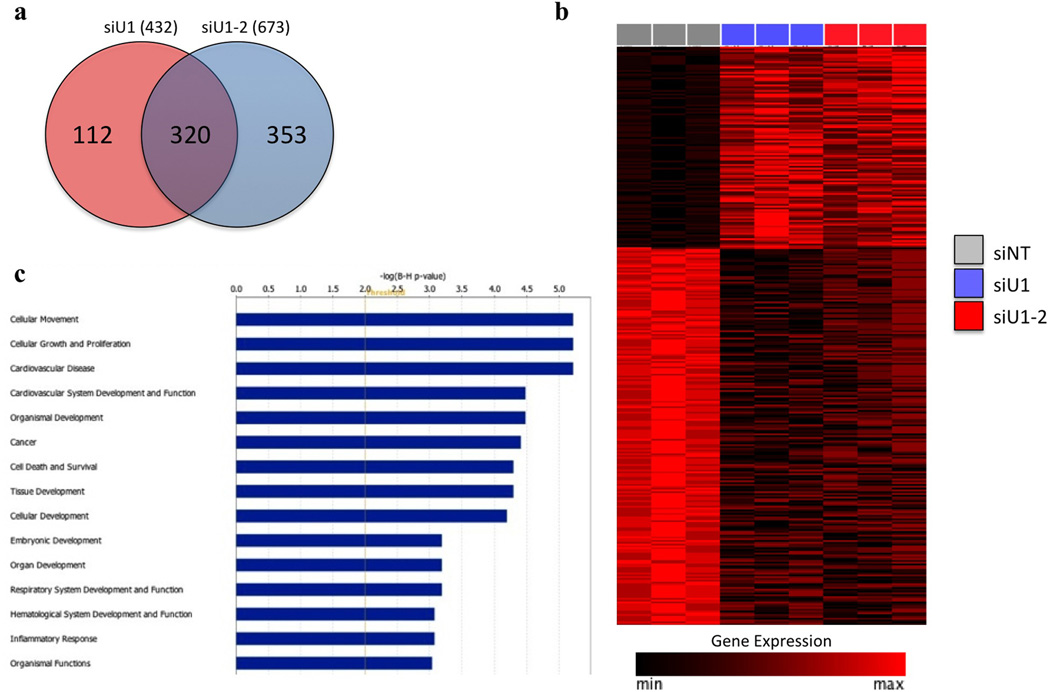
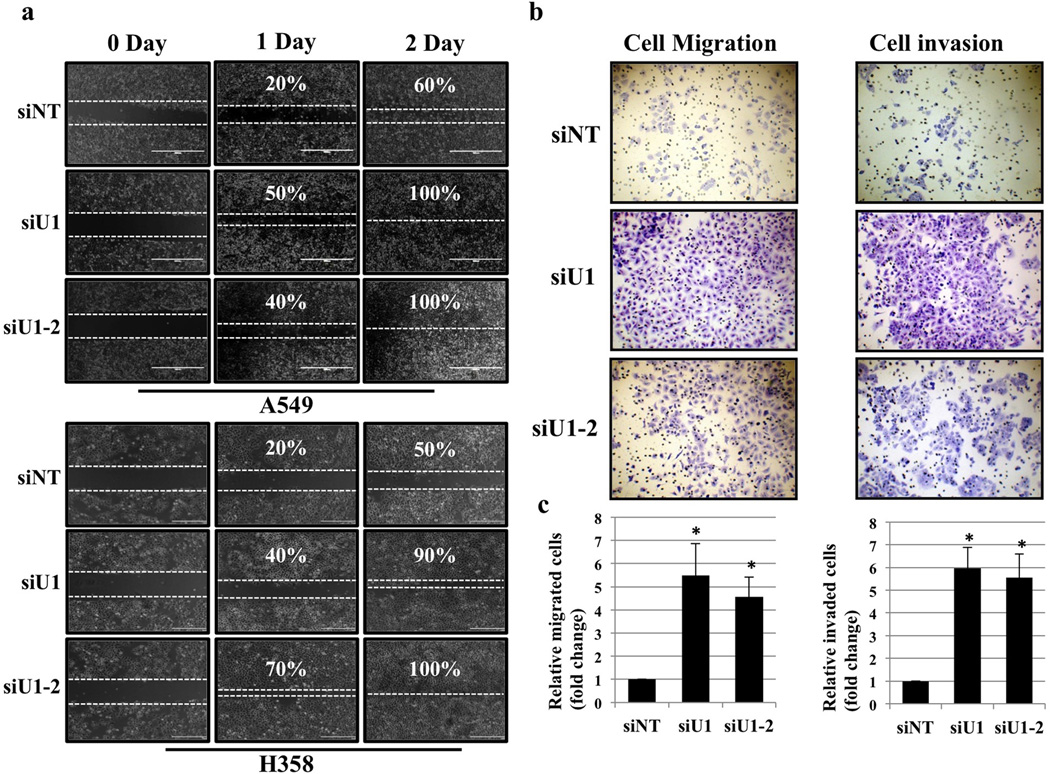
We did not observe activation of ER stress or alterations in ERAD following loss of UBQLN1. In addition, potential cellular processes regulated by UBQLN1 in cancer cells are completely unknown. Therefore, we performed gene expression analysis of cells after siRNA-mediated reduction of UBQLN1 to gain insight into the potential processes regulated by UBLQN1 (fig. 2a and 2b). Interestingly, pathway analysis of the UBQLN1 regulated gene signature demonstrated a significant enrichment for genes involved in cell movement, cell growth and proliferation and cancer. (fig. 2c, Table S1 and S2). As cell movement was the most significantly affected cellular function, we performed wound healing in vitro assays to determine whether loss of UBQLN1 increases cell migration, A549 and H358 cells were transfected with non-targeting siRNA (siNT) or with siRNAs targeting UBQLN1 (siU1 and siU1-2). After 24hrs of transfection, a “wound” was formed and cells were examined after 24 hrs and 48hrs post wound formation. Loss of UBQLN1 through U1 and U1-2 siRNAs showed nearly complete healing of the wound after 48 hrs compared with cells transfected with non-targeting (siNT) siRNA (fig. 3a), which had comparatively fewer cells migrated into the gap suggesting that UBQLN1 expression suppresses tumor cell migration. To determine if UBQLN1 is also capable of inhibiting invasiveness, we performed ‘Boyden chamber’ cell migration and invasion assay using A549 cells. Interestingly, cells transfected with UBQLN1 siRNAs (siU1 and siU1-2) acquired more migratory and invasive phenotype as determined by the number of cells that invaded through matrigel compared with cells transfected with non-targeting siRNA (siNT), further confirming that loss of UBQLN1 resulted in increased cell migration and invasion (fig. 3b and 3c).
