Abstract
Several linkage studies on anxiety have been carried out in samples ascertained through probands with panic disorder. The results indicated that using a broad anxiety phenotype instead of a DSM-IV anxiety disorder diagnosis might enhance the chance of finding a linkage signal. In the current study, a genome-wide linkage analysis was performed on anxiety measured with a self-report questionnaire whose scores are highly correlated with DSM-IV anxiety disorders. The self-report questionnaire was included in five surveys of a longitudinal study of the Netherlands Twin Register. Genotype and phenotype data were available for 1,602 twins and siblings. To estimate Identity By Descent (IBD), additional genotype data for 564 parents and 22 siblings were used. Linkage analyses were carried out using MERLIN-Regress on the average anxiety scores across time. A linkage signal (LOD-score 3.4, empirical p-value 0.07) was obtained at chromosome 14 for marker D14S65 at 105 cM (90% confidence interval 99 cM - 115 cM bounded by markers D14S1434 and D14S985). This finding replicates a linkage finding for a broad anxiety phenotype in a clinically based sample, indicating that the region might harbor a QTL associated with the whole spectrum of general anxiety, i.e. from the normal to the clinical range. Moreover, genome-wide linkage and association studies on emotionality in mice obtained significant results in a syntenic region on mouse chromosome 12. Two homolog genes lie in this region –Dlk1 (delta-like 1 homolog, Drosophila) and Rtl1 (retrotransposon-like 1). Future association studies of these genes are warranted.
Keywords: anxiety, genomewide linkage, family study, stai, genetics
Introduction
Anxiety disorders are common, often with a chronic and disabling course, and cause significant mental health care problems. In epidemiological samples in the Netherlands and in the United States lifetime prevalence rates between 19% and 28% were reported for any anxiety disorder.1,2 Genetic epidemiological studies have shown that the etiology of these disorders is in part genetic with heritability estimates varying between 23% and 50%.3–5 Several genome-wide linkage studies have tried to identify genomic regions harboring QTLs that influence the vulnerability for anxiety disorders. All these studies,6–15 ascertained the pedigrees through probands with panic disorder, except for one study16 that included pedigrees with at least one member having an anxiety disorder or somatoform pain. Panic disorder is the most frequently analyzed phenotype.6–8,13,14 Two studies focused on panic disorder with comorbid medical conditions i.e. renal/bladder conditions, thyroid irregularities, mitral valve prolapse and severe headaches.11,15 Simple phobia and social phobia have each been investigated once.9,10 Other anxiety phenotypes.12,14,16 have been based on findings from genetic epidemiological studies indicating a common genetic background for anxiety disorders,4,17 Thorgeirsson et al.16 considered anxiety disorder or somatoform pain as indicators of affection status. Smoller et al.14 and Kaabi et al.12 constructed new phenotypes of anxiety proneness.
An overview of the results7,9,10 from these studies shows that no genomic region has been unequivocally identified to harbor a QTL involved in anxiety. Table 1 summarizes the most promising results so far. One region on chromosome 7 and one region on chromosome 1 showed linkage in two scans6,8,13,14 with LOD scores from 1.71 to 2.23. A linkage signal on chromosome 14 was observed for panic disorder, simple and social phobia in a sample of 20 families with 153 individuals.8–10 The highest LOD scores (>4) have been reported on chromosome 1315 when analyzing panic disorder with comorbid medical conditions and on chromosomes 412 and 916 when analyzing a broad anxiety phenotype. The results of the latter two studies suggest that using a broad anxiety phenotype might enhance the chance of finding a linkage signal.
Table 1.
Most promising linkage results for anxiety phenotypes (Regions with a significant linkage signal or a suggestive linkage signal found at least twice)
| Location | LOD | N subjects/families | Phenotype | Sample ascertainment |
|---|---|---|---|---|
| Chromosome 1, 218–234 cM8,14 | 2.04 | 153/20 | Panic disorder | Probands with panic disorder |
| 2.05* | 99/1 | Aniety proneness | Probands with panic disorder | |
| Chromosome 4, 157cM12 | >4 | 153/20 | Broad anxiety phenotype | Probands with panic disorder |
| Chromosome 7, 47–57cM6,13 | 1.71 | -/23 | Panic disorder | Probands with panic disorder |
| 2.23 | 253/23 | Panic disorder | Probands with panic disorder | |
| Chromosome 9, 105cM16 | 4.18 | -/25 | Anxiety/Panic disorder | Probands with panic attacks, GAD or phobias |
| Chromosome 13, 96cM15 | 4.2 | -/34 | Panic disorder combined with bladder/kidney problems | Probands with panic disorder |
| Chromosome 14, 36–45cM8–10 | 3.7 | 129/14 | Simple phobia | Probands with panic disorder |
| 2.93** | 163/17 | Social Phobia | Probands with panic disorder | |
| 2.38* | 153/20 | Panic disorder | Probands with panic disorder |
This is the NPL score, not the LOD score
This is the Zlr score, not the LOD score
In the current study, linkage analyses were carried out on longitudinal self-report measures of anxiety, i.e. the State Trait Anxiety Inventory (STAI) – Trait version.18,19 The sample consisted of 1,602 siblings with phenotype and genotype data. Additional genotype data, available for 564 parents and 22 siblings, were used to estimate Identity by Descent (IBD). The families were selected from a Dutch population based twin-family sample. Earlier analyses have shown that, in a subsample, the scores on the STAI are strongly associated with DSM-IV20 diagnoses of panic disorder and/or agoraphobia, social phobia and generalized anxiety disorder.21 Consequently, it can be considered a broad anxiety phenotype.
Materials and methods
Subjects
This study is part of a longitudinal survey study of the Netherlands Twin Register (NTR) that has assessed twin families roughly every two years since 1991. Sample selection and response rates are described in detail in Boomsma et al..22 A sub-sample was approached to provide DNA. For this linkage study, genotype and STAI data were available for 539 dizygotic male twins and brothers, 758 dizygotic female twins and sisters, 108 monozygotic male and 197 monozygotic female twins. DNA was also available for 249 fathers and 315 mothers and for 22 siblings for whom anxiety data were missing. In total, 566 families and 2,188 genotyped subjects were included in the linkage analyses.
Instruments
In 1991, 1993, 1997, 2000 and 2002, anxiety was measured with the STAI-trait version 18,19., which consist of 20 items that are scored on 4-point scales (Cronbach’s alpha is 0.92). Examples of items are “I worry too much over something that really does not matter”, “I feel nervous and restless”, “I feel secure”. The answer categories were “almost never”, “sometimes”, “often” and “almost always”. As the distribution of the scores was slightly skewed, the scores were log transformed according to earlier analyses.23
The association between STAI scores and DSM-IV mood and anxiety disorders, assessed with the Composite International Diagnostic Interview 24 was analyzed in a sub-sample of 1248 family members, 521 men and 727 women. Prevalence rates of lifetime major depression, panic disorder and/or agoraphobia, generalized anxiety disorder and social phobia were respectively 15%, 6.4%, 5.8% and 4.5%, with higher prevalence rates in men than in women. Subjects with these panic disorders scored significantly higher on the STAI than subjects without a mood or anxiety disorder. The difference was more than one standard deviation.21
Analyses of the data obtained in 1991, 1993 and 1997 showed that STAI scores are related to self-report neuroticism25,26 and depression scores.27–29 with correlations between 0.60 and 0.7521.Genetic analyses of the STAI scores showed a heritability of around 45% without any evidence that different genes influence variation in men and women, as correlations between same-sex and opposite-sex twin and sibling pairs were the same.23
Genotyping
DNA from the siblings and their parents was extracted from either whole blood or buccal swaps following standard protocols.30,31 Samples were genotyped by the Marshfield Mammalian Genotyping Service and/or the Molecular Epidemiology Section, Leiden University Medical Centre. The genotype data from these screens were combined. Allele calling and binning were equalized between markers that were present in multiple scans, using ~ 30 control samples. In case there were inconsistencies, the data were set to unknown for tested markers (binning and allele calling inconsistencies) and persons (genotyping errors). Sex and prior measured zygosity were checked with the marker data. Pedigree relations in the entire samples were checked with the GRR program.32 Errors of Mendelian inheritance were detected with Pedstats.26 Markers and samples were removed if their total error rate was more than 1%; in all other cases the specific erroneous genotypes were set as unknown. Unlikely recombinants were detected using Merlin and erroneous genotypes were removed with Pedwipe.33
After cleaning, only sib-pairs were selected that had at least 280 autosomal markers genotyped for each individual. The average number of markers genotyped in siblings was 366 (range 280–761) and the average heterozygosity of autosomal markers was 75% with an average spacing of 7 cM. For the statistical analyses, the Haldane mapping function was used. The marker positions were interpolated via locally weighted linear regression from the National Center for Biotechnology Information (NCBI) build 35.1 physical map positions and the Rutgers genetic map.34,35 The information of the parents and siblings without phenotypes was used to obtain estimates of the IBD status of the sibling pairs (see below).
Statistical analysis
The sib-pair strategy to map loci influencing quantitative traits is based on identifying marker alleles that are inherited IBD. If a marker is co-segregating with a trait, then siblings whose trait values are more alike, are more likely to receive the same alleles IBD at a closely linked marker locus than siblings whose resemblance for the trait is less. IBD refers to the fact that siblings share a particular allele because they received it from the same parent. The number of alleles that are IBD in sib-pairs is thus 0, 1 or 2, but unless both parents are heterozygous for different marker alleles, it is not always possible to determine IBD status exactly. In families in which the parents are not genotyped, IBD is estimated using population allele frequencies for the markers. Allele frequencies were obtained from the observed genotype data.
Linkage analysis was carried out in MERLIN -regress.36 The regression analysis implemented in this software package is based on a modified method initially proposed by Haseman and Elston to investigate linkage in sibling pairs.37 In brief, the multipoint IBD sharing is regressed on trait squared sums and squared differences, among all pairs of siblings.36 The trait squared sums and differences indicate the resemblance between the siblings. The method takes into account incomplete IBD information. Mean, variance and heritability are user-specified.
Anxiety scores were adjusted for age and sex, as women score higher and older subjects score somewhat lower. The sex and age adjusted scores were averaged over the five measurement occasions. In MERLIN-regress, repeated measures can be entered in the analysis as an average across occasions with further specification of the test-retest correlation and the number of measurements for each individual (in this case varying form 1 to 5) in order to readjust the trait variance.38 The intra-class correlation between the five measures was estimated in SPSS using a mixed model procedure, which allows using all available data, even of the subjects who did not participate every occasion.
The 1-LOD-drop support interval was used as an estimate for the 90% confidence interval of any QTL locations,32 and was used to examine linkage replication compared to previously reported locations and candidate genes (using Ensembl: www.Ensembl.org and OMIM: www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM).
Empirically derived P-values were calculated from 1000 simulated data sets using the gene-dropping method implemented in MERLIN -regress.39
Results
Mean age and anxiety scores across the five occasions were calculated for the total sample (N=11,336) of twins and siblings and for the linkage sample (N=1606). The subjects who provided DNA were slightly older than the total sample of twins and siblings in (31.6 years versus 27.1 years). The anxiety scores were comparable (33.6 and 33.5 with standard deviations of 8.0 and 8.5 respectively). After the log-transformation, the scores were normally distributed in both samples.
The heritability was specified at 42%. This estimate was based on analyses in MERLIN-regress using the option “random sample” and was comparable to the results of earlier genetic epidemiological analyses of STAI scores.23 Subjects had participated on average three times. The intra-class correlation of the five anxiety measures was 0.58.
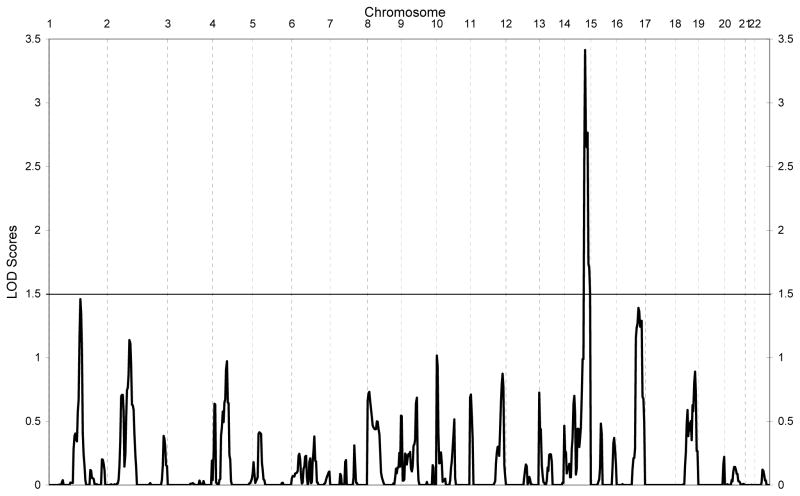
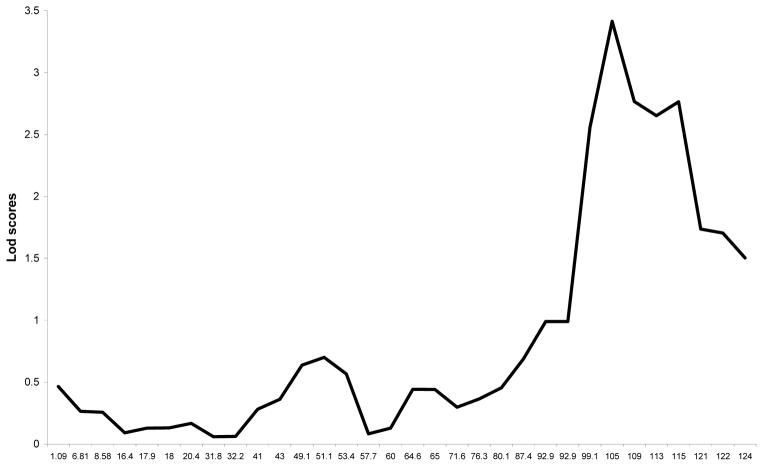
Figure 1 shows the results of the linkage analysis. A LOD score of 3.4 (empirical p-value = 0.07) was found on chromosome 14 at 105cM (90% confidence interval 99cM-115cM). There was no other region with suggestive or significant linkage. Figure 2 shows the linkage results for chromosome 14 indicating that marker D14S65 showed the highest LOD-score and the linkage peak was bounded by markers D14S1434 and D14S985.
Figure 1.
Results of the genome wide linkage analysis of the anxiety scores averaged across the five occasions.
Figure 2.
Results of the genome wide linkage analysis for chromosome 14 with on the x-axis the position of the markers in cM (Haldane)
Discussion
A genomewide linkage study on a broad anxiety phenotype was performed in a sample of 1,602 twins and siblings with additional genotype data of 564 parents and 22 siblings used to estimate the IBD status of the sibling pairs. There was evidence for linkage on chromosome 14 at 105cM with a LOD score of 3.4 and an empirical p-value of 0.07.
For the same region at chromosome 14, Kaabi et al.12 reported a significant linkage result (nominal p = 0.005 and empirical p = 0.04). However, after adjusting for sib-pair age differences, the signal disappeared (nominal p = 0.07, empirical p not given). Possible explanations for the loss of linkage signal after age adjustment, as given by the authors, are chance due to power loss or differential gene expression at different ages. The first explanation seems to be the most probable. First, for the sib-pair age difference to be a confounding factor in a linkage analysis, it is necessary that it is associated both with the amount of resemblance between siblings and with the IBD-status. It seems unlikely that sib-pair age difference is associated with IBD-status.
Second, this region on chromosome 14 is syntenic with a region showing a significant association with emotionality in mice measured in two previous linkage studies (chromosome 12, 41cM-61cM)40,41 and in a recent genome-wide association analysis.42,43. The linkage studies were carried out in inbred mouse strains whose open field activity (OFA) was measured. Genome-wide association was analyzed in heterogeneous mice stocks. Multiple complex phenotypes were investigated in the heterogeneous mice stocks and the results are freely available on http://gscan.well.ox.ac.uk.42,43 Unconditioned anxiety was measured with the elevated plus maze (EPM), OFA and food hyponeophagia (FN). Genomewide significance was found in the region syntenic with our linkage signal for the open arm latency in the EPM. Two homolog genes lie in this region, i.e. Dlk1: delta-like 1 homolog (Drosophila) and Rtl1: retrotransposon-like 1. The function of these genes is still unknown, although for Dlk1, it has become clear that this gene is over expressed in brain tumors.44,45 None of these genes have ever been related to anxiety, but the results of the studies in mice together with our results indicate that it may be worthwhile to investigate the association. A further search did not reveal other genes under this peak that have been related to symptoms of anxiety. This does not exclude that they play a role, but it seems more obvious to start investigating Dlk1 or Rtl1.
