Summary
Working memory, the capacity of actively maintaining task-relevant information during a cognitive task, is a heritable trait. Working memory deficits are characteristic for many psychiatric disorders. We performed genome-wide gene-set enrichment analyses in multiple independent data sets of young and aged cognitively healthy subjects (n = 2’824), and in a large schizophrenia case-control sample (n = 32’143). The voltage-gated cation channel activity gene-set, consisting of genes related to neuronal excitability, was robustly linked to performance in working memory-related tasks across ages, and to schizophrenia. Functional brain imaging in 707 healthy participants linked this gene-set also to working memory-related activity in the parietal cortex and the cerebellum. Gene-set analyses may help to dissect the molecular underpinnings of cognitive dimensions, brain activity and psychopathology.
Introduction
The study of the genetic underpinnings of human cognition, emotion and personality impacts substantially on the understanding of physiological and pathophysiological processes relevant to mental health and psychiatric disease. Working memory (WM), which represents a limited-capacity neural network capable of actively maintaining task-relevant information during the execution of a cognitive task (Shah and Miyake, 1999), is a key cognitive trait well amenable to behavioral genetic studies: WM is heritable (Karlsgodt et al., 2011), can be assessed in a valid and reliable manner, and has well-defined neural correlates as shown in functional brain imaging studies (D’Esposito, 2007). Deficits in WM are a key component of psychiatric disorders such as schizophrenia (Barch, 2005), bipolar disorder (Balanza-Martinez et al., 2008) and attention deficit hyperactivity disorder (ADHD) (Doyle, 2006).
Genome-wide association studies (GWAS) employing single-marker statistics have been successful in identifying cognitive trait-associated single-gene loci (Papassotiropoulos and de Quervain, 2011). It is, however, widely accepted that single-marker-based analyses have limited power to identify the genetic basis of a given trait, as for example, many loci will fail to reach stringent genome-wide significance threshold, despite the fact that they may be genuinely associated with the trait. Triggered by statistical approaches for the analysis of gene expression and protein-protein interaction, gene-set-based analytical methods have recently become available. These methods aim at identifying biologically meaningful sets of genes associated with a certain trait, rather than focusing on a single GWAS gene locus (Wang et al., 2010). By taking into account prior biological knowledge, gene-set-based approaches examine whether test statistics for a group of related genes have consistent deviation from chance (Wang et al., 2010). As shown recently in studies on autism (Voineagu et al., 2011), bipolar disorder (Holmans et al., 2009; Sklar et al., 2011), ADHD (Stergiakouli et al., 2012) and schizophrenia (O’Dushlaine et al., 2011), such approaches can identify convergent molecular pathways relevant to neuropsychiatry and provide initial evidence, which can serve as starting point for testable hypotheses addressing functionality within the indicated pathways. However, the methodological heterogeneity of different pathway analytical tools makes it necessary to demonstrate the methodological invariance and replicability of the results (for reviews see (Holmans et al., 2009; Wang et al., 2010; Wang et al., 2011)).
We performed genome-wide gene-set enrichment analyses of WM performance in multiple independent data sets of young and aged cognitively healthy subjects. In a large case-control sample we also performed genome-wide gene-set enrichment analyses of the risk for schizophrenia. We show that the voltage-gated cation channel activity gene-set, consisting of genes related to neuronal excitability, is robustly linked to WM performance across ages, and to schizophrenia. In addition, this gene-set is associated to WM-related activity in brain regions known for their involvement in psychiatric disease.
Results
Gene-set enrichment analysis (GSEA) in young healthy adults
Discovery sample (n = 905)
After genome-wide calculation of P values for association with WM performance (n-back task, see Experimental Procedures) under the additive genetic model, we ran GSEA using MAGENTA (Segre et al., 2010). Q-O-plotting indicated that the statistical power of the sample was sufficient to detect loci reaching genome-wide significance (P< 5 × 10 -8) (Figure S1A). Five SNPs exceeded genome-wide significance (Figure 1). Four SNPs (rs711159, rs2049590, rs7300145, rs10861993) mapped to chromosome 12 and spanned PAWR, which encodes a neuronally expressed transcriptional regulator. One SNP (rs12028754) mapped to chromosome 1, specifically to an intron of OLFM3, which encodes olfactomedin 3 (alias noelin 3). This protein is known to interact with the AMPA receptor complex.
Figure 1.
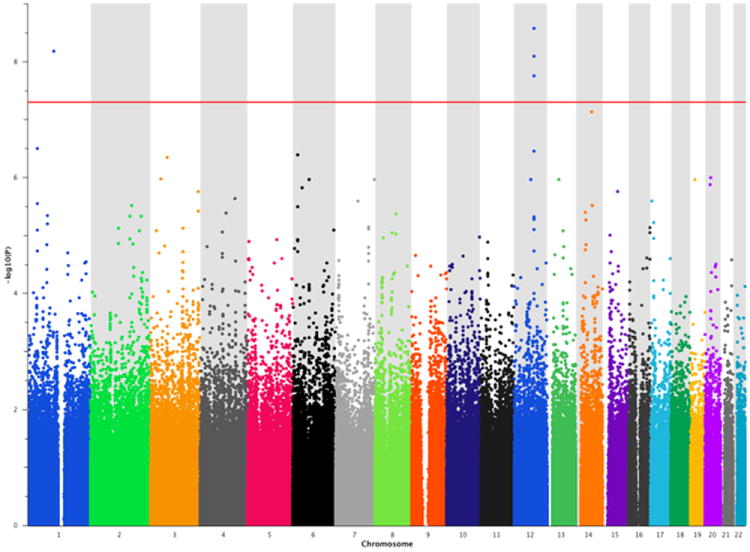
Manhattan plot of the GWAS in the discovery sample. The red horizontal line indicates genome-wide Bonferroni correction.
Among the 1’411 database-derived gene-sets which served as input, MAGENTA identified multiple testing-corrected significant enrichment (PFDR<0.05) for two gene-sets: “voltage-gated cation channel activity” (GO:0022843; gene-set database: gene ontology) and “transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds”, (gene-set database: Reactome; Pnominal= 9.9 × 10-5 and 9.9 × 10-5, respectively; see Table 1). The contribution to the overall significance of the gene sets was balanced across significant gene set SNPs (Figure 2, Table S1). No additional gene-set withstood FDR correction for testing 1’411 gene sets.
Table 1.
Pathway associations in four GWAS data sets of cognitively healthy adults(PFDR< 0.05 and the corresponding uncorrected Pnominal are shown).
| Discovery sample (n = 905) | Replication sample (n = 746) | Zurich sample (n = 410) | AgeCoDe sample (n = 763) | |||||
|---|---|---|---|---|---|---|---|---|
| Database | Gene-set | Number of significant genes | Pnominal | PFDR | Pnominal | PFDR | Pnominal | Pnominal |
| GO | Voltage gated cation channel activity | 28 | 9.90E-05 | 3.19E-02* | 9.90E-05 | 1.28E-02* | 2.00E-03 | 1.68E-02 |
| REACTOME | Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds | 35 | 9.90E-05 | 3.10E-02* | 1.90E-01 | 5.65E-01 | 1.48E-01 | 2.08E-01 |
| GO | Voltage gated channel activity | 28 | 1.00E-04 | 9.10E-02 | 9.90E-05 | 1.85E-02* | 6.20E-03 | 1.11E-02 |
| GO | Metal ion transmembrane transporter activity | 51 | 3.00E-04 | 5.69E-02 | 1.80E-03 | 1.45E-01 | 9.00E-04 | 2.20E-02 |
| KEGG | Arrhythmogenic right ventricular cardiomyopathy | 29 | 7.00E-04 | 7.64E-02 | 5.35E-02 | 4.23E-01 | 2.90E-02 | 2.99E-01 |
| GO | Cation channel activity | 40 | 1.40E-03 | 1.71E-01 | 9.90E-05 | 1.28E-02* | 9.40E-03 | 2.50E-02 |
| GO | Calcium channel activity | 15 | 1.70E-03 | 1.64E-01 | 4.06E-02 | 5.72E-01 | 8.67E-02 | 6.12E-01 |
| REACTOME | Signaling by Rho GTPases | 36 | 2.20E-03 | 1.59E-01 | 2.30E-03 | 1.23E-01 | 1.00E-03 | 3.98E-02 |
| GO | Gated channel activity | 39 | 2.40E-03 | 2.00E-01 | 1.00E-03 | 1.22E-01 | 1.53E-02 | 1.90E-03 |
| GO | Transmembrane receptor protein kinase activity | 21 | 2.50E-03 | 2.09E-01 | 3.95E-01 | 9.11E-01 | 1.79E-02 | 3.01E-01 |
| GO | Ion channel activity | 45 | 2.80E-03 | 2.46E-01 | 7.00E-04 | 9.30E-02 | 5.64E-02 | 2.31E-02 |
| REACTOME | Signaling by robo receptor | 14 | 3.10E-03 | 1.73E-01 | 5.49E-02 | 2.98E-01 | 1.18E-01 | 2.06E-01 |
| KEGG | Dilated cardiomyopathy | 31 | 3.40E-03 | 1.39E-01 | 3.03E-02 | 4.95E-01 | 2.51E-02 | 2.45E-01 |
| GO | Calmodulin binding | 12 | 3.40E-03 | 1.72E-01 | 9.37E-01 | 9.76E-01 | 4.74E-01 | 3.03E-01 |
| KEGG | Adherens junction | 28 | 3.80E-03 | 1.08E-01 | 2.09E-01 | 6.27E-01 | 2.31E-01 | 6.32E-01 |
| REACTOME | Integrin cell surface interactions | 29 | 4.00E-03 | 2.18E-01 | 8.40E-02 | 3.96E-01 | 3.92E-01 | 2.99E-02 |
| GO | Lamellipodium | 12 | 4.00E-03 | 1.83E-01 | 1.99E-01 | 8.58E-01 | 1.06E-01 | 1.86E-01 |
| REACTOME | Regulation of IFNA signaling | 6 | 4.30E-03 | 1.83E-01 | 6.34E-01 | 7.61E-01 | 1.63E-01 | 3.92E-01 |
| REACTOME | Transport of inorganic cations anions and amino acids oligopeptides | 31 | 4.80E-03 | 2.76E-01 | 8.08E-02 | 3.97E-01 | 9.92E-02 | 1.97E-01 |
| REACTOME | Cell surface interactions at the vascular Wall | 29 | 5.00E-03 | 2.73E-01 | 6.33E-01 | 8.10E-01 | 4.16E-01 | 4.06E-02 |
| GO | Cell cortex part | 12 | 5.10E-03 | 1.93E-01 | 1.08E-01 | 6.99E-01 | 1.96E-01 | 1.98E-01 |
| GO | Leading edge | 18 | 6.70E-03 | 2.30E-01 | 3.46E-01 | 9.70E-01 | 3.11E-01 | 1.22E-01 |
| GO | Protein tyrosine kinase activity | 23 | 6.90E-03 | 2.33E-01 | 3.82E-01 | 9.02E-01 | 1.45E-01 | 2.18E-01 |
| GO | Cortical cytoskeleton | 10 | 7.60E-03 | 2.41E-01 | 1.78E-01 | 8.19E-01 | 3.35E-01 | 3.40E-01 |
| REACTOME | L1cam interactions | 28 | 8.60E-03 | 3.08E-01 | 2.33E-02 | 2.25E-01 | 6.30E-03 | 1.80E-03 |
Pnominal: uncorrected values; PFDR: genome-wide corrected values (FDR);
Asterisks denote significance after genome-wide correction
Figure 2.
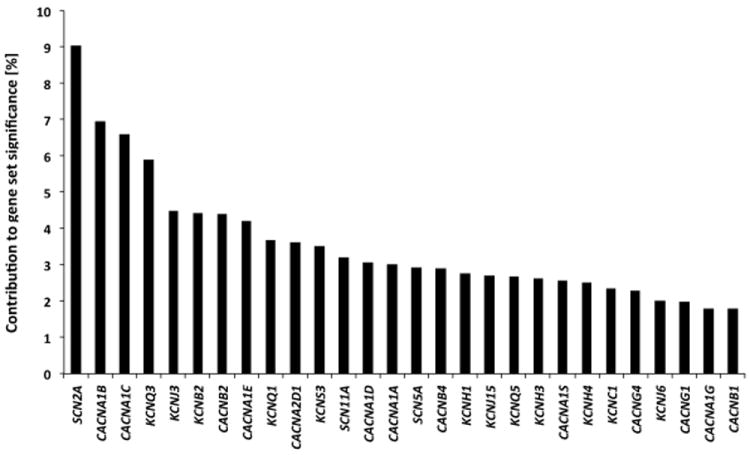
Visualization of each significant SNP’s percent contribution (in descending order) to the overall significance of the voltage-gated cation channel activity gene set in the discovery sample.
Replication sample (n = 746)
A second independent gene-set analysis on the identical phenotype (n-back task) was performed by using the same MAGENTA settings as in the discovery sample. Among the 1’411 database-derived gene-sets tested, the voltage-gated cation channel activity gene-set (GO:0022843) was the best hit, showing significant enrichment at a genome-wide FDR corrected level (Pnominal= 9.9 × 10-5, PFDR = 1.28 × 10-2). Furthermore, the cation channel activity gene-set and the voltage gated channel activity gene-set, two gene-sets highly overlapping with GO:0022843 (59% and 90% overlap, respectively), also showed significant enrichment in the replication sample (PFDR = 1.28 × 10-2 and 1.85 × 10-2, respectively; see Table 1).
Use of different GSEA algorithms
To assess whether the identified gene set is detected independently of the algorithm used, we used different gene-set analysis tools in the combined discovery and replication samples (n = 1’651), for which the identical cognitive task served as phenotype.
GSA-SNP is a gene-set tool that uses SNP P values as input and identifies pathways in a competitive way (Nam et al., 2010). Importantly, GSA-SNP offers the option to set the kth (k = 2, 3, 4 or 5) best P value as proxy for the respective gene instead of using only the best P value. We ran four analyses, and for each option the voltage-gated cation channel activity gene-set (GO:0022843) showed a highly significant enrichment score, with FDR corrected P values ranging between PFDR = 1 × 10-8 and 4 × 10-8.
INRICH (Lee et al., 2012), another gene-set tool, implements a permutation-based algorithm to correct for possible biases induced by marker density, linkage disequilibrium (LD) structure and gene size. Of 449 independent intervals that contained the best genome-wide association signals, 9 intervals overlapped with the 66 target genes of GO:0022843. Subsequent permutation analysis (100’000 permutations) showed significant enrichment for GO:0022384 (Pempirical = 0.018).
We also performed additional genome-wide analyses and subsequent GSEA for non-WM phenotypes, such as performance in the 0-back task (related to attentional processes), picture and verbal delayed free recall (related to episodic memory), and delayed free recall for positive and negative pictures (related to emotional memory, see Experimental Procedures). GO:0022843 did not show any enrichment for these phenotypes (all Pnominal> 0.05).
Zurich sample (n = 410)
In an independent sample of Swiss young adults we performed GSEA on another WM-related phenotype (digit span forward). Despite the smaller sample size, we detected significant enrichment of the voltage-gated cation channel activity gene-set (GO:0022843) (Pnominal = 2.0 × 10-3; see Table 1).
GSEA in non-demented elderly subjects
AgeCoDe sample (n = 763)
This sample of cognitively healthy elderly individuals was included to study whether the observed association of the voltage-gated cation channel activity gene-set with WM-related cognitive performance depends on age. In analogy to the discovery and replication samples we used MAGENTA to calculate gene-set enrichment for immediate verbal free recall performance. Immediate verbal free recall does not represent WM. Nevertheless, WM capacity is one of the cognitive components necessary to perform this task. The voltage-gated cation channel activity gene-set showed significant enrichment (Pnominal = 1.6 × 10-2; see Table 1). No gene-set was significant at an FDR corrected level of significance (all PFDR> 0.05).
GSEA in schizophrenia
Because WM deficits are commonly observed in schizophrenia, we investigated whether GSEA would identify the voltage-gated cation channel activity gene-set as related to the risk for schizophrenia.
Psychiatric GWAS Consortium (PGC) and Sweden 1-6 case-control study, schizophrenia
9.8 million P values of autosomal SNPs from a publicly available meta-analysis (Sweden 1-6 case-control study with the 2011 PGC SCZ report (Ripke et al., 2013; Ripke et al., 2011) served as input for MAGENTA. MAGENTA was run with the identical parameters as before, except for the annotation, which was done with genome build NCBI36 (hg18). In this data set of patients with schizophrenia, we observed a robust replication of the voltage-gated cation channel activity gene-set, both on a nominal and a genome-wide FDR corrected level of significance (Pnominal = 9.9 × 10-5; ; PFDR = 1.48 × 10-2, see Table S2).
Functional brain imaging in healthy individuals
In an additional experiment, conducted in a subgroup (n = 707) of the replication sample, functional magnetic resonance imaging (fMRI) was used to identify gene set-independent and gene set-dependent differences in brain activity related to WM (i.e. n-back task, see Experimental Procedures). Independently of genotype, we detected highly robust task-related activations in the premotor cortex, prefrontal cortex, frontal poles, parietal cortex, cerebellum, thalamus and insula (Figure 3, Figure 4, Table S3) being consistent with meta-analyses of fMRI studies using the n-back task (Owen et al., 2005; Wager and Smith, 2003).
Figure 3.
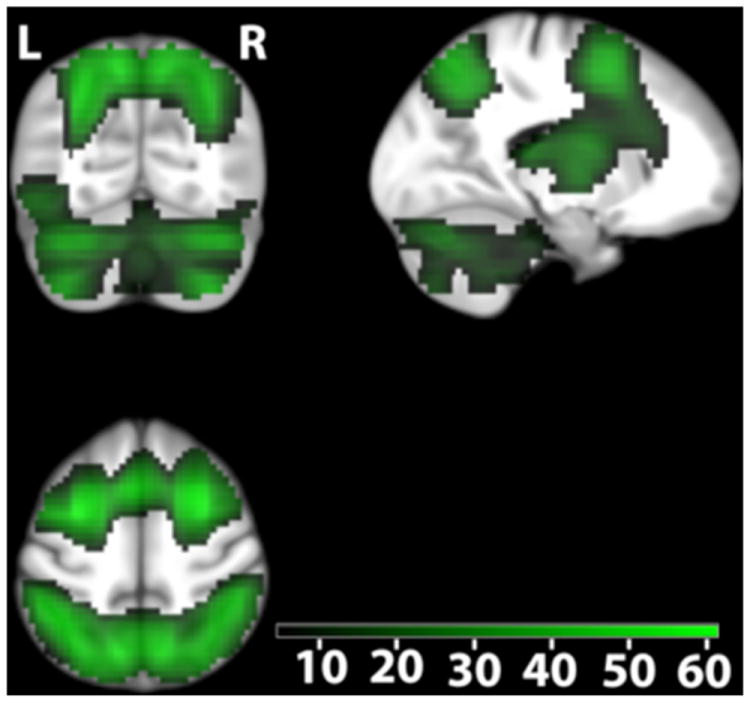
Parietal activity related to working memory independent of genotype. Color-coded t values (Pwhole-brain -FWE-corrected < 0.05; n=707). The map is centered at [-17 -66 52] in the left superior parietal cortex. Activations are overlaid on coronal (upper left), sagital (upper right) and axial (lower left) sections of the study specific group template (see Experimental procedure). L, left side of the brain; R, right side of the brain.
Figure 4.
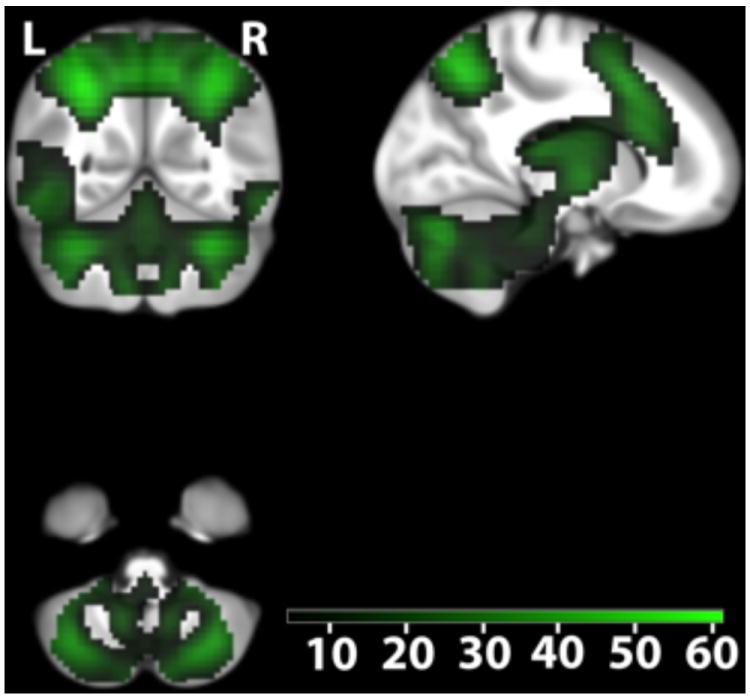
Cerebellar activity related to working memory independent of genotype. Color-coded t values (Pwhole-brain-FWE-corrected < 0.05; n=707). The map is centered at [14 -55 -48] to illustrate activations in the cerebellum. Activations are overlaid on coronal (upper left), sagital (upper right) and axial (lower left) sections of the study specific group template (see Experimental procedure). L, left side of the brain; R, right side of the brain.
To capture the multi-allelic effect of GO:0022843 on brain activity related to WM, PLINK was used to generate an individual multilocus genetic score. The score comprises 29 SNPs from an equal number of the significant genes from the voltage-gated cation channel activity gene-set (see Table S4). As in the GSEA, genes were represented by one SNP. SNPs were not intercorrelated. The genetic score of individuals carrying none of the reference alleles was set equal to 0, which corresponds to the lowest possible score value. Homozygosity for the reference allele (i. e. minor) was given a value of 2, heterozygous state was given a value of 1. The score was then weighted by the direction of effect (see the Experimental Procedures section for a detailed description).
Genetic score-dependent analysis revealed a family-wise error (FWE) multiple comparison corrected positive correlation between genetic score values and a large activity cluster in the superior parietal and supramarginal cortex (peak activation at [-16.5 -66 52]; t = 5.04; Puncorrected = 5.9 × 10-7, Pwhole-brain-FWE-corrected = 0.0063, Figure 5A, Table 3; genotype-independent task-related activation at this coordinate t = 43.50) as well as a positive correlation with activity in the right cerebellum (peak activation at [13.75 -55 -48]; t = 4.55; Puncorrected = 6.31 × 10-6, Pwhole-brain-FWE-corrected = 0.049, Figure 5B; genotype-independent task-related activation at this coordinate t = 18). At a lower statistical threshold (Puncorrected < 0.001), 768 voxels showed positive correlation with the multilocus genetic score (Table S5). No voxel correlated with the genetic score in the opposite direction at an FWE-corrected level (3 voxels at Puncorrected < 0.001).
Figure 5.
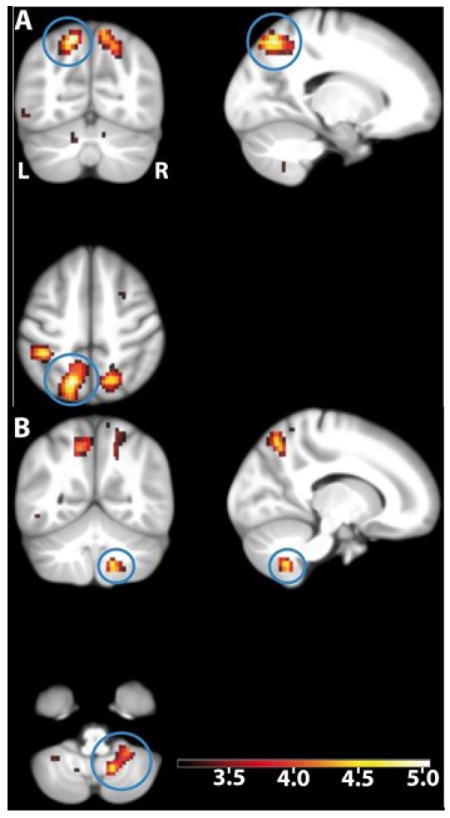
Genetic score-dependent increases in WM-related brain activity (n=707). The blue circles show the activation in (A) the left parietal cortex and (B) the right cerebellum. The maximum is located at [-16.5 -66 52] in the left superior parietal cortex; t = 5.04, Pwhole-brain-FWE-corrected = 0.0063, Puncorrected = 5.9×10-7. The local maximum in the cerebellar cortex is located at [13.75 -55 -48]; t = 4.55, Pwhole-brain-FWE-corrected = 0.049, Puncorrected = 6.31×10-6. Activations are overlaid on coronal (upper left), sagital (upper right) and axial (lower left) sections of the study specific group template (see Experimental Procedures); displayed at Puncorrected = 0.001, using color-coded t values. L, left side of the brain; R, right side of the brain.
Table 3.