Figure 2. Inhibition of UBQLN1 in A549 cells induces a gene expression signature related to EMT.
(a) Genes differentially expressed upon UBQLN1 knockdown in A549 cells. Comparison of gene expression profiles between A549 cells transfected with either siU1 or siU1-2 and siNT identified a set of 320 unique overlapping genes differentially expressed upon UBQLN1 knockdown (FDR<0.05, fold-change > or < 2). (b) Expression levels of the 320 UBQLN1 regulated genes in siU1, siU2 and siNT treated A549 cells. Genes are arranged in descending order of the average fold change of siU1 and siU1-2 treated cells vs siNT. Expression values are displayed scaled to the maximum expression for each individual gene across the samples. (c) Cellular functions and disease processes enriched within the UBQLN1 gene signature. IPA analysis of the 320 UBQLN1 regulated genes identified cellular movement as the most significantly enriched function. The top 15 diseases and disorders, molecular and cellular functions, and physiological development and function processes are indicated (B-H corrected p-value <0.01).
Figure 3. UBQLN1 loss induces cell migration and invasion.
(a) Migration assay in A549 and H358 cells. Cells were transfected with either non targeting (siNT) siRNA or two different siRNAs targeting UBQLN1 (siU1 and siU1-2). 24 hrs post siRNA transfections a pipette tip was used to scratch the dish to make a “wound”. Cells were examined after wound has been formed and successively for 24hr and 48hr post wound formation and photographed. (b) Invasion assay in A549 cells. Cells were transfected with either non targeting (siNT) siRNA or two different siRNAs targeting UBQLN1 (siU1 and siU1-2). 24 hours after siRNA transfection cells were seeded into Boyden chambers without (left) or with (right) matrigel. The lower chamber contained media with serum, whereas the upper chamber containing the cells was without serum. 48 hrs later cells on the underside of the membrane were fixed and stained. (c) Quantification of relative number of cells migrated or invaded through matrigel (*P < 0.05).
Loss of UBQLN1 induces EMT
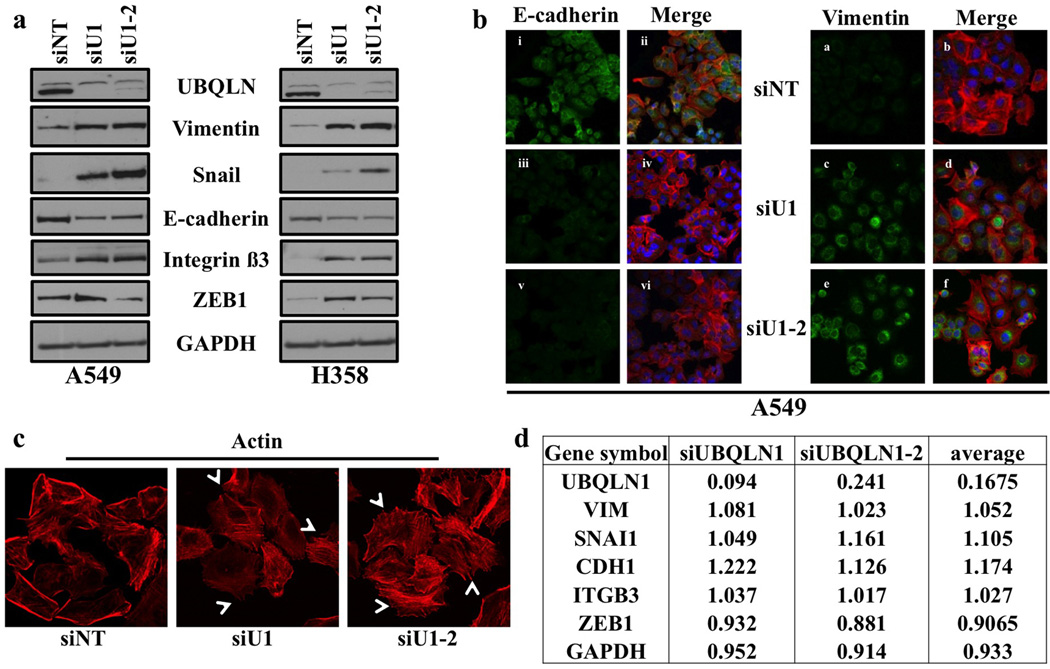
Increased cell migration and invasion is often associated with epithelial-to-mesenchymal transition (EMT) (19). We wondered if the increased migration and invasion observed following loss of UBQLN1 was concomitant with the acquisition of an EMT-like state. EMT has been shown to be controlled by several transcription factors including Twist, Snail, Slug along with ZEB1 family members ZEB1 and ZEB2 (11). To determine whether reduced UBQLN1 expression in non-small cell lung cancer cells induces EMT by changing the expression levels of key EMT markers and regulators, we first performed western blot analysis for E-cadherin, Vimentin, Snail, ZEB1 and Integrin beta3 following knockdown of UBQLN1 with two different siRNAs (siU1 and siU1-2). Consistent with increased EMT, we found that A549 and H358 cells showed decreased expression of E-cadherin, whereas the expression levels of Snail, Vimentin and ZEB1 proteins were significantly increased (fig. 4a). Moreover, we found increased expression of integrin β3 which has been previously reported to be involved in EMT (20), indicating a role for UBQLN1 in regulating EMT (fig. 4a). These results were further confirmed by performing immunofluorescence staining for E-cadherin and Vimentin following knockdown of either UBQLN1, using two siRNAs targeting UBQLN1 (siU1 and siU1-2), or with non-targeting (siNT) siRNA. We found significantly decreased expression of E-cadherin (fig. 4b, iii and v) after loss of UBQLN1 (siU1 and siU1-2) compared to non-targeting (siNT) siRNA (fig 4b, i). The loss of E-cadherin was concurrent with an increased expression of Vimentin in cells depleted for UBQLN1 (siU1 and siU1-2) (fig. 4b, c and e) compared to non-targeting (siNT) siRNA (fig. 4b, a). Similar results were obtained when we performed immunofluorescence on H358 cells following loss of UBQLN1 (fig. S2a). EMT induced cancer cells have been reported to show membrane extension and formation of cellular protrusions acquiring more invasive phenotype (21). Consistent with this findings, immunofluorescence analysis of A549 cells following loss of UNQLN1 (siU1 and siU1-2) revealed re-organization of actin cytoskeleton through destruction and cellular protrusion formation compared with non-targeting (siNT) siRNA supporting the role of UBQLN1 in regulating EMT (fig. 4c and S2b). We were curious to know whether the regulation of EMT-associated proteins following UBQLN1 loss was at the level of transcription or post-transcription. Examination of our microarray data did not suggest transcriptional alterations of any EMT-associated genes following UBQLN1 loss (fig. 4d). These data fit with the dogma for UBQLN1 function that loss of UBQLN1 likely alters the post-translational stability of proteins.
Figure 4. Loss of UBQLN1 induces EMT.
(a) UBQLN1 loss induces EMT in A549 and H358 cells. A549 and H358 cells were transfected with either with non-targeting siRNA (siNT) or siRNAs targeting UBQLN1 (siU1, siU1-2). After 72 hrs of transfection cells were harvested and analyzed for protein expression using the indicated antibodies. (b) Fluorescence staining for E-cadherin and Vimentin in A549. After 24 hrs of transfection either with non-targeting siRNA (siNT) or with siRNAs targeting UBQLN1 (siU1 and siU1-2) cells were trypsinized and plated on chamber slides and stained for EMT markers. i, iii and v: E-cadherin was detected using Alexa Fluor 488 goat anti-rabbit IgG (green). ii, iv and vi: overlay of respective E-cadherin and F-actin (Alexa Fluor 568 Phalloidin; red) staining with DAPI counter stain. a, c and e: Vimentin was detected using Alexa Fluor 488 goat anti-rabbit IgG (green). b, d and f: overlay of respective E-Vimentin and F-actin (Alexa Fluor 568 Phalloidin; red) staining with DAPI counter stain. (c) Cells were prepared as described in B and F-actin was detected with Alexa Fluor 568 Phalloidin (red). Re-organization of actin cytoskeleton through destruction and cellular protrusion formation is indicated by arrows. (d) Table indicating the fold change of mRNA following siRNA mediated knockdown of UBQLN1 for EMT-associated genes, as compared to non-targeting siRNA transfected cells. Values are in fold change and each value is the average of the triplicate samples for each siRNA.
UBQLN1 and ZEB1 coordinately regulate EMT
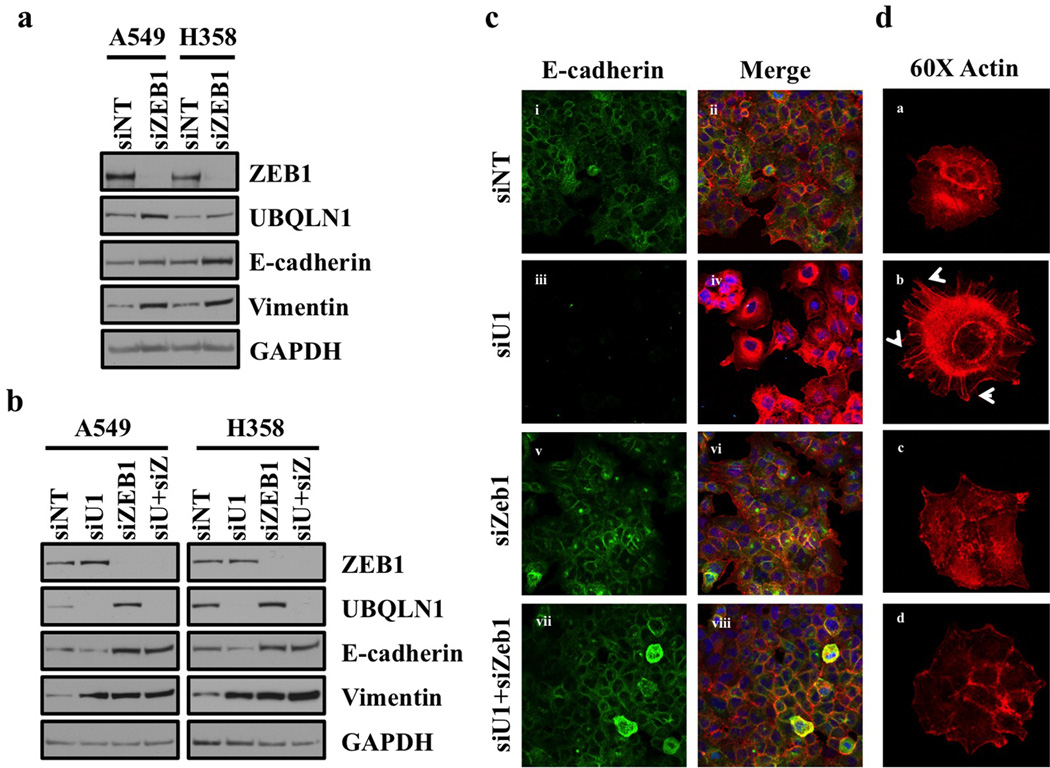
ZEB proteins function as transcriptional repressors and ZEB1 has been shown to be direct suppressor of E-cadherin during EMT (22). As we observed increased expression of ZEB1 following loss of UBQLN1 during EMT, we were interested to see if there was a reciprocal regulation, such that loss of ZEB1 alters expression of UBQLN1. We performed western blot analysis following knockdown of ZEB1 in both A549 and H358 cell lines and interestingly we observed increased expression of UBQLN1 following loss of ZEB1 (fig. 5a). We also observed increased expression of epithelial markers, whereas expression of mesenchymal markers was slightly altered in both A549 and H358 cell lines (fig. 5a). As we observed a significant increase in expression of UBQLN1 following loss of ZEB1 and vice versa, we were interested to see if there was a requirement of ZEB1 for EMT induced by the loss of UBQLN1. We performed western blot analysis following combined loss of UBQLN1 and ZEB1 and observed an increase in the expression of epithelial markers following loss of ZEB1 alone or combined loss of UBQLN1 and ZEB1 (fig. 5b). Furthermore, the requirement for ZEB1 expression in the down-regulation of e-cadherin and alteration of actin cytoskeleton following UBQLN1 loss was demonstrated using immunofluorescence staining (fig. 5c, 5d and fig. S3). This finding indicates that ZEB1 expression is required for induction of EMT following loss of UBQLN1 in non-small cell lung cancer cells.
Figure 5. Coordinate regulation of EMT by UBQLN1 and ZEB1.
(a) Loss of Zeb1 increases expression of UBQLN1 and increases epithelial markers in A549 and H358 cells. Western blot analysis of ZEB1, UBQLN1 and EMT markers in A549 and H358 cells. Cells were transfected with either non-targeting siRNA (siNT) or with siRNA targeting ZEB1 (siZEB1). After 72 hrs of transfection, cells were harvested and subjected to western blot for protein expression analysis for UBQLN1, ZEB1 along with other EMT markers (b) UBQLN1 loss requires ZEB1 to induce EMT in A549 and H358 cells. Cells were transfected with non-targeting siRNA, with siUBQLN1, siZEB1 or the combination of siUBQLN1 and siZEB1. After 72 hrs of transfection, cells were harvested. Western blot analysis confirming knockdown of UBQLN1 and ZEB1 along with different EMT markers. (c) Fluorescence staining for E-cadherin in A549. After 24 hrs of transfection either with non-targeting siRNA (siNT) or with siRNAs targeting UBQLN1 (siU1), siZeb1 or combination of siU1 and siZeb1, cells were trypsinized and plated on chamber slides and stained for E-cadherin. i, iii, v and vii: E-cadherin was detected using Alexa Fluor 488 goat anti-rabbit IgG (green). ii, iv, vi and viii: overlay of respective E-cadherin and F-actin (Alexa Fluor 568 Phalloidin; red) staining with DAPI counter stain. (d) A549 cells were prepared as described in (c) and F-actin was detected with Alexa Fluor 568 Phalloidin (red) with 60x objective. Re-organization of actin cytoskeleton through destruction and cellular protrusion formation is indicated by arrows.
UBQLN1 is lost, mutated and under-expressed in human cancers
As our data demonstrates a significant role for UBQLN1 in regulating the migration of lung cancer cells, we next aimed to determine if UBQLN1 is disrupted in cancer cell lines and cancer patient samples. Analysis of copy number status for 877 cancer cell lines from the Cancer Cell Line Encyclopedia found that 243 (27.7%) had loss or deletion of UBQLN1 (fig. 6a). Furthermore, cancer cell lines with loss or deletion of UBQLN1 demonstrated significantly lower expression (p<0.0001) than those without loss or deletion suggesting that copy number alteration is a mechanism of UBQLN1 inactivation in cancer cells. Likewise, examination of 230 lung adenocarcinoma tumors from The Cancer Genome Atlas revealed a high frequency of copy number loss (50.8%) with a corresponding significant decrease in gene expression levels (p<0.0001) (fig. 6b). Through examination of various available datasets, including COSMIC (Sanger Institute) and cBioPortal (MSKCC) we found multiple confirmed somatic mutations in UBQLN1 in human cancers. We wondered if mutations in UBLQN1 would lead to loss of UBQLN1 function. We cloned two nonsense mutations (UBQLN1G573X and UBQLN1G499X) and determined if they were stable proteins. Interestingly, these mutations were not detected following transfection into 293T cells, whereas the GFP marker, which is expressed from the same mRNA following an internal ribosomal entry sequence, was readily detected (Fig. 6c). Following treatment of the transfected cells with MG132, to inhibit the proteasome, we observed rapid stabilization of the mutant UBQLN1 proteins. This data demonstrates that there are UBQLN1 mutations found in human patients that are likely expressed at normal mRNA levels, however the protein is not stable and therefore not functional. This suggests that the number of human cancers with loss of UBQLN1 activity may be more common that even the mRNA or genomic analysis would indicate. Together, this data show that UBQLN1 is commonly disrupted in human tumor samples indicating a potential role in tumorigenesis.
Figure 6. UBQLN1 is lost and under-expressed in human cancers.
(a) UBQLN1 is frequently lost and underexpressed in cancer cell lines. Cancer cell lines are separated by UBQLN1 copy number status and their corresponding UBQLN1 expression plotted. Box-plots depict the median group expression (middle line), the 25th and 75th percentiles (bottom and top of box, respectively), with the whiskers indicating the 10th and 90th percentiles. Values for all samples outside this range are represented by dots. The UBQLN1 locus is lost in 243 of the 877 cell lines examined. UBQLN1 is significantly under-expressed in samples with UBQLN1 loss or deletion (p<0.0001, Mann-Whitney U-Test). (b) UBQLN1 is frequently lost and underexpressed in human lung adenocarcinomas. Samples are plotted as in A. The UBQLN1 locus is lost in 117 of 230 lung cancers examined. UBQLN1 is significantly underexpressed in tumors with loss or deletion (p<0.0001, Mann-Whitney U-Test). (c) Cancer relevant nonsense mutations in UBQLN1 are degraded in a proteasome dependent manner. Two nonsense mutations found in UBQLN1 in human cancers (UBQLN1G573X and UBQLN1G499X) were introduced into wild-type UBQLN1 (UBQLN1wt). Constructs expressing the indicated gene or an empty vector (V) were transfected into 293t cells and 36 hours post transfection they were treated with MG132 for the indicated times. GFP is expressed from the same mRNA as the UBQLN1 genes following and IRES. (d) Recurrent mutations observed in both UBQLN1 and UBQLN2 which have been identified in human lung cancers. (e) Loss of UBQLN2 induces EMT. UBQLN2 loss induces EMT in A549 and H358 cells. A549 and H358 cells were transfected with either with non-targeting siRNA (siNT) or siRNAs targeting UBQLN2 (siU2-5, siU2-6). After 72 hrs of transfection cells were harvested and analyzed for protein expression using the indicated antibodies. Arrow indicates the UBQLN2 band.
UBQLN2 also regulates EMT and is mutated in human cancer
We were curious to know whether UBQLN1 was unique in its ability to regulate EMT and cancer biology. By mining the available databases of sequenced human cancers, including COSMIC (Sanger Institute) and cBioPortal (MSKCC) we were able to identify recurrent non-synonymous mutations in all five Ubiquilin family members in human cancers (fig. 6d). For the most part mutations were not found in the same samples, thus we wondered if loss of other Ubiquiln family members, such as UBQLN2, could also drive EMT and cell migration. Knocking down UBQLN2 with two independent siRNA in both A549 and H358 cells demonstrated a marked increase in markers of EMT (fig. 6e). Furthermore, loss of UBQLN2 was capable of increasing migration in a wound healing assay, similar to loss of UBQLN1 (fig. S4a). As further support of a non-redundant role of Ubiquilin family members we performed microarray analysis on cells following siRNA-mediated loss of UBQLN1 or UBQLN2 and found an extremely similar pattern of transcriptional alterations (fig. S4b). Taken together these data strongly suggest that the entire Ubiquilin family will need to be interrogated in much more detail in order to understand exactly how this family may be affecting human tumorigenesis.
Discussion
Over the past decade (EMT) has been described as a biological process, which is involved in wound healing, organ fibrosis, cancer progression, drug resistance and metastasis of wide variety of cancers including lung cancer (23–25). Recently, UBQLN1 has been shown to play an important role in a wide variety of other cellular processes including ERAD (1, 2), but the role of UBQLN1 in EMT has not been demonstrated. Several studies have reported that cells undergoing EMT reveal a migratory/invasive phenotype (26, 27). Consistent with these findings, we observed that loss of UBQLN1 resulted in increased cell migration in multiple non-small cell lung cancer cell lines (fig. 3a). Furthermore, loss of UBQLN1 causes cells to acquire a more invasive phenotype (fig. 3b) indicating for the first time that loss of UBQLN1 in non-small cell lung cancer cells induces EMT, cell migration and invasion. Loss of expression of epithelial protein E-cadherin and gain of mesenchymal proteins including Vimentin are considered hallmarks of EMT (10). Interestingly, there are five Ubiquilin family members, which are all closely related. Reports suggest that family members are capable of interacting as hetero-dimers. We show that loss of the most similar family member UBQLN2 also causes EMT-like alterations in lung cancer cells (fig. 6e, S4). Consistent with these findings, immunofluorescence analysis of A549 and H358 cells for E-cadherin and Vimentin revealed a significantly higher level of protein expression for Vimentin in cells transfected with UBQLN1 or UBQLN2 siRNA compared with non-targeting siRNA and decreased protein expression of E-cadherin (fig. 4b, 6e and S2). During metastasis, cells lose their polarity, cell-to-cell adhesive properties and become more motile forming cellular protrusions (21). Immunofluorescence analysis of A549 and H358 cells following loss of UNQLN1 revealed re-organization of actin cytoskeleton through destruction and cellular protrusion formation compared with non-targeting siRNA indicating potential role of UBQLN1 in induction of EMT (fig. 4c and S2).
Furthermore, following loss of UBQLN1 or UBQLN2 we observed dramatic increase in expression of ZEB1, which has been shown to be a direct repressor of E-cadherin (fig. 4a and 6e) (28). Dohadwala et al. in their study using non-small cell lung cancer (NSCLC) reported ZEB1 and Snail in the cyclooxygenase-2-dependent down regulation of E-cadherin (29). Recently, high level expression of ZEB1 was correlated with liver metastasis and poor prognosis in colorectal cancer (30) and shown to be associated with metastasis and invasion in osteosarcoma (31). Consistent with these findings, in our model of non-small cell lung cancer, we observed increase expression of ZEB1 following loss of UBQLN1 and vice versa (fig. 5) supporting that an increase in expression of ZEB1 is important factor in driving EMT. Furthermore, we show that ZEB1 expression is critical for the induction of EMT following loss of UBQLN1 (fig. 5c)
In a recent study involving Caenorhabditis elegans, Ubiquilin1 and erasin were reported as ER stress-response genes. Furthermore they demonstrated that Ubiquilin1 and VCP bind erasin, forming a complex involved in ERAD (1). In order to tease apart the functional similarities and differences of UBQLN1 and its associated protein VCP we performed a variety of experiments. First we observed loss of VCP, but not UBQLN1, caused ER stress as revealed by western blot analysis of the different proteins including Bip and Chop involved in ER stress (fig. 1a). Interestingly, by combining Thapsigargin (TH), a known inducer of ER stress, (32) with loss of UBQLN1, we did not observe any increase in ER stress, confirming that loss of UBQLN1 does not appear to block ER stress in lung cancer cells (fig. 1b). Furthermore, using 293T cells expressing TCRypf a known substrate involved in ERAD, we observed the loss of VCP but not UBQLN1 altered ERAD (fig. 1d). These findings beg the question: does UBQLN1 regulate ER stress and ERAD in lung cancer cells? Our data herein suggest that modulating UBQLN1 expression, by either knocking down with siRNA or over-expressing the cDNA does not alter ER-stress response or degradation of an ERAD substrate. This finding is in conflict with previously published literature (1). The first potential explanation for these discrepancies is that our experiments were performed in lung cancer cell lines, whereas previous experiments were done in alternative cell types (1). As far as the stability of the ERAD substrate goes, it may be possible that there is cell type specificity for individual ERAD substrates and perhaps UBQLN1 expression does not regulate TCR destruction in 293T cells. The final possible explanation is that there is functional redundancy among Ubiquilin family members. As mentioned, there are five closely related Ubiquilin proteins and simply modulating UBQLN1 may not be sufficient to induce ER stress or alter ERAD. It is thought that Ubiquilin proteins can form hetero-dimers and we have shown that loss of either UBQLN1 or UBQLN2 can induce EMT, therefore we propose that Ubiquilin family members are non-redundant in controlling EMT in lung cancer cells. Whether or not they are redundant in controlling ER stress and ERAD remains to be studied.
Can expression of UBQLN1 serve as a potential predictive biomarker for therapeutic efficacy in patients of non-small cell lung cancer? In the present study, we show that UBQLN1 locus is lost and under-expressed in many human cancer cell lines and primary human lung cancer samples and all five of the Ubiqulin family member harbor recurrent non-synonymous mutations in human cancers (fig. 6a and b). Interestingly, our previous work demonstrated that patients with higher mRNA levels of UBQLN1 have a decreased survival rate compared to patients with a lower mRNA expression of UBQLN1. Given the present study this finding seems a bit counterintuitive since decreased UBQLN1 causes more migratory and invasive disease. However, since metastatic disease is often treated with aggressive cytotoxic chemotherapeutics it may be that cells with decreased UBQLN1 expression are actually more sensitive to these regimens,. This prediction will be the focus of our future endeavors, as the appropriate databases and patient information are currently not available to us. In addition, our previous work demonstrated a role for UBQLN1 in regulating the stability of the anti-apoptotic BCL2 family member, BCLb/BCL2L10. In limited attempts, we did not observe any alterations in BCLb following manipulation of UBQLN1 expression, suggesting that this regulation is not largely responsible for the phenotypes we observed in this study.
In conclusion, our results suggest for the first time that UBQLN1 plays important and novel role in the induction of EMT and potentially cancer progression (fig. 7). Future work will explore the pre-clinical potential of UBQLN1 as a predictive biomarker of patient response, as well as to determine the exact molecular mechanisms by which UBQLN1 expression is capable of suppressing EMT, migration and cancer progression.
Figure 7. Schematic presentation of regulation of EMT by UBQLN1.
See text for details.
Materials and methods
Antibodies used for study
E-cadherin #3195, Vimentin #5741, ZEB1 #3396, Claudin1 #4933, Snail #3879, β-catenin #9562, Bip #3183, CHOP #2895, eIF2α #9722 and p-eIF2α #9721 (Cell Signaling Technologies Inc. Danvers, MA 01923); Tubulin #B512 (Sigma); GAPDH #FL335 (Santa Cruz); Ubqln polyclonal was made by inoculating rabbits with a peptide specific to Ubqln1 (Yenzym Antibodies LLC); Alexa Fluor 488 goat anti-rabbit IgG #A11034 (Molecular Probes, Invitrogen detection technologies, Eugene, OR. USA); Alexa Fluor 568 Phalloidin #A12380 (Life technologies Eugene, OR. USA).
Cell culture, siRNA transfection and protein analysis
Human non-small cell lung carcinoma cell line A549 and H358 were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured in RPMI medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and 1% antibiotic/antimycotic (Sigma, St Louis, MO, USA). The cell lines were routinely subcultured every 3–4 days. All siRNA transfections were performed using Dharmafect1 #T-2001-03 (Thermo Fisher Scientific Inc, Pittsburgh, PA, USA) as per manufacturer’s protocol. After total 72 hrs of transfection cells were harvested in CHAPS lysis buffer (1% CHAPS detergent, 150mM NaCl, 50mM Tris pH 7, 5mM EDTA). Protein was quantitated by using Pierce's BCA Protein Assay Reagent Kit (# 23227) from Pierce Biotechnology, Rockford, IL, USA as per manufacturer’s protocol.
Immunofluorescence staining
Cells were cultured in 60mm culture plates and transfected with either non targeting siRNA (siNT) or with siRNAs targeting UBQLN1 (siU1 and siU1-2). After total 24 hrs of transfections, cells were trypsinized and plated in chamber slides (Millicell EZ Slide: Millipore, EMD Millipore Corporation, Billerica, MA, USA). After 72 hrs of transfection, cells were fixed with 4.0% paraformaldehyde in PBS for 30 min and then permeabilized with 0.2% Triton X-100 for 15 min at room temperature. Cells were rinsed twice with PBS, and then incubated for 1-2 hrs with anti-Vimentin antibody (at a dilution of 1:1000) or anti-E-cadherin (at a dilution of 1:1000). After successive washes, cells were then incubated with Alexa Fluor 488 goat anti-rabbit IgG (A11034: Molecular Probes, Invitrogen detection technologies, Eugene, OR. USA) at a dilution of 1:1000. After incubation with secondary antibody for 45 min, cells were rinsed with PBS and incubated with Alexa Fluor 568 Phalloidin (A12380: Life technologies Eugene, OR. USA) at the dilution of 1:1000 to stain actin filaments. After successive washing with washing buffer, nuclei were counterstained with DAPI (diluted 1:000) for 10 min at room temperature followed by three washes (5–10 min each) with washing buffer. The cells were then mounted with polymount antifade solution (Sigma) and observed under MRC 600 confocal laser scanning microscope (Bio-Rad).
FACS analysis
FACS analysis was performed by using FACScan Flow Cytometer from Cytek Development, Fremont, CA. USA. USA. Briefly, 293T cells expressing TCRyfp (a kind gift from Yihong Ye, NIDDK, NIH, Bethesda, USA), a well-known ERAD substrate, were untreated (NT), treated with MG132, or transfected with two different siRNAs targeting UBQLN1 (siU1 and siU1-2) and two different siRNAs targeting VCP1 (siVCP1 and siVCP1-2). After 48 hours of transfection cells were treated with MG132 for 24 hrs and after total 72 hrs post siRNA transfection, cells were subjected to flow cytometry to determine the intensity of yfp signal. Data were analyzed with FlowJo software.
Cell migration assay or Wound healing assay
Cells were plated on 6-well plates and after 24 hrs of siRNA transfections, wound were made using the pipette tip following replacement with fresh media. Cells were examined successively after 24 hrs and 48 hrs of wound formation and photographed.
Boyden Chamber cell migration and cell invasion assay
BD BioCoat™ Matrigel Invasion Chambers (#354480), with 8.0 µm PET Membrane 24-well Cell Culture Inserts packaged ready-to-use in BD Falcon™ Companion Plates (#40480) and Growth Factor Reduced BD Matrigel™ Matrix, a solubilized basement membrane (#354230) (BD Biosciences, San Jose, CA, USA) were used in Matrigel invasion assay. Assay was performed according to manufacturer’s protocol (for more detail see supplemental methods). Briefly, cells were allowed to grow on Transwells for 72 hrs and then the total number of cells migrated or invaded through matrigel were photographed, counted and relative number of cells following loss of UBQLN1 (siU1 and siU1-2) were calculated.
Drug treatments
Thapsigargin- A549 cells were either transfected with non-targeting siRNA (siNT) or with siRNA targeting UBQLN1 (siU1 and siU1-2). After 48 hrs of transfection, cells were either treated with vehicle or with indicated concentration of Thapsigargin (TH) for 30 min. After 30 min, TH containing media was removed and fresh complete media was added to the cells, then cells were harvested after 24 hrs.
siRNA sequences used for study
All siRNAs used for study were ordered from Thermo Fisher Scientific Biosciences Inc. Lafayette, CO 80026, USA.
| Non targeting siRNA | (siNT): UAAGGCUAUGAAGAGAUACAA |
| UBQLN1 siRNA | (siU1): GAAGAAAUCUCUAAACGUUUUUU |
| (siU1-2): GUACUACUGCGCCAAAUUU | |
| VCP siRNA | (siVCP1): GUAAUCUCUUCGAGGUAUA |
| (siVCP2): CAAAUUGGCUGGUGAGUCU | |
| ZEB1 pool siRNA | (siZEB1) Cat. No. L-006564-01 |
| UBQLN2 | (siU2-5): CCUGGUAUCUCUAAGUAUAUU |
| (siU2-6): GUAGAAUCUGAGUGUAAUAUU |
Gene expression microarray analyses
After 72 hrs of transfection of A549 cells either with non-targeting siRNA (siNT) or with siRNAs targeting UBQLN1 (siU1 and siU1-2), cells were harvested and total RNA was purified using TRI reagent (#9424) (Sigma-Aldrich, Inc. St. Louis, MO. USA), according to the manufacturer’s protocol. Three biological replicates were performed for each siRNA. Affymetrix (Santa Clara, CA) microarray and data analysis was performed using standard methods (for detailed description see supplemental methods). Microarray data has been deposited in GEO database, NCBI, NIH.
UBQLN1 copy number and expression status
Gene expression (V2 RSEM values for RNA-seq and expression z-scores for microarray data) and copy number (GISTIC alteration status) data for UBQLN1 were downloaded for 230 lung adenocarcinomas and 877 cancer cell lines from the MSKCC cBIO Cancer Genomics Portal (http://www.cbioportal.org/, accessed July 31, 2013). For each dataset, the expression of UBQLN1 was compared between samples with UBQLN1 copy number loss or deletion versus those without using the Mann-Whitney U-Test (two-tailed).
Statistical analysis
Differences between two groups were statistically analyzed using unpaired Student's t-test. Differences were considered significant when P<0.05.
Supplementary Material
Acknowledgments
We thank Lavona Casson and current members of the Beverly lab, for technical assistance and Dr. Mohammad T. Malik for valuable comments, discussion and technical advice.
The work was supported by the start-up funds from James Graham Brown Cancer Center, University of Louisville and Kosair Pediatric Cancer Program and Molecular Targets COBRE 8P20GM103482-10 from NIH, an award from the Lung Cancer Research Foundation to LJB, an award from the Rounsavall Foundation to LJB, an award from the Wendy Will Case Cancer Fund to LJB, R25-CA-134283 from the National Cancer Institute to SPS. Part of this work was performed with assistance of the UofL Genomics Facility, which is supported by NIH/NIGMS Phase III COBRE 1P30 GM106396-01, NIH/NIGMS KY-INBRE P20GM103436, the James Graham Brown Foundation, and user fees.