We have not replicated any other of the previous promising linkage results as summarized in Table 1. An explanation for this divergence in results could be the ascertainment of the sample. In contrast to the other studies in which the pedigrees are ascertained through probands with panic disorder or another anxiety disorder, our sample is selected from a Dutch population based twin-family sample. An advantage of a clinical based sample is that the chance is high that the most extreme subjects from the population, who have the largest burden of disease, are included in the analysis. A disadvantage, on the other hand, is that a clinical sample can be biased by, for example, help seeking behavior or a high prevalence of a comorbid disorder. This signifies that a linkage signal can be due to the disease, but also to other characteristics of a clinical sample. As our significant linkage result was also previously found in a clinical sample,12 the region on chromosome 14 is possible associated with the whole spectrum of general anxiety, i.e. from the normal to the clinical range.
Another reason that no replication to earlier results was seen could lie in the definition of phenotype across different studies. Scores on the STAI are highly associated with a diagnosis of generalized anxiety disorder, but less with panic disorder and/or agoraphobia and intermediately with social phobia.21 Furthermore, it has been suggested that the STAI not only measures anxiety, but also depression.46 However, results of linkage studies of major depression did not overlap with our findings either.46
A review on the comorbidity between anxiety and depression showed that anxiety and depression might be influenced by shared genes, possibly expressed in the personality trait neuroticism.17 Linkage findings for neuroticism were summarized by Fullerton et al47 and do not include findings on chromosome 14 in humans.
To summarize, the current linkage study suggests that the region on chromosome 14, around 105cM, could harbor a gene associated with anxiety. The region is also syntenic with regions found to be associated with emotionality in mice (40,41 and http://gscan.well.ox.ac.uk). Further association studies could focus on the homolog genes Dlk1 and Rtl1.
Acknowledgments
The study was supported by the Netherlands Organization for Scientific Research NWO/ZonMW (400-05-717, 911-03-016, 904-61-193, 985-10-002, 575-25-006), NIH R01 HL55976 and NHBLI Mammalian Genotyping Service (Marshfield). Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) which is financially supported by the Netherlands Scientific Organization (NWO 480-05-003). CM was supported by the Hersenstichting Nederland (13F05(2).47).
References
- 1.Bijl RV, Ravelli A, van Zessen G. Prevalence of psychiatric disorder in the general population: results of The Netherlands Mental Health Survey and Incidence Study (NEMESIS) Soc Psychiatry Psychiatr Epidemiol. 1998;33:587–595. doi: 10.1007/s001270050098. [DOI] [PubMed] [Google Scholar]
- 2.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 3.Hettema JM, Neale MC, Kendler KS. A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am J Psychiatry. 2001;158:1568–1578. doi: 10.1176/appi.ajp.158.10.1568. [DOI] [PubMed] [Google Scholar]
- 4.Hettema JM, Prescott CA, Myers JM, Neale MC, Kendler KS. The structure of genetic and environmental risk factors for anxiety disorders in men and women. Arch Gen Psychiatry. 2005;62:182–189. doi: 10.1001/archpsyc.62.2.182. [DOI] [PubMed] [Google Scholar]
- 5.Middeldorp CM, Birley AJ, Cath DC, Gillespie NA, Willemsen G, Statham DJ, et al. Familial clustering of major depression and anxiety disorders in Australian and Dutch twins and siblings. Twin Res Hum Genet. 2005;8:609–615. doi: 10.1375/183242705774860123. [DOI] [PubMed] [Google Scholar]
- 6.Crowe RR, Goedken R, Samuelson S, Wilson R, Nelson J, Noyes R., Jr Genomewide survey of panic disorder. Am J Med Genet. 2001;105:105–109. [PubMed] [Google Scholar]
- 7.Fyer AJ, Hamilton SP, Durner M, Haghighi F, Heiman GA, Costa R, et al. A third-pass genome scan in panic disorder: evidence for multiple susceptibility loci. Biol Psychiatry. 2006;60:388–401. doi: 10.1016/j.biopsych.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 8.Gelernter J, Bonvicini K, Page G, Woods SW, Goddard AW, Kruger S, et al. Linkage genome scan for loci predisposing to panic disorder or agoraphobia. Am J Med Genet. 2001;105:548–557. doi: 10.1002/ajmg.1496. [DOI] [PubMed] [Google Scholar]
- 9.Gelernter J, Page GP, Bonvicini K, Woods SW, Pauls DL, Kruger S. A chromosome 14 risk locus for simple phobia: results from a genomewide linkage scan. Mol Psychiatry. 2003;8:71–82. doi: 10.1038/sj.mp.4001224. [DOI] [PubMed] [Google Scholar]
- 10.Gelernter J, Page GP, Stein MB, Woods SW. Genome-wide linkage scan for loci predisposing to social phobia: evidence for a chromosome 16 risk locus. Am J Psychiatry. 2004;161:59–66. doi: 10.1176/appi.ajp.161.1.59. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton SP, Fyer AJ, Durner M, Heiman GA, Baisre dL, Hodge SE, et al. Further genetic evidence for a panic disorder syndrome mapping to chromosome 13q. Proc Natl Acad Sci U S A. 2003;100:2550–2555. doi: 10.1073/pnas.0335669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaabi B, Gelernter J, Woods SW, Goddard A, Page GP, Elston RC. Genome scan for loci predisposing to anxiety disorders using a novel multivariate approach: strong evidence for a chromosome 4 risk locus. Am J Hum Genet. 2006;78:543–553. doi: 10.1086/501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles JA, Fyer AJ, Vieland VJ, Weissman MM, Hodge SE, Heiman GA, et al. Results of a genome-wide genetic screen for panic disorder. Am J Med Genet. 1998;81:139–147. doi: 10.1002/(sici)1096-8628(19980328)81:2<139::aid-ajmg4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 14.Smoller JW, Acierno JS, Jr, Rosenbaum JF, Biederman J, Pollack MH, Meminger S, et al. Targeted genome screen of panic disorder and anxiety disorder proneness using homology to murine QTL regions. Am J Med Genet. 2001;105:195–206. doi: 10.1002/ajmg.1209. [DOI] [PubMed] [Google Scholar]
- 15.Weissman MM, Fyer AJ, Haghighi F, Heiman G, Deng Z, Hen R, et al. Potential panic disorder syndrome: clinical and genetic linkage evidence. Am J Med Genet. 2000;96:24–35. doi: 10.1002/(sici)1096-8628(20000207)96:1<24::aid-ajmg7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 16.Thorgeirsson TE, Oskarsson H, Desnica N, Kostic JP, Stefansson JG, Kolbeinsson H, et al. Anxiety with panic disorder linked to chromosome 9q in Iceland. Am J Hum Genet. 2003;72:1221–1230. doi: 10.1086/375141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Middeldorp CM, Cath DC, van Dyck R, Boomsma DI. The co-morbidity of anxiety and depression in the perspective of genetic epidemiology. A review of twin and family studies. Psychol Med. 2005;35:611–624. doi: 10.1017/s003329170400412x. [DOI] [PubMed] [Google Scholar]
- 18.Spielberger CD, Gorsuch RL, Lushene RE. STAI Manual for the State-Trait Anxiety Inventory. Consulting Psychologists Press; Palo Alto, CA: 1970. [Google Scholar]
- 19.Van der Ploeg H, Defares PB, Spielberger CD. Zelfbeoordelingsvragenslijst STAI, versie DY-1 en DY-2. Swets & Zeitlinger; Lisse: 1979. [Google Scholar]
- 20.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4. APA; Washington DC: 1994. [Google Scholar]
- 21.Middeldorp CM, Cath DC, van den BM, Beem AL, van Dyck R, Boomsma DI. The Association of Personality with Anxious and Depressive Psychopathology. In: Canli T, editor. The Biological Basis of Personality and Individual Differences. Guilford Press; New York: 2006. pp. 251–272. [Google Scholar]
- 22.Boomsma DI, Vink JM, Van Beijsterveldt TC, de Geus EJ, Beem AL, Mulder EJ, et al. Netherlands Twin Register: a focus on longitudinal research. Twin Res. 2002;5:401–406. doi: 10.1375/136905202320906174. [DOI] [PubMed] [Google Scholar]
- 23.Boomsma DI, Beem AL, van den BM, Dolan CV, Koopmans JR, Vink JM, et al. Netherlands twin family study of anxious depression (NETSAD) Twin Res. 2000;3:323–334. doi: 10.1375/136905200320565300. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization. Composite International Diagnostic Interview (version 2.1) WHO; Geneva: 1992. [Google Scholar]
- 25.Eysenck HJ, Eysenck SBG. Eysenck Personality Inventory. Educational Industrial Testing Service; San Diego, Ca: 1964. [Google Scholar]
- 26.Wilde GJS. Neurotische labiliteit gemeten volgens de vragenlijstmethode (The questionnaire method as a means of measuring neurotic instability) Van Rossen; Amsterdam: 1970. [Google Scholar]
- 27.Achenbach TM. The Young Adult Self Report. University of Vermont, Dept of Psychiatry; Burlington, VT: 1990. [Google Scholar]
- 28.Beck AT, Rial WY, Rickels K. Short form of depression inventory: cross-validation. Psychol Rep. 1974;34:1184–1186. [PubMed] [Google Scholar]
- 29.Verhulst FC, Ende Jv, Koot HM. Handleiding voor de Youth Self Report. Afdeling Kinder-en Jeugdpsychiatrie, Sophia Kinderziekenhuis/Academisch Ziekenhuis Rotterdam/Erasmus Universiteit Rotterdam; Rotterdam: 1997. [Google Scholar]
- 30.Meulenbelt I, Droog S, Trommelen GJ, Boomsma DI, Slagboom PE. High-yield noninvasive human genomic DNA isolation method for genetic studies in geographically dispersed families and populations. Am J Hum Genet. 1995;57:1252–1254. [PMC free article] [PubMed] [Google Scholar]
- 31.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 33.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 34.Duffy DL. An integrated genetic map for linkage analysis. Behav Genet. 2006;36:4–6. doi: 10.1007/s10519-005-9015-x. [DOI] [PubMed] [Google Scholar]
- 35.Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–1148. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71:238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haseman JK, Elston RC. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 1972;2:3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- 38.Nash MW, Huezo-Diaz P, Williamson RJ, Sterne A, Purcell S, Hoda F, et al. Genome-wide linkage analysis of a composite index of neuroticism and mood-related scales in extreme selected sibships. Hum Mol Genet. 2004;13:2173–2182. doi: 10.1093/hmg/ddh239. [DOI] [PubMed] [Google Scholar]
- 39.Sawcer S, Jones HB, Judge D, Visser F, Compston A, Goodfellow PN, et al. Empirical genomewide significance levels established by whole genome simulations. Genet Epidemiol. 1997;14:223–229. doi: 10.1002/(SICI)1098-2272(1997)14:3<223::AID-GEPI1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 40.Flint J, Corley R, DeFries JC, Fulker DW, Gray JA, Miller S, et al. A simple genetic basis for a complex psychological trait in laboratory mice. Science. 1995;269:1432–1435. doi: 10.1126/science.7660127. [DOI] [PubMed] [Google Scholar]
- 41.Plomin R, McClearn GE, Gora-Maslak G, Neiderhiser JM. Use of recombinant inbred strains to detect quantitative trait loci associated with behavior. Behav Genet. 1991;21:99–116. doi: 10.1007/BF01066330. [DOI] [PubMed] [Google Scholar]
- 42.Solberg LC, Valdar W, Gauguier D, Nunez G, Taylor A, Burnett S, et al. A protocol for high-throughput phenotyping, suitable for quantitative trait analysis in mice. Mamm Genome. 2006;17:129–146. doi: 10.1007/s00335-005-0112-1. [DOI] [PubMed] [Google Scholar]
- 43.Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, et al. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- 44.Altenberger T, Bilban M, Auer M, Knosp E, Wolfsberger S, Gartner W, et al. Identification of DLK1 variants in pituitary- and neuroendocrine tumors. Biochem Biophys Res Commun. 2006;340:995–1005. doi: 10.1016/j.bbrc.2005.12.094. [DOI] [PubMed] [Google Scholar]
- 45.Yin D, Xie D, Sakajiri S, Miller CW, Zhu H, Popoviciu ML, et al. DLK1: increased expression in gliomas and associated with oncogenic activities. Oncogene. 2006;25:1852–1861. doi: 10.1038/sj.onc.1209219. [DOI] [PubMed] [Google Scholar]
- 46.Camp NJ, Cannon-Albright LA. Dissecting the genetic etiology of major depressive disorder using linkage analysis. Trends Mol Med. 2005;11:138–144. doi: 10.1016/j.molmed.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Fullerton J. New approaches to the genetic analysis of neuroticism and anxiety. Behav Genet. 2006;36:147–161. doi: 10.1007/s10519-005-9000-4. [DOI] [PubMed] [Google Scholar]