Genotype-dependent activation. Voxels demonstrating a positive relationship with the genetic score of 29 polymorphic positions (Pwhole-brain-FWE-corrected< 0.05).
| Number of cluster | Maximum t value within cluster | Regional correspondence of the maximum | MNI coordinates at maximum | Number of voxels | ||
|---|---|---|---|---|---|---|
|
| ||||||
| X | Y | Z | ||||
|
| ||||||
| 1 | 5.041 | ctx-lh-superiorparietal (51%) | -16.5 | -66 | 52 | 20 |
| ctx-lh-precuneus (2%) | ||||||
|
| ||||||
| 2 | 4.9071 | ctx-lh-supramarginal (28%) | -41.25 | -38.5 | 48 | 9 |
| ctx-lh-superiorparietal (19%) | ||||||
| ctx-lh-postcentral (10%) | ||||||
|
| ||||||
| 3 | 4.6694 | ctx-rh-superiorparietal (4%) | 16.5 | -60.5 | 48 | 1 |
| ctx-rh-precuneus (2%) | ||||||
|
| ||||||
| 4 | 4.5526 | Right-Cerebellum-Cortex (81%) | 13.75 | -55 | -48 | 1 |
| Right-Cerebellum-White-Matter (19%) | ||||||
| Lobule IX (51%)1 | ||||||
Regions and probabilities in accordance to the in-house atlas. Abbreviations: ctx = cortex; lh = left hemisphere; rh = right hemisphere.
The anatomical location of the voxel has been furthermore determined by using a probabilistic atlas of the cerebellum as implemented in the SPM anatomy toolbox (Diedrichsen, 2006).
We also conducted a mediator analysis (Baron and Kenny, 1986) to examine whether activity in the brain regions identified as being related to the multilocus genetic score mediated the genetic association between voltage-gated cation channel genes and WM performance. The association between genetic score and WM performance was lower after correcting for brain activity (i.e. individual activation values (beta values) at the left superior parietal and supramarginal cortex [-16.5 -66 52] and at the right cerebellum [13.75 -55 -48], see also Experimental Procedures and Figure S2).
We also analyzed the correlation between genetic score values and activity during an episodic memory task (subsequent memory analysis, Dm, see Experimental Procedures). We did not observe any FWE-corrected correlations. We also performed a voxel-based morphometry (VBM) structural analysis. We did not observe a significant effect of the genetic score on VBM, neither at a whole brain- nor at a small volume-corrected significance threshold. Small volume correction was applied for (i) all regions and clusters showing significant, score-independent or (ii) score-dependent activations in the WM task. In addition, we used psychophysiological interaction (PPI) as described previously (Rasch et al., 2009) to analyze the functional connectivity between a seed region and all other areas of the brain. Seed regions were (i) the superior parietal cortex and (ii) the cerebellum. The resulting connectivity maps were then associated with the genetic score. In both analyses, connectivity did not significantly correlate with genetic score values at a whole brain-corrected level.
To further investigate the score-independent relation of brain activity and WM performance, we conducted a regression analysis in the fMRI sample. Whole brain analysis revealed that only one activity cluster (MNI coordinates at maximum: -19.25 -66 52, Table S6) was associated significantly with WM performance at a whole brain corrected threshold (t=5.2, Pwhole-brain-FWE-corrected < 0.05). The correlation was positive. Importantly, this activity cluster was located at the left superior parietal cortex, the same region, which also showed significant gene score-dependent activations. No voxels correlated negatively with performance at an FWE-corrected level. Use of a liberal significance threshold (Puncorrected < 0.001) revealed the existence of several activation clusters correlating with WM performance in both directions (Table S6).
Discussion
We detected a robust association of the voltage-gated cation channel activity gene-set (Gene Ontology ID: GO:0022843) with WM and WM-dependent traits in both young and elderly cognitively healthy individuals in 4 independent samples. Despite substantial methodological differences between gene-set enrichment tools (e.g. statistical power and control for type I error), we could also demonstrate that the association of the voltage-gated cation channel activity gene-set with WM is significant independently of the applied algorithm. This finding is compatible with the physiological role of voltage-gated cation channel activity: Voltage-gated ion channels, and particularly cation channels, are the major determinants of the active electrical properties of neurons, such as excitability (Llinas, 1988), which are crucial for the proper function of the WM network (Goldman-Rakic, 1996). In accordance with our findings, single variation in genes of the voltage-gated cation channel activity gene-set, such as KCNH2, KCNB2 and CACNA1C has been recently associated with WM performance (Cirulli et al., 2010; Erk et al., 2010; Huffaker et al., 2009; Zhang et al., 2012). We would like to stress that the assumption of an exclusive role of this gene set in WM would be erroneous, given the properties of the encoded molecules and their localization throughout the human brain. On the one hand, we found evidence for genetic association of the voltage-gated cation channels only with WM-related phenotypes. However, we did not cover the entire spectrum of heritable neurocognitive traits. Thus, we estimate that the probability of the voltage-gated cation channels being associated with other neurocognitive traits, not tested herein, is high.
Interestingly, the voltage-gated cation channel activity gene-set was also associated with the risk for schizophrenia in a large dataset. WM deficits have been repeatedly observed in both psychiatric conditions (Barch, 2005). Importantly, neuronal excitability, which significantly depends on the proper function of voltage-gated cation channels, is a pharmacological target for schizophrenia (Rogawski and Loscher, 2004). Interestingly, recent GWAS identified single variations in calcium channel genes to be robustly associated with schizophrenia (2012; Ripke et al., 2013; Ripke et al., 2011). Furthermore, GSEA in a large sample of 33’332 cases and 27’888 controls support a role for the calcium channel gene-set for five major psychiatric disorders, including schizophrenia (Smoller et al., 2013). Interestingly, our study also identified a substantial overlap of significant genes encoding voltage-gated calcium channels across samples: CACNA1A, CACNA1B, CACNA1C, CACNA1D, CACNA1E, CACNA1S, CACNB1, CACNB2, and CACNB4 were significant in each of the studied cohorts, including the schizophrenia cohort (Table 2).
Table 2.
Overlap of significant voltage-gated cation channel genes between samples. Only genes found to be significant in the discovery sample are shown.
| Discovery sample | Replication sample | Zurich sample | AgeCoDe sample | PGC SchizophreniaandSweden 1-6 case-controlstudy |
|---|---|---|---|---|
| CACNA1A | CACNA1A | CACNA1A | CACNA1A | CACNA1A |
| CACNA1B | CACNA1B | CACNA1B | CACNA1B | CACNA1B |
| CACNA1C | CACNA1C | CACNA1C | CACNA1C | CACNA1C |
| CACNA1D | CACNA1D | CACNA1D | CACNA1D | CACNA1D |
| CACNA1E | CACNA1E | CACNA1E | CACNA1E | CACNA1E |
| CACNA1G | ||||
| CACNA1S | CACNA1S | CACNA1S | CACNA1S | CACNA1S |
| CACNA2D1 | CACNA2D1 | CACNA2D1 | CACNA2D1 | |
| CACNB1 | CACNB1 | CACNB1 | CACNB1 | CACNB1 |
| CACNB2 | CACNB2 | CACNB2 | CACNB2 | CACNB2 |
| CACNB4 | CACNB4 | CACNB4 | CACNB4 | CACNB4 |
| CACNG1 | CACNG1 | CACNG1 | ||
| CACNG4 | CACNG4 | |||
| KCNB2 | KCNB2 | |||
| KCNC1 | KCNC1 | KCNC1 | ||
| KCNH1 | KCNH1 | KCNH1 | ||
| KCNH3 | KCNH1 | |||
| KCNH4 | KCNH3 | |||
| KCNJ15 | ||||
| KCNJ3 | KCNJ3 | |||
| KCNJ6 | KCNJ6 | KCNJ6 | ||
| KCNQ1 | KCNQ1 | KCNQ1 | ||
| KCNQ3 | KCNQ3 | KCNQ3 | ||
| KCNQ5 | KCNQ5 | KCNQ5 | ||
| KCNS3 | KCNS3 | KCNS3 | ||
| SCN11A | SCN11A | |||
| SCN2A | ||||
| SCN5A | SCN5A |
fMRI data revealed that the individual genetic load of gene-set alleles associated with better WM performance correlated with WM-related activity in the parietal cortex and in the cerebellum. The location of the genotype-dependent activations is physiologically plausible, as extensive evidence from imaging studies indicates that both regions are crucially involved in working memory (Owen et al., 2005). Moreover, our finding supports the hypothesis that the cerebellum is involved in high-order cognitive tasks (Leiner et al., 1991; O’Halloran et al., 2012; Owen et al., 2005). Interestingly, abnormal activation of both brain regions reported herein has been observed in schizophrenia as compared to healthy controls (Barch, 2005; Konarski et al., 2005; Wiser et al., 1998).
In summary, our study supports a role of voltage-gated cation channels in WM and in psychiatric conditions characterized by WM deficits and points to a genetic link between voltage-gated cation channels and WM-related activation of the parietal cortex and the cerebellum. Gene-set analyses may help to uncover cognitive dimensions with different genetic and brain activation patterns, a subtyping that may be necessary to improve understanding and treatment of psychopathology.
Experimental procedures
Samples
Discovery sample
We recruited 905 healthy young Swiss adults (66.2% female; mean age: 22.53 ± 3.55) for a behavioral genetics study in the city of Basel, Switzerland. Subjects were free of any neurological or psychiatric condition and did not take medication at the time of the experiment. All participants gave written informed consent before participation and completed the 0- and 2-back version of the n-back task (Gevins and Cutillo, 1993). The task consists of 12 blocks (six 0-back, six 2-back), in which 14 test stimuli (letters) were presented. The 0-back condition required participants to respond to the occurrence of the letter “x” in a sequence of letters (e. g. N − l − X − g…) and served as a nonmemory-guided control condition, measuring general attention, concentration and reaction time. The 2-back condition required subjects to compare the currently presented letter with the penultimate letter to decide whether they are identical or not (e. g. S − f − s − g…). This task requires online monitoring, updating and manipulation of remembered information and is therefore assumed to involve key working memory-related processes. Performance was recorded as number of correct responses (accuracy), and the difference between 2-back and the 0-back condition served as core phenotype. Performance in the 0-back condition (mean accuracy and d’ values) served as phenotype reflecting attentional processes. Participants also performed verbal and picture delayed free recall tasks reflecting episodic and emotional memory performance. In the verbal task, subjects viewed six series of five semantically unrelated nouns presented at a rate of one word per second with the instruction to learn the words immediately after each series. Then, subjects underwent an unexpected delayed free-recall test of the learned words after 5 min. The number of correctly recalled words (hits) was the relevant output. In the picture task, subjects were presented with 24 neutral, 24 positive and 24 aversive photographs in a random order. The photographs were taken from the international affective picture system (IAPS) and were presented for 2.5 s each. Immediately following the presentation of each photograph, subjects were asked to rate it for valence and arousal using the IAPS rating scales. Free recall was tested 10 min after presentation of all photographs. To document performance for the delayed recall of positive, negative, and neutral pictures, subjects had to describe in writing each picture with a few words. A picture was judged as correctly recalled if the rater could identify the presented picture based on the subject’s description. Two blinded investigators independently rated the descriptions for recall success (inter-rater reliability > 99%). For the pictures, which were judged differently by the two raters (i.e. a particular picture was judged as correctly recalled by one rater but not the other), a third independent and blinded rater made a final decision with regard to whether the particular picture could be considered as successfully recalled.
Replication sample
We recruited 746 healthy young Swiss adults (59.7% female; mean age: 22.44 ± 3.49) for a functional brain imaging study in the city of Basel, Switzerland. Subjects were free of any neurological or psychiatric condition and did not take medication at the time of the experiment. All participants gave written informed consent before participation and completed the 0- and 2-back version of the n-back task (Gevins and Cutillo, 1993) (see also hypothesis testing sample). Participants of the replication sample performed the n-back task in the brain scanner. Complete imaging data was available for 707 participants of this sample. The ethics committee of the canton Basel approved the experiments of both the hypothesis testing and the replication sample.
Zurich sample
We recruited 410 healthy young Swiss adults (72.2% female; mean age: 21.19 ± 1.94) for a behavioral genetics study in the city of Zurich, Switzerland. Subjects were free of any neurological or psychiatric condition and did not take medication at the time of the experiment. All participants gave written informed consent before participation and completed the Wechsler digit span forward task (Wechsler, 1987). In this task, increasing numbers of orally presented digits at a rate of one per second had to be repeated in the same order. The test started with three digits. Every other trial the number of digits was increased by one. When errors in two consecutive trials were made, the test was ended and the number of correctly recalled trials was counted.
The ethics committee of the canton of Zurich approved the study protocol.
German AgeCoDe sample
This sample consisted of elderly participants of the German Study on Ageing, Cognition and Dementia in primary care patients (AgeCoDe). The AgeCoDe study is an ongoing primary care-based prospective longitudinal study on early detection of mild cognitive impairment and dementia established by the German Competence Network Dementia. The sampling frame and sample selection process of the AgeCoDe study have been described in detail previously (Luck et al., 2007). Briefly, participants were recruited between January 2003 and November 2004 in six German study centers (Bonn, Düsseldorf, Hamburg, Leipzig, Mannheim, Munich) via general practitioners (GP) connected to the respective study sites. Inclusion criteria were age of 75 years and older, absence of dementia (according to the GP’s judgment) and at least one contact with the GP within the last 12 months. Exclusion criteria were GP consultations by home visits only, residence in a nursing home, presence of a severe illness with an anticipated fatal outcome within three months, insufficient German language abilities, deafness or blindness, lack of ability to provide an informed consent and status as being only an occasional patient of the participating GP. A total of 3’327 subjects were successfully contacted and assessed with structured clinical interviews at their homes. A total of 110 individuals were excluded after the first interview due to presence of dementia or an actual age below 75 (falsely classified as 75 or older in the sample selection process). For the present analyses, data from baseline and three follow-up measurements with 18 months intervals were available. In a primary care-based sample of older individuals, conditions can be present that affect cognition and the reliability of neuropsychological tests. In order to generate a sample of healthy elderly individuals we further employed the following selection criteria at baseline: Age between 75 and 90 years, German as native language, at least school-leaving certificate, absence of severe hearing or vision impairments, absence of insufficient test motivation as judged by the interviewer, absence of disturbing factors during neuropsychological testing and absence of all of the following comorbid conditions: Parkinson’s disease, epilepsy, alcohol abuse, stroke, multiple sclerosis, evidence of depression (a score of 6 or higher on the Geriatric Depression Scale), traumatic brain injury with unconsciousness of more than 30 minutes, visible neurological malfunctions and dementia according to DSM-IV criteria. In addition, we excluded subjects who converted to dementia up to the third follow-up or without neuropsychological test data available on baseline and all follow-up visits. After application of these selection criteria, a total of 1244 subjects remained in the sample. Sufficient DNA-samples for genome-wide genotyping were available for 782. Further 19 subjects were excluded due to sex-check inconsistencies. The final sample comprises 763 subjects (67.8% female; mean age: 79.5 ± 3.02). Immediate recall performance as quantified by the Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) battery (Welsh et al., 1994) served as phenotype. Subjects were presented a list of 10 words three times (presentation per word: 2 seconds), each time presented in a different order. After each run, subjects freely recalled as many words as possible. The number of correctly remembered items over the three runs served as the phenotypic measure. Immediate verbal free recall does not represent WM. Nevertheless, WM capacity is one of the cognitive components necessary to perform this task.
Genetic heterogeneity
For each of the four cognitively healthy samples with genotypes available, the genomic control inflation factor lambda (λGC) was calculated to assess admixture in our samples. Lambda is defined as the median χ2 association test statistic divided by the theoretical distribution under the null distribution (Devlin and Roeder, 1999). λGC showed a range between 0.993 and 1.047, indicating the absence or only minor admixture in our samples (see Figure S1, A-D; (Ge et al., 2008)). We also performed additional analyses in these cohorts to exclude the possibility that genetic admixture biased the results of the GSEA. Hence, we ran principal component analysis (PCA) with the GCTA software on the full GWAS data sets and extracted 20 principal components (PCs). The algorithm implemented in GCTA is equivalent to the one used in EIGENSTRAT. Each subject was assigned an individual load for each of the 20 PC axes. These values together with the individual genetic score served as independent variables in forward and backward linear regression analyses, with phenotypic performance as the dependent variable. In none of these analyses did we detect any effect of genetic stratification (as represented by the PCs) on the correlation between allele load and memory performance.
Array-based SNP genotyping
Samples were processed as described in the Genome-Wide Human SNP Nsp/Sty 6.0 User Guide (Affymetrix). Briefly, genomic DNA concentration was determined by using a Nano-Drop ND-1000 and adjusted to 50 ng/μl in water. 250 ng of DNA was digested in parallel with 10 units of Sty I and Nsp I restriction enzymes (New England Biolabs, Beverly, MA) for 2 hours at 37°C. Enzyme specific adaptor oligonucleotides were then ligated onto the digested ends with T4 DNA Ligase for 3 hours at 16°C. After adjustment to 100 μl with water, 10 μl of the diluted ligation reactions were subjected to PCR. Three PCR reactions of 100 μl were performed for Sty digested products and four PCR reactions for Nsp. PCR was performed with Titanium Taq DNA Polymerase (Clontech, Mountain View, CA) in the presence of 4.5 μM PCR primer 002 (Affymetrix), 350 μM each dNTP (Clontech), 1M G-C Melt (Clontech), and 1X Titanium Taq PCR Buffer (Clontech). Cycling parameters were as follows: initial denaturation at 94°C for 3 minutes, amplification at 94°C for 30 seconds, 60°C for 45 seconds and extension at 68°C for 15 seconds repeated a total of 30 times, final extension at 68°C for 7 minutes. Reactions were then verified to migrate at an average size between 200-1100 bps using 2% TBE gel electrophoresis. PCR products were combined and purified with the Filter Bottom Plate (Seahorse Bioscience, North Billerica, MA) using Agencourt Magnetic Beads (Beckman Coulter, Fullerton, CA). Purified PCR products were quantified on a Zenith 200rt microplate reader (Anthos-Labtec, Cambridge, UK). 4 to 5 μg/μl were obtained on average for each sample. From this stage on, the SNP Nsp/Sty 5.0/6.0 Assay Kit (Affymetrix) was used. Around 250 μg of purified PCR products were fragmented using 0.5 units of DNAse I at 37°C for 35 minutes. Fragmentation of the products to an average size less than 180 bps was verified using 4% TBE gel electrophoresis. Following fragmentation, the DNA was end labeled with 105 units of terminal deoxynucleotidyl transferase at 37°C for 4 hours. The labeled DNA was then hybridized onto Genome-Wide Human SNP 6.0 Array at 50°C for 18 hours at 60 rpm. The hybridized array was washed, stained, and scanned according to the manufacturer’s (Affymetrix) instructions using Affymetrix GeneChip Command Console (AGCC, version 3.0.1.1214). Generation of SNP calls and Array quality control were performed using the command line programs of the Affymetrix Power Tools package (version: apt-1.12.0). According to the manufacturer’s recommendation, Contrast QC was chosen as QC metric, using the default value of greater or equal than 0.4. Mean Call Rate for all samples averaged >98.5%. This value refers to per sample (i.e. individual) call rate, and ranged from 95.1% to 99.7%. Thus, no individual with a SNP call rate below 95% was included. All samples passing QC criteria were subsequently genotyped using the Birdseed (v2) algorithm.
fMRI data acquisition and processing (see also replication sample)
Measurements were performed on a Siemens Magnetom Verio 3 T wholebody MR unit equipped with a twelve-channel head coil. Functional time series were acquired with a single-shot echo-planar sequence using parallel imaging (GRAPPA). We used the following acquisition parameters: TE (echo time) = 35 ms, FOV (field of view) = 22 cm, acquisition matrix = 80 × 80, interpolated to 128 × 128, voxel size: 2.75 × 2.75 × 4 mm3, GRAPPA acceleration factor R = 2.0. Using a midsaggital scout image, 32 contiguous axial slices placed along the anterior–posterior commissure (AC–PC) plane covering the entire brain with a TR = 3000 ms (α = 82°) were acquired using an ascending interleaved sequence. A high-resolution T1-weighted anatomical image was acquired using a magnetization prepared gradient echo sequence (MPRAGE, TR = 2000 ms; TE=3.37 ms; TI=1000 ms; flip angle=8; 176 slices; FOV= 256 mm; voxel size = 1 × 1 × 1 mm3).
Preprocessing and data analysis was performed using SPM8 (Statistical Parametric Mapping, Wellcome Trust Centre for Neuroimaging, London, UK; http://www.fil.ion.ucl.ac.uk/spm/) implemented in Matlab R2011b (The Mathworks Inc., Natick, MA, USA). Volumes were slice-time corrected to the first slice and realigned using the ‘register to mean’ option. A mean image was generated from the realigned series and coregistered to the structural image. This ensured that functional and structural images were spatially aligned.
The functional images and the structural images were spatially normalized by applying DARTEL, which leads to an improved registration between subjects (Ashburner, 2007; Klein et al., 2009). Normalization incorporated the following steps: 1. Structural images of each subject were segmented using the “New Segment” procedure in SPM8. 2. The resulting gray and white matter images were used to derive a study-specific group template. The template was computed from a larger population of 1’000 subjects, which included the 707 subjects of the present study (Basel replication sample). 3. An affine transformation was applied to map the group template to MNI space. 4. Subject-to-template and template-to-MNI transformations were combined to map the functional images to MNI space. The functional images were smoothed with an isotropic 8 mm full width at half maximum (FWHM) Gaussian filter.
Intrinsic autocorrelations were accounted for by AR(1) and low-frequency drifts were removed via high-pass filter (time constant 128 s). Separate regressors were constructed for the 0- and 2-back conditions comprising a boxcar reference waveform convolved with a canonical hemodynamic response function. Events during the presentation of the instruction as well as movement regressors from spatial realignment were modeled separately.
fMRI contrasts and analyses
To investigate neural correlates of association with working memory load, the contrast between brain activity during performance of the 2-back versus 0-back was calculated individually using a fixed effects model (first level analysis). The resulting contrast parameters were then used for genotype-dependent analyses in a random effects model (second level analysis). Specifically, we used a regression model to analyze differences in brain activity, whereas the individual genetic multilocus score served as covariate in our analysis. We controlled for the effect of sex by including it as a covariate. The significance threshold for score-dependent analysis was set at P < 0.05, FWE-corrected for multiple comparisons in the whole brain.
To investigate neural correlates of association with episodic memory load, subjects also performed a picture task. Stimuli consisted of 72 pictures that were selected from the IAPS as well as from in-house standardized picture sets that allowed us to equate the pictures for visual complexity and content (e.g., human presence). On the basis of normative valence scores, pictures were assigned to emotionally negative, emotionally neutral, and emotionally positive conditions, resulting in 24 pictures for each emotional valence. Negative and positive pictures were equated for valence extremity. Four additional pictures showing neutral objects were used to control for primacy and recency effects in memory. Two of these pictures were presented in the beginning and two at the end of the picture task. They were not included in the analysis. In addition, 24 scramble pictures were used. The background of the scramble pictures contained the color information of all pictures used in the experiment (except primacy and recency pictures), over layed with a crystal and distortion filter (Adobe Photoshop CS3). In the foreground, a mostly transparent geometrical object (rectangle or ellipse of different sizes and orientations) was shown. Pictures were presented in the scanner by using MR-compatible LCD goggles (Visuastim XGA; Resonance Technology). Eye correction was used when necessary. At the encoding phase, pictures were presented for 2.5 seconds in a quasirandomized order so that at maximum, four pictures of the same category occurred consecutively. A fixation cross appeared on the screen for 500 msec before each picture presentation. Trials were separated by a variable intertrial period of 9–12 sec (jitter) that was equally distributed for each stimulus category. During the intertrial period, participants subjectively rated the picture showing scenes according to valence (negative, neutral, positive) and arousal (large, medium, small) on a three-point scale (Self Assessment Manikin) by pressing a button with a finger of their dominant hand. For scramble pictures, participants rated form (vertical, symmetric, or horizontal) and size (large, medium, small) of the geometrical object in the foreground. The encoding phase of the picture had a total duration of 22 min. Participants were not told that they had to remember the pictures for later recall. Participants were instructed and trained on the picture task before being positioned in the scanner. Training consisted of presentation and rating of five pictures, including scenes and scrambled pictures, which were not used during scanning. The event-related fMRI procedure allowed for investigation of brain regions involved in memory formation by analyzing differential activity during encoding of subsequently remembered vs. subsequently forgotten events (subsequent memory analysis, “difference due to memory,” or Dm). The contrast used was Dmremembered vs. Dmforgotten.
fMRI group statistics
EPI sequences suffer from signal loss in the presence of magnetic field in homogeneities that can occur close to air-tissue boundaries. The normalization procedure applied in DARTEL accurately transforms both voxels with signal and voxels with signal loss to MNI space. In SPM8, signal loss at a MNI coordinate in a functional image of only one subject leads to the exclusion of the voxel at this coordinate from the group level analysis. Therefore, the probability of a voxel being excluded increases with sample size. GLM Flex circumvents this problem by allowing a variable number of subjects at each voxel, (Martinos Center & Mass General Hospital, Charlestown, MA, USA; http://nmr.mgh.harvard.edu/harvardagingbrain/People/AaronSchultz/Aarons_Scripts.html). The minimum number of subjects per voxel was set to be 600.
Construction of a population-average anatomical probabilistic atlas
Automatic segmentation of the subjects’ T1-weighted images was used to build a population-average probabilistic anatomical atlas. More precisely, each participant’s T1-weighted image was first automatically segmented into cortical and subcortical structures using FreeSurfer (version 4.5, http://surfer.nmr.mgh.harvard.edu/) (Fischl et al., 2002). Labeling of the cortical gyri was based on the Desikan-Killiany Atlas (Desikan et al., 2006), yielding 35 regions per hemisphere. The segmented T1 image was then normalized to the study-specific anatomical template space using the subject’s previously computed warp field, and affine-registered to the MNI space. Nearest-neighbor interpolation was applied, in order to preserve labeling of the different structures. The normalized segmentations were finally averaged across subjects, in order to create a population-average probabilistic atlas. Each voxel of the template could consequently be assigned a probability of belonging to a given anatomical structure, based on the individual information from 1’000 subjects.
Mediator analysis
Mediator analysis (Baron and Kenny, 1986) was performed in R to examine whether activity in the brain regions identified as being related to the multilocus genetic score mediated the genetic association between voltage-gated cation channel genes and WM performance. Of note, the correlation between the genetic score and brain activity (i.e. individual activation values (beta values) at the reported peaks) was highly significant (r=0.18, P=3.2 × 10-6 in the left superior parietal and supramarginal cortex with peak activation at [-16.5 -66 52]; r=0.17, P=6.1 × 10-6 in the right cerebellum with peak activation at [13.75 -55 -48]), but at a considerably lower effect size than that of the correlation between the score and WM performance (r=0.43, P=3.2 × 10-31, see also Figure S2). The effect size of the latter correlation is due to the nature of the multilocus genetic score, which represents the total load with alleles selected a priori for their significant association with WM performance. Because of this large difference in effects sizes, the prerequisites for a mediator analysis are sub-optimal. The analysis combined a series of linear models thereby testing whether the influence of the independent variable gene-score on the dependent variable working memory performance (model c) is mediated by the brain (fMRI) activity (model c’). In all models sex was included as covariate.
Statistical analysis
Genome-wide association analyses
For each genome-wide analysis, P values were obtained using the Wald test as implemented in PLINK (Purcell et al., 2007). We applied the following quality control criteria: Non-significant deviation from Hardy-Weinberg equilibrium (HWE; p(HWE>0.0001)), a minor allele frequency (MAF)>0.01 and a per SNP genotype call rate>90%. Mean per SNP call rate was 99.4% with 1.7% of the SNPs reaching a call rate < 95%. The resulting P values of the GWAS (one list per sample) served as input for the GSEA analysis described beneath. Importantly, the mean per SNP call rate of the significant SNPs constituting the voltage-gated cation channel gene set was 99.7% with only 1 SNP reaching a call rate < 95%. Thus, there is no evidence for bias towards lower quality SNPs.
GSEA analysis
GSEA was performed using MAGENTA developed by Broad Institute (Segre et al., 2010). Briefly, the method first maps SNPs onto genes and then assigns each gene a SNP association score (i.e. the maximum SNP p-value within ±0kb of the annotated gene). By applying a step-wise multiple linear regression analysis, the analysis is corrected for the following confounders: gene size, number of SNPs, number of independent SNPs, number of recombination hotspots, linkage disequilibrium and genetic distance (measured in centiMorgans). Lastly, a gene-set enrichment-like statistical test is applied to determine if a gene set is enriched for highly ranked P values compared to a gene-set of identical size, randomly drawn from the genome. False-discovery rate (FDR) based on the 75th percentile of association P values from all genes was used for multiple testing correction. As recommended in the original paper, we used the 75th percentile cutoff instead of the 95th percentile cutoff, since it has the optimal power for weak genetic effects that are expected for highly polygenic traits (e.g., WM performance) (Segre et al., 2010). The utilized gene sets are extracted and curated from the MSigDB v3.1 database (http://www.broadinstitute.org/gsea/msigdb), including gene-sets from different online databases (KEGG, Gene Ontology GO, BioCarta and Reactome (Ashburner et al., 2000; Ogata et al., 1999)). We used a gene set size ranging between 20 and 200 genes to avoid both overly narrow and broad functional gene-set categories, resulting in 1’411 to be analyzed gene-sets. Furthermore, to reduce bias, genes mapping in the high linkage disequilibrium (LD) extended major histocompatibility complex were excluded from the respective gene-sets (Horton et al., 2004).
Alternative gene-set identification tools
GSA-SNP
We also applied GSA-SNP (http://gsa.muldas.org/) for the identification of gene-sets. GSA-SNP is a JAVA application that, comparable with i-GSEA4 GWAS, uses SNP P values as input and identifies gene-sets in a competitive way (Nam et al., 2010). GSA-SNP calculates gene set enrichment scores by calculating a Z-statistic out of effect size measures (i.e. GWAS p-values). Instead of using the maximum effect per gene as a proxy for the respective gene, GSA-SNP has the option to choose the 2nd, third, fourth and fifth best p-value to represent the effect of that gene. The application allocates Gene Ontology gene-sets as the default database. Similar to the MAGENTA analyses, we set the gene-set size between 20 and 200 genes.
INRICH
INRICH (http://atgu.mgh.harvard.edu/inrich/) is a software tool that examines enrichment of association signals for genetic gene-sets (Lee et al., 2012). INRICH is robust to potential confounders (i.e. varying gene and gene-set sizes and LD structure of GWAS SNPs). The INRICH algorithm counts the number of overlaps between independent intervals (clumping threshold in PLINK was set to P < 0.001, interval size ≤ 250kb and r2 = 0.2) of LD, harboring the strongest association signals of a GWAS and predefined gene-sets. This overlap (i.e. a real number) is then corrected with permutation-based methods, by randomly drawing intervals, which match the observed intervals in size, and then calculating an empirical p-value as percentage of random intervals showing at least same overlap as the observed overlap. The empirical p-value is corrected for the number of gene-sets that have been tested in the same analysis via bootstrapping. First, we used the – clump option in PLINK to define independent intervals harboring the best GWAS association signals. We ran INRICH with 100’000 permutations and 10’000 bootstrapping samples. As in MAGENTA, the target gene borders were set to ±0kb, and the gene-set size ranged between 20 and 200 target genes. Finally, 3’170 gene-sets provided by the INRICH software (MsigDB v3.0) were tested.
Multilocus genetic score calculation
To capture the multi-allelic effect of GO:0022843, we generated an individual multilocus genetic score using the scoring procedure implemented in PLINK (Purcell et al., 2007). The score comprises 29 SNPs from an equal number of those pathway genes, which were significant in the replication sample (see Table S4). The PLINK algorithm calculates the score by summing up the individual number of reference alleles over all (i.e. 29) SNPs, weighted by the direction of effect on WM performance with “1” (the reference allele enhances WM performance) or “-1” (the reference allele decreases WM performance), and finally averages the score by the number of non-missing SNPs.
Supplementary Material
Highlights.
The voltage-gated cation channel activity gene-setis linked to working-memory.
The association is robust in both young and elderly cognitively healthy individuals.
The association is also observed in schizophrenia.
The gene-set is linked to brain activity in the parietal and cerebellar cortex.
Acknowledgments
We thank Elmar Merkle, Christoph Stippich and Oliver Bieri for granting access to the fMRI facilities of the University Hospital Basel.
Funding: This work was funded by the Swiss National Science Foundation (Sinergia grant CRSI33_130080 to D.Q. and A.P.), by the 7th framework programme of the European Union (ADAMS project, HEALTH-F4-2009-242257), by the National Center for Competence in Research SYNAPSY, and by the German Research Network on Dementia (KND) and the German Research Network on Degenerative Dementia (KNDD), German Federal Ministry of Education and Research grants 01GI0420 and 01GI0711.
Footnotes
Author contributions: A.H., M.F., H.B., F.J., H.K., W.M., P.S., S.R., S.R-H, M.W., S.We., D. J.-F. d. Q. & A.P. designed and conceptualized the study. A.H., M.F., B.A., D.C., L.G., F.J., H.K., A.M., M.P., S.R., S.R-H, P.S., C.V., M.W., S. We., S. Wo. & A.P. did the data analysis. S.A., H.B., W.M., M.P., K.S., M.W., S.We. & S.Wo. were responsible for data collection and preparation. D. J.-F. d. Q. & A.P. supervised the project. All authors contributed to the writing of the manuscript.
References and Notes
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balanza-Martinez V, Rubio C, Selva-Vera G, Martinez-Aran A, Sanchez-Moreno J, Salazar-Fraile J, Vieta E, Tabares-Seisdedos R. Neurocognitive endophenotypes (endophenocognitypes) from studies of relatives of bipolar disorder subjects: a systematic review. Neurosci Biobehav Rev. 2008;32:1426–1438. doi: 10.1016/j.neubiorev.2008.05.019. [DOI] [PubMed] [Google Scholar]
- Barch DM. The cognitive neuroscience of schizophrenia. Annu Rev Clin Psychol. 2005;1:321–353. doi: 10.1146/annurev.clinpsy.1.102803.143959. [DOI] [PubMed] [Google Scholar]
- Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986;51:1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- Cirulli ET, Kasperaviciute D, Attix DK, Need AC, Ge D, Gibson G, Goldstein DB. Common genetic variation and performance on standardized cognitive tests. Eur J Hum Genet. 2010;18:815–820. doi: 10.1038/ejhg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Esposito M. From cognitive to neural models of working memory. Philos Trans R Soc Lond B Biol Sci. 2007;362:761–772. doi: 10.1098/rstb.2007.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Doyle AE. Executive functions in attention-deficit/hyperactivity disorder. J Clin Psychiatry. 2006;67(Suppl 8):21–26. [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Schnell K, Opitz von Boberfeld C, Esslinger C, Kirsch P, Grimm O, Arnold C, Haddad L, Witt SH, et al. Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch Gen Psychiatry. 2010;67:803–811. doi: 10.1001/archgenpsychiatry.2010.94. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Ge D, Zhang K, Need AC, Martin O, Fellay J, Urban TJ, Telenti A, Goldstein DB. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevins A, Cutillo B. Spatiotemporal dynamics of component processes in human working memory. Electroencephalogr Clin Neurophysiol. 1993;87:128–143. doi: 10.1016/0013-4694(93)90119-g. [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic PS. Regional and cellular fractionation of working memory. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13473–13480. doi: 10.1073/pnas.93.24.13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Owen MJ, O’Donovan MC, Craddock N. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- Huffaker SJ, Chen J, Nicodemus KK, Sambataro F, Yang F, Mattay V, Lipska BK, Hyde TM, Song J, Rujescu D, et al. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat Med. 2009;15:509–518. doi: 10.1038/nm.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt KH, Bachman P, Winkler AM, Bearden CE, Glahn DC. Genetic influence on the working memory circuitry: behavior, structure, function and extensions to illness. Behavioural brain research. 2011;225:610–622. doi: 10.1016/j.bbr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarski JZ, McIntyre RS, Grupp LA, Kennedy SH. Is the cerebellum relevant in the circuitry of neuropsychiatric disorders? J Psychiatry Neurosci. 2005;30:178–186. [PMC free article] [PubMed] [Google Scholar]
- Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner T. Genome-Wide Association Study Implicates HLA-C*01:02 as a Risk Factor at the Major Histocompatibility Complex Locus in Schizophrenia. Biological Psychiatry. 2012;72:620–628. doi: 10.1016/j.biopsych.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiner HC, Leiner AL, Dow RS. The human cerebro-cerebellar system: its computing, cognitive, and language skills. Behavioural brain research. 1991;44:113–128. doi: 10.1016/s0166-4328(05)80016-6. [DOI] [PubMed] [Google Scholar]
- Llinas RR. The intrinsic electrophysiological properties of mammalian neurons: insights into central nervous system function. Science. 1988;242:1654–1664. doi: 10.1126/science.3059497. [DOI] [PubMed] [Google Scholar]
- Luck T, Riedel-Heller SG, Kaduszkiewicz H, Bickel H, Jessen F, Pentzek M, Wiese B, Koelsch H, van den Bussche H, Abholz HH, et al. Mild cognitive impairment in general practice: age-specific prevalence and correlate results from the German study on ageing, cognition and dementia in primary care patients (AgeCoDe) Dement Geriatr Cogn Disord. 2007;24:307–316. doi: 10.1159/000108099. [DOI] [PubMed] [Google Scholar]
- Nam D, Kim J, Kim SY, Kim S. GSA-SNP: a general approach for gene set analysis of polymorphisms. Nucleic Acids Res. 2010;38:W749–754. doi: 10.1093/nar/gkq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Dushlaine C, Kenny E, Heron E, Donohoe G, Gill M, Morris D, Corvin A. Molecular pathways involved in neuronal cell adhesion and membrane scaffolding contribute to schizophrenia and bipolar disorder susceptibility. Mol Psychiatry. 2011;16:286–292. doi: 10.1038/mp.2010.7. [DOI] [PubMed] [Google Scholar]
- O’Halloran CJ, Kinsella GJ, Storey E. The cerebellum and neuropsychological functioning: a critical review. J Clin Exp Neuropsychol. 2012;34:35–56. doi: 10.1080/13803395.2011.614599. [DOI] [PubMed] [Google Scholar]
- Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen AM, McMillan KM, Laird AR, Bullmore E. N-back working memory paradigm: a meta-analysis of normative functional neuroimaging studies. Hum Brain Mapp. 2005;25:46–59. doi: 10.1002/hbm.20131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papassotiropoulos A, de Quervain DJ. Genetics of human episodic memory: dealing with complexity. Trends Cogn Sci. 2011;15:381–387. doi: 10.1016/j.tics.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasch B, Spalek K, Buholzer S, Luechinger R, Boesiger P, Papassotiropoulos A, de Quervain DJ. A genetic variation of the noradrenergic system is related to differential amygdala activation during encoding of emotional memories. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19191–19196. doi: 10.1073/pnas.0907425106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature genetics. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, et al. Genome-wide association study identifies five new schizophrenia loci. Nature genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogawski MA, Loscher W. The neurobiology of antiepileptic drugs for the treatment of nonepileptic conditions. Nat Med. 2004;10:685–692. doi: 10.1038/nm1074. [DOI] [PubMed] [Google Scholar]
- Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Miyake A. Models of Working Memory: Mechanisms of Active Maintenance and Executive Control. In: Miyake A, Shah P, editors. Models of Working Memory: An Introduction. Cambridge: Cambridge University Press; 1999. pp. 1–27. [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, Edenberg HJ, Nurnberger JI, Jr, Rietschel M, Blackwood D, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature genetics. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Craddock N, Kendler K, Lee PH, Neale BM, Nurnberger JI, Ripke S, Santangelo S, Sullivan PF. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiakouli E, Hamshere M, Holmans P, Langley K, Zaharieva I, Hawi Z, Kent L, Gill M, Williams N, Owen MJ, et al. Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry. 2012;169:186–194. doi: 10.1176/appi.ajp.2011.11040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD, Smith EE. Neuroimaging studies of working memory: a meta-analysis. Cogn Affect Behav Neurosci. 2003;3:255–274. doi: 10.3758/cabn.3.4.255. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- Wang L, Jia P, Wolfinger RD, Chen X, Zhao Z. Gene set analysis of genome-wide association studies: methodological issues and perspectives. Genomics. 2011;98:1–8. doi: 10.1016/j.ygeno.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. Wechsler Memory Scale-Revised Manual. New York, NY: Psychological Corp; 1987. [Google Scholar]
- Welsh KA, Butters N, Mohs RC, Beekly D, Edland S, Fillenbaum G, Heyman A. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part V. A normative study of the neuropsychological battery. Neurology. 1994;44:609–614. doi: 10.1212/wnl.44.4.609. [DOI] [PubMed] [Google Scholar]
- Wiser AK, Andreasen NC, O’Leary DS, Watkins GL, Boles Ponto LL, Hichwa RD. Dysfunctional cortico-cerebellar circuits cause ‘cognitive dysmetria’ in schizophrenia. Neuroreport. 1998;9:1895–1899. doi: 10.1097/00001756-199806010-00042. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Shen Q, Xu Z, Chen M, Cheng L, Zhai J, Gu H, Bao X, Chen X, Wang K, et al. The effects of CACNA1C gene polymorphism on spatial working memory in both healthy controls and patients with schizophrenia or bipolar disorder. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2012;37:677–684. doi: 10.1038/npp.2011.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