Footnotes
Conflict of Interest: the authors declare no conflict of interest
References
- 1.Lim PJ, Danner R, Liang J, Doong H, Harman C, Srinivasan D, et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol. 2009;187:201–217. doi: 10.1083/jcb.200903024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu A, Hiltunen M, Romano DM, Soininen H, Hyman BT, Bertram L, et al. Effects of ubiquilin 1 on the unfolded protein response. J Mol Neurosci. 2009;38:19–30. doi: 10.1007/s12031-008-9155-6. [DOI] [PubMed] [Google Scholar]
- 3.Kim H, Kim HY, Choi MR, Hwang S, Nam KH, Kim HC, et al. Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasometargeting factor, ubiquilin 1. J Biol Chem. 2009;284:8083–8092. doi: 10.1074/jbc.M808064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias-Vásquez A, de Lau L, Pardo L, Liu F, Feng BJ, Bertoli-Avella A, et al. Relationship of the Ubiquilin 1 gene with Alzheimer's and Parkinson's disease and cognitive function. Neurosci Lett. 2007;424:1–5. doi: 10.1016/j.neulet.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Olzmann JA, Li L, Chin LS. Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr Med Chem. 2008;15:47–60. doi: 10.2174/092986708783330692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford NJ, Lewis J, Clippinger AK, Thomas MA, Adamson J, Cruz PE, et al. Unbiased screen reveals ubiquilin-1 and-2 highly associated with huntingtin inclusions. Brain Res. 2013;1524:62–73. doi: 10.1016/j.brainres.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beverly LJ, Lockwood WW, Shah PP, Erdjument-Bromage H, Varmus H. Ubiquitination, localization, and stability of an anti-apoptotic BCL2-like protein, BCL2L10/BCLb, are regulated by Ubiquilin1. Proc Natl Acad Sci USA. 2012;109:E119–E126. doi: 10.1073/pnas.1119167109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G, Wang X, Yu J, et al. Autoantibody profiles reveal ubiquilin 1 as a humoral immune response target in lung adenocarcinoma. Cancer Res. 2007;67:3461–3467. doi: 10.1158/0008-5472.CAN-06-4475. [DOI] [PubMed] [Google Scholar]
- 9.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwatsuki M, Mimori K, Yokobori T, Ishi H, Beppu T, Nakamori S, et al. Epithelialmesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;101:293–299. doi: 10.1111/j.1349-7006.2009.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y. New Insights into Epithelial-Mesenchymal Transition in Kidney Fibrosis. J Am Soc Nephrol. 2010;21:212–222. doi: 10.1681/ASN.2008121226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 15.Wójcik C, Rowicka M, Kudlicki A, Nowis D, McConnell E, et al. Valosin-containing Protein (p97) Is a Regulator of Endoplasmic Reticulum Stress and of the Degradation of N-End Rule and Ubiquitin-Fusion Degradation Pathway Substrates in Mammalian Cells. Molecular Biology of the Cell. 2006;17:4606–4618. doi: 10.1091/mbc.E06-05-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bevilacqua E, Wang X, Majumder M, Gaccioli F, Yuan CL, Wang C, et al. eIF2-α phosphorylation tips the balance to apoptosis during osmotic stress. J Biol Chem. 2010;285:17098–17111. doi: 10.1074/jbc.M110.109439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Li L, Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Biol Chem. 2006;174:963–971. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soetandyo N, Wang Q, Ye Y, Li L. Role of intramembrane charged residues in the quality control of unassembled T-cell receptor alpha-chains at the endoplasmic reticulum. J Cell Sci. 2010;123:1031–1038. doi: 10.1242/jcs.059758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 20.Shah PP, Fong MY, Kakar SS. PTTG induces EMT through integrin αVβ3-focal adhesion kinase signaling in lung cancer cells. Oncogene. 2011;31:3124–3135. doi: 10.1038/onc.2011.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oyanagi J, Ogawa T, Sato H, Higashi S, Miyazaki K. Epithelial-mesenchymal transition stimulates human cancer cells to extend microtubule-based invasive protrusions and suppresses cell growth in collagen gel. PLoS One. 2012;7(12):e53209. doi: 10.1371/journal.pone.0053209. Epub 2012 Dec 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grooteclaes ML and Frisch SM. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene. 2000;19:3823–3828. doi: 10.1038/sj.onc.1203721. [DOI] [PubMed] [Google Scholar]
- 23.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial– mesenchymal transitions: the importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nurwidya F, Takahashi F, Murakami A, Takahashi K. Epithelial mesenchymal transition in drug resistance and metastasis of lung cancer. Cancer Res Treat. 2012;244:151–156. doi: 10.4143/crt.2012.44.3.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keshamouni VG, Michailidis G, Grasso CS, Anthwal S, Strahler JR, Walker A, et al. Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype . J Proteome Res. 2006;5:1143–1154. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- 27.Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66:8319–8326. doi: 10.1158/0008-5472.CAN-06-0410. [DOI] [PubMed] [Google Scholar]
- 28.Vandewalle C, Van Royb F, Berxa G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dohadwala M, Yang SC, Luo J, Sharma S, Batra RK, Huang M, et al. Cyclooxygenase-2- dependent regulation of E-cadherin: rostaglandin E(2) induces transcriptional repressors ZEB1 and snail in non-small cell lung cancer. Cancer Res. 2006;66:5338–5345. doi: 10.1158/0008-5472.CAN-05-3635. [DOI] [PubMed] [Google Scholar]
- 30.Zhang GJ, Zhou T, Tian HP, Liu ZL, Xia SS. High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncology Letters. 2013;5:564–568. doi: 10.3892/ol.2012.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen A, Zhang Y, Yang H, Xu R, Huang G. Overexpression of ZEB1 relates to metastasis and invasion in osteosarcoma. J Surg Oncol. 2012;105:830–834. doi: 10.1002/jso.23012. [DOI] [PubMed] [Google Scholar]
- 32.Kamiya T, Obara A, Hara H, Inagaki N, Adachi T. ER stress inducer, thapsigargin, decreases extracellular-superoxide dismutase through MEK/ERK signaling cascades in COS7 cells. Free Radical Research. 2011;45:692–698. doi: 10.3109/10715762.2011.567985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.