Abstract
Monascorubrin and its derivatives are polyketides used as natural colorants for a wide range of food for more than one thousand years. Since the biosynthetic pathway for this ancient chemical compound is unknown and genome sequence unavailable for any Monascus species, monascorubrin production has relied on extraction from fungal cultures of Monascus species. In vitro synthesis and genetic manipulation are not possible. Here we report the polyketide gene cluster and pathway for monascorubrin biosynthesis in Penicillium marneffei, a diffusible red pigment-producing, thermal dimorphic fungus, taking advantage of available genome sequence and faster growth rate than Monascus species. We also documented that the red pigment of P. marneffei is a mixture of more than 16 chemical compounds, which are amino acid conjugates of monascorubrin and rubropunctatin, and showed that this polyketide gene cluster and pathway are also responsible for biosynthesis of ankaflavin and citrinin, a mycotoxin with nephrotoxic activity in mammals. The present study on elucidation of the biosynthetic pathway of monascorubrin is a proof-of-the-concept study that serves as a cornerstone for future studies on monascorubrin biosynthesis pathway dissection in Monascus species.
Monascorubrin and its related compounds are polyketides that have been used as natural red colorants for a wide range of food, such as red wines, tofu and meats for more than one thousand years in China, Japan and other Southeast Asian countries1. The earliest known reference to use these red colorants was in a recipe for red pot-roast lamb, in which meat was simmered with hong qu (red rice koji, made with Monascus purpureus), recorded in the Qing Yilu in AD 9652. In Pen Ts'ao Kang Mu (Compendium of Materia Medica), the most complete and comprehensive herbal medicine book ever written in the history of traditional Chinese medicine, complied by Li Shi-zhen in the 16th century, he described the utilization of these red pigments as a coloring agent and as a medicine in the treatment of medical diseases3. Subsequently, these red pigments were also used in the western world in the meat industry, for coloring sausages and hams4. In 2005, more than 50 patents have been issued in Japan, America, France and Germany, on the use of Monascus pigments for food; with an annual consumption of these pigments in Japan increased from 100 tons in 1981 to 600 tons by the end of the 1990s5. Throughout the years, production of monascorubrin and its related compounds has relied on extraction from cultures of fungi of the genus Monascus, most commonly M. purpureus. The chemical pathway for monascorubrin biosynthesis has never been elucidated in any fungal species. Moreover, no genome sequence is available for any Monascus species. Therefore, in vitro synthesis and manipulation by genetic engineering are not possible. Citrinin, another polyketide and mycotoxin with nephrotoxic activity in mammals, was found to be present in the cultures of many Monascus species, including M. purpureus4,6,7. Regulations have been set up to limit the content of citrinin in the pigments.
Penicillium marneffei is the most important thermal dimorphic fungus causing respiratory, skin and systemic mycosis8,9,10,11,12,13. Biologically, it is one of the most important causes of opportunistic mycosis in HIV positive patients in China and Southeast Asia. P. marneffei produces black, yellow and red pigments. In addition to its thermal dimorphic property, the production of the characteristic diffusible red pigment by its mold form is the most important characteristic for laboratory identification of P. marneffei. Recently, we have sequenced the genome of P. marneffei14,15,16. Interestingly, 23 putative PKS genes and two putative PKS-non-ribosomal peptide synthase hybrid genes were identified in the P. marneffei genome, a diversity much higher than those of other pathogenic thermal dimorphic fungi, such as Histoplasma capsulatum (one PKS gene) and Coccidioides immitis (10 PKS genes)17. We have also characterized the black and yellow pigments of P. marneffei, which are synthesized by one and two of the 25 PKS genes respectively17,18. Due to its relatively fast growth rate, which is twice as fast as M. purpureus, and the availability of genome sequence, it is an ideal model fungus for fungal genetic studies.
Since monascorubramine has been found in cultures of P. marneffei6, we hypothesized that it is the red diffusible pigment of P. marneffei and it is synthesized by one of the PKS gene clusters in its genome. To test the hypothesis, we systematically knocked down all 25 PKS genes of P. marneffei. We also knocked down genes upstream and downstream to the PKS gene responsible for red pigment production and characterized the pathway for biosynthesis of the red pigment. Uniquely, the diffusible red pigment of P. marneffei is a mixture of more than 16 chemical compounds and these compounds as well as ankaflavin and citrinin were synthesized by the same PKS gene cluster.
Results
Red pigment biosynthesis gene cluster in P. marneffei is composed of pks3 and four neighboring genes
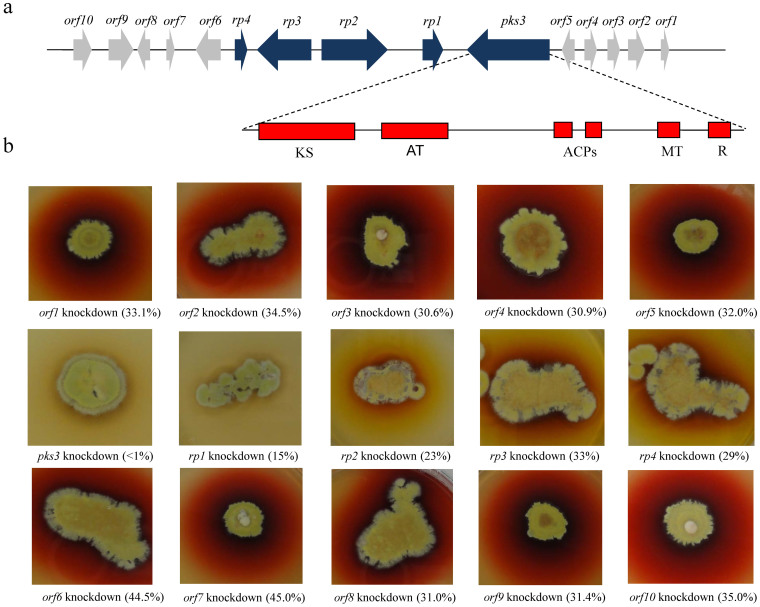
All 25 PKS genes of P. marneffei were systematically knocked down. A loss of the red pigment was observed exclusively in the mold form of the pks3 knockdown mutant (Fig. 1), which has a pks3 transcription level < 1% of that in wild type. The pks3 gene is 8104 bp in length. It has one intron of 52 bp (from 619 to 670 bp). The resultant putative mRNA encodes 2684 amino acid residues with a predicted molecular mass of 293.68 kDa. PKS3 has one ketosynthase, one acyltransferase, two acyl carrier proteins, one methyltransferase and one thiolester reductase domains (Fig. 1). This domain organization is consistent with clade III multi-aromatic ring PKS among fungal non-reducing PKSs under Kroken et al. classification19.
Figure 1. Red pigment biosynthesis gene cluster in P. marneffei.
(a) Each arrow indicates the direction of transcription and relative sizes of the ORFs. The domain structure of pks3 is shown. (b) Each knockdown mutant was grown on Sabouraud dextrose agar after 7 days incubation at 25°C. The degrees of the silencing transcriptional levels of the respective genes in knockdown mutants are indicated in brackets. ACP, acyl carrier protein; AT, acyltransferase; KS, ketosynthase; MT, methyltransferase; R, thioester reductase.
Five ORFs upstream (orf1 to orf5) and nine ORFs downstream (rp1 to rp4 and orf6 to orf10) to pks3 were systematically knocked down (Fig. 1). RT-PCR confirmed that the 14 ORFs were successfully knocked down (Supplementary Fig. S1). A loss of the red pigment was observed in the mold form of four of the knockdown mutants (Fig. 1). These four ORFs (rp1 to rp4) were immediately downstream to pks3, encoding putative transcriptional activator, fatty synthase beta subunit, 3-oxoacyl-[acyl-carrier-protein] synthase and oxidoreductase respectively (Table 1). Notably, knockdown mutants of rp2, rp3 and rp4 possessed diffusible yellow pigment (Fig. 1). Quantitative real-time RT-PCR showed that the expression levels of the pks3, rp1, rp2, rp3 and rp4 genes in the pks3, rp1, rp2, rp3 and rp4 knockdown mutants were <1%, 15%, 23%, 33% and 29% respectively (Supplementary Table S1).
Table 1. Sequence analysis of genes identified in red pigment biosynthetic cluster in Penicillium marneffei.
| Gene | Size (bp/aa) | Plausible function* | Match from BLASTP search at NCBI (accession number) | Amino acid identity (%) |
|---|---|---|---|---|
| rp1 | 2039/602 | Transcriptional activator | Monascus purpureus citrinin transcriptional activator (BAE95337.1) | 44 |
| rp2 | 6335/2045 | Fatty acid synthase beta subunit | Talaromyces stipitatus fatty acid synthase beta subunit (XP 002340041.1) | 80 |
| rp3 | 5104/1629 | 3-oxoacyl-[acyl-carrier-protein] synthase in fatty acid synthesis | Talaromyces stipitatus 3-oxoacyl-[acyl-carrier-protein] synthase (XP 002340042.1) | 81 |
| rp4 | 1110/369 | Oxidoreductase | Exophiala dermatitidis oxidoreductase (EHY54423.1) | 52 |
*The plausible function of each encoded protein was deduced from the protein homolog of the BLASTP search and its functional domains.
By UV-Vis spectroscopic analysis, four absorption maxima, 373, 405, 499 and 553 nm, were recognized in the culture medium growing the mold form of wild type P. marneffei (Supplementary Table S2). The absorption maximum at 499 nm, which indicated the presence of red color, was not observed in the culture medium growing the mold form of the pks3, rp1, rp2, rp3 and rp4 knockdown mutants and the RPMI medium control. Furthermore, the absorption maxima at 373 nm and 405 nm, which indicated the presence of yellow color, were found in the wild type and rp2, rp3 and rp4 knockdown mutants but not observed in the culture medium growing pks3 and rp1 knockdown mutants. The results were in line with the color observed when the strains were cultured on Sabouraud dextrose agar (Fig. 1). For the other ten knockdown mutants (orf1-orf10), the measured absorption maxima were the same as that of wild type P. marneffei.
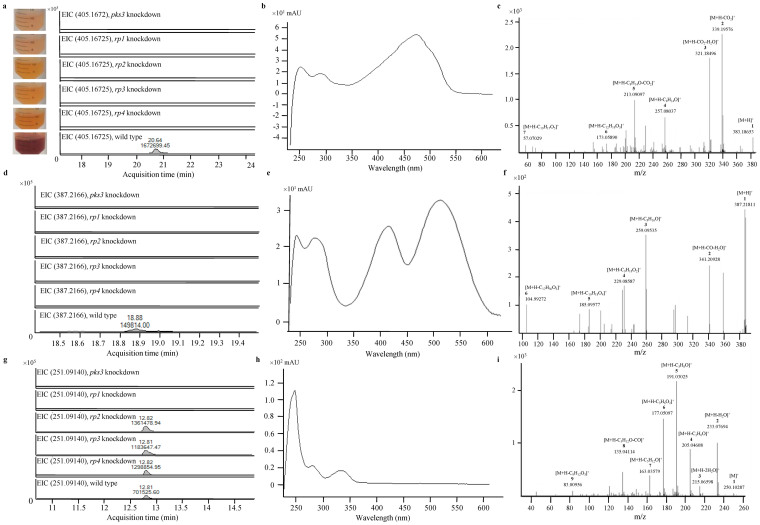
Red pigment of P. marneffei is composed of monascorubrin, rubropunctatin and their amino acid conjugates. To elucidate the identity of the red pigment, ultra-high performance liquid chromatography (UHPLC) profiles of wild type and pks3, rp1, rp2, rp3 and rp4 knockdown mutants of P. marneffei were monitored using DAD at scan range from 200 nm to 640 nm. Peaks that were present in the wild type but not in pks3, rp1, rp2, rp3 and rp4 knockdown mutants were recorded and subjected to mass spectrometry (MS) and tandem mass spectrometry (MS/MS) analysis. All the MS and MS/MS data were adjusted to mass accuracies within 10 ppm and 20 ppm respectively. At retention time 20.6 min, a peak corresponding to a compound with m/z 405.1672 and UV absorption maximum at 475 nm was observed in the wild type, but not in pks3, rp1, rp2, rp3 and rp4 knockdown mutants in positive ionization mode of the UHPLC-MS analysis (Fig. 2a and b). The molecular formula of this compound, C23H26O5Na, matched that of the sodium adduct of monascorubrin. The protonated monascorubrin ion, with m/z 383.1865, was selected for further confirmation of its identity by MS/MS analysis operated in positive ionization mode (Fig. 2c). The peak at m/z 339.1958 was a result of a neutral loss of carbon dioxide from [M + H]+. A further loss of a water molecule yielded the fragment ion [M-CO2-H2O + H]+ with m/z 321.1850. Another [M-C8H14O + H]+ peak at m/z 257.0804, was generated from losing the long keto-aliphatic chain, C8H14O, of the precursor ion. A further loss of a carbon dioxide molecule yielded the fragment ion [M-C8H14O-CO2 + H]+ of m/z 213.0910. The fragment ion [M-C12H18O3 + H]+ at m/z 173.0589 corresponded to the loss of carbon dioxide, keto-aliphatic chain and alkene branch chains from [M + H]+. Another fragment ion, butane ion, gave the peak at m/z 57.0703. All the UHPLC-MS and MS/MS data matched those using the monascorubrin standard, confirming that this compound observed in the red pigment was monascorubrin.
Figure 2. Detection of monascorubrin, ankaflavin and citrinin in P. marneffei.
(a) Extracted ion chromatogram, (b) UV absorption spectrum and (c) MS/MS fragmentation pattern showing the presence of monascorubrin detected and identified in wild type P. marneffei but not in pks3 or rp1 to rp4 knockdown mutants. (d) Extracted ion chromatogram, (e) UV absorption spectrum and (f) MS/MS fragmentation pattern showing the presence of ankaflavin detected and identified in wild type P. marneffei but not in pks3 or rp1 to rp4 knockdown mutants. In the MS/MS fragmentation pattern, the neutral loss of a water and a carbon monoxide molecule from the protonated ion yielded a fragment ion [M-H2O-CO + H]+ with m/z 341.2093. Another [M-C8H16O + H]+ peak at m/z 259.0954 was generated from losing the long keto-aliphatic chain of the precursor ion. Another three fragment ions, [M-C9H18O2 + H]+, [M + H-C10H18O4]+ and [M-C17H30O3 + H]+ gave the peaks at m/z 229.0859, m/z 185.0958 and m/z 104.9927 respectively were generated from losing long keto-aliphatic branch chains of the precursor ion. (g) Extracted ion chromatogram, (h) UV absorption spectrum and (i) MS/MS fragmentation pattern showing the presence of citrinin detected and identified in wild type and rp2 to rp4 knockdown mutants of P. marneffei but not in pks3 or rp1 knockdown mutants. In the MS/MS fragmentation pattern, a neutral loss of one and two water molecules respectively from the protonated ion yielded fragment ions [M-H2O + H]+ and [M-2H2O + H]+ with m/z 233.0807 and 215.0714. The peak at m/z 205.0461 represented the ion [M-C2H6O + H]+ which was resulted from the loss of a water molecule and two CH2 group from the precursor ion. A further loss of a CH2 group from [M-C2H6O + H]+ yielded fragment ion [M-C3H8O + H]+ with m/z 191.0303. Another [M-C3H6O2 + H]+ peak at m/z 177.0510 was generated by the loss of a carboxyl group and two CH2 groups from the precursor ion. Another three fragment ions, [M-C5H12O + H]+, [M-C5H12O-CO + H]+ and [M-C9H12O3 + H]+ gave the peaks at m/z of 163.0358, 135.0411 and 83.0096 respectively.
In addition to monascorubrin, peaks were also observed in wild type but not pks3, rp1, rp2, rp3 and rp4 knockdown mutants of P. marneffei at retention time 16.037, 16.913, 18.901, 19.205, 19.348, 19.533, 19.584, 20.696, 22.157, 22.279, 22.941, 23.197 and 23.209 min. Using UHPLC-MS and MS/MS, these peaks were identified to be the arginine, lysine, asparagine, serine, glutamic acid, tyrosine, glycine, aspartic acid, tryptophan, valine, methionine, leucine, isoleucine and phenylalaine conjugates of monascorubrin respectively (Supplementary Fig. S2–S15 and Supplementary Table S3). Notably, the peak at 18.901 min represented two amino acid (asparagine and serine) conjugates of monascorubrin. Furthermore, peaks were observed at 13.407 and 19.730 min. Using UHPLC-MS and MS/MS, these peaks were identified to be the arginine and phenylalaine conjugates of rubropunctatin (the molecular formula of which is that of monascorubrin minus C2H4) respectively (Supplementary Fig. S16–S17 and Supplementary Table S3).
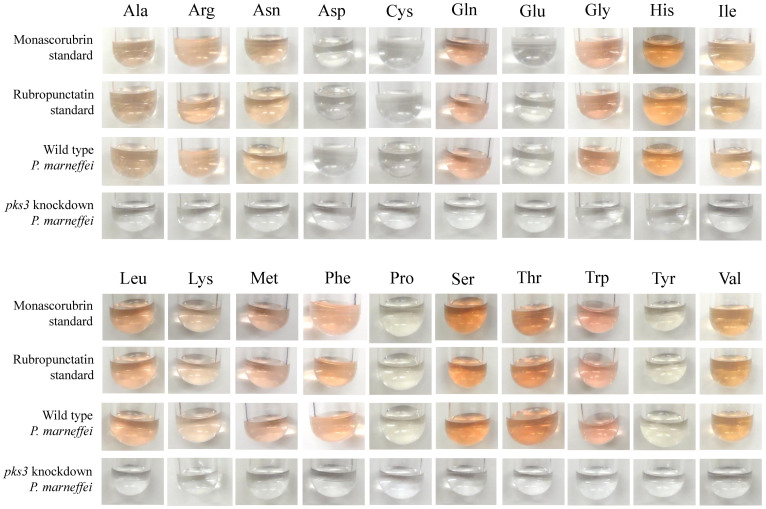
To verify that the red pigment of P. marneffei is composed of monascorubrin, its amino acid conjugates and the amino acid conjugates of rubropunctatin, wild type P. marneffei was cultured in Aspergillus minimal medium supplemented with each of the 20 essential amino acids. The colors of the cultures were identical to those of monascorubrin or rubropunctatin conjugated with the corresponding amino acids (Fig. 3).
Figure 3. Colors of P. marneffei cultured with different amino acids match those of monascorubrin/rubropunctatin conjugated with different amino acids.
Wild type P. marneffei and pks3 knockdown mutant were grown in Aspergillus minimal medium supplemented with one of the 20 essential amino acids. Monascorubrin and rubropunctatin standard conjugated with the corresponding amino acids were used as controls.
Red pigment biosynthesis pathway in P. marneffei is also responsible for ankaflavin and citrinin biosynthesis
To determine whether the red pigment biosynthesis pathway in P. marneffei is also responsible for synthesis of other polyketides, additional peaks that were present in wild type but not in some or all of the pks3, rp1, rp2, rp3 and rp4 knockdown mutants of P. marneffei were searched for.
At retention time 18.9 min, a peak corresponding to a compound with m/z 387.2166 and UV absorption maxima at 285 nm, 390 nm and 510 nm was observed in wild type, but not in pks3, rp1, rp2, rp3 and rp4 knockdown mutants in positive ionization mode of the UHPLC-MS analysis (Fig. 2d and e). The molecular formula of this compound, C23H30O5, was compatible with ankaflavin. The identity was confirmed by MS/MS analysis (Fig. 2f).
At retention time 12.8 min, a peak corresponding to a compound with m/z 251.0914 and UV absorption maxima at 240 nm, 282 nm and 333 nm was observed in wild type and rp2, rp3 and rp4 knockdown mutants but not in pks3 and rp1 knockdown mutants in positive ionization mode of the UHPLC-MS analysis (Fig. 2g and h). The molecular formula of this compound, C13H14O5, was compatible with citrinin. The identity was confirmed by MS/MS analysis (Fig. 2i). All the UHPLC-MS and MS/MS data matched those using the citrinin standard, confirming that this compound observed was citrinin.
Intermediates of the pathway for red pigment, ankaflavin and citrinin biosynthesis in P. marneffei
To elucidate the biochemical pathway for red pigment, ankaflavin and citrinin biosynthesis in P. marneffei, intermediates accumulated in the pks3, rp1, rp2, rp3 and rp4 knockdown mutants were searched for.
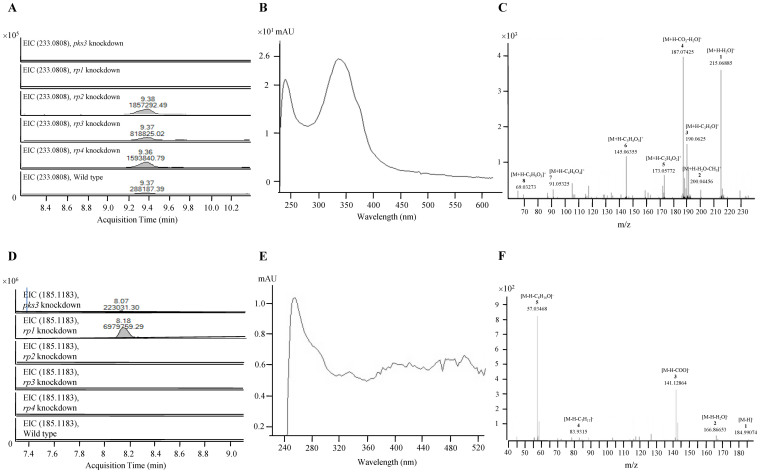
At retention time 9.4 min, a peak corresponding to a compound with m/z 233.0808 and UV absorption maxima at 240 nm and 338 nm, implying that the compound was yellow in color, was observed in wild type and rp2, rp3 and rp4 knockdown mutants but not in pks3 and rp1 knockdown mutants in positive ionization mode of the UHPLC-MS analysis (Fig. 4a and b). This compound, corresponding to protonated compound 1, with the molecular formula C13H12O4 was observed. The identity was confirmed by MS/MS analysis (Fig. 4c). The loss of a water molecule from the protonated ion yielded fragment ion [M-H2O + H]+ with m/z 215.0689. A further loss of a methyl group yielded [M-H2O-CH3 + H]+ with m/z 200.0446, whereas a further loss of a carbonyl group resulted in [M-H2O-CO + H]+ with the peak at m/z 187.0743. The peak at m/z 190.0625 represented [M-C2H3O + H]+, which was formed by the cleavage of the carbon-carbon bond between the two keto- groups in the ring structure. Ionized side groups [M-C2H4O2 + H]+ and [M-C3H4O3 + H]+ yielded the peaks at m/z of 173.0577 and m/z of 145.06355 respectively. Compound 1 was accumulated in rp2, rp3 and rp4 knockdown mutants as compared to wild type as its levels were 2.8 to 6.4 folds higher in rp2, rp3 and rp4 knockdown mutants than in wild type (Fig. 4a).
Figure 4. Detection of intermediates in red pigment biosynthesis pathway in P. marneffei.
(a) Extracted ion chromatogram, (b) UV absorption spectrum and (c) MS/MS fragmentation pattern showing the presence of compound 1 detected and identified in wild type and rp2 to rp4 knockdown mutants of P. marneffei but not in pks3 or rp1 knockdown mutants. (d) Extracted ion chromatogram, (e) UV absorption spectrum and (f) MS/MS fragmentation pattern showing the presence of 3-oxo-decanoic acid detected and identified in pks3 and rp1 knockdown mutants of P. marneffei but not in wild type or rp2 to rp4 knockdown mutants.
At retention time 8.2 min, a peak corresponding to a compound with m/z 185.1183 and UV absorption maxima at 400 nm and 485 nm was observed in pks3 and rp1 knockdown mutants, but not in wild type and rp2, rp3 and rp4 knockdown mutants in negative ionization mode of the UHPLC-MS analysis (Fig. 4d and e). The molecular formula of this compound was compatible with the deprotonated fatty acid 3-oxo-decanoic acid, C10H18O3. The identity was confirmed by MS/MS analysis (Fig. 4f). A loss of a water molecule or a carbonyl group from the [M-H]− ion resulted in [M-H2O-H]− and [M-COO-H]− respectively, giving peaks at m/z 166.8665 and 141.1286. The peak at m/z 83.9315 corresponded to the ion [M-C7H17-H]−, which was formed by the cleavage of the carbon-carbon bond between the alkyl carbon chain and the oxo-group, confirming the oxo-group at the C3-position of the fatty acid molecule. The peak at m/z 57.0347 represented [M-C8H16O-H]−, which was a result of the cleavage of the carbon-carbon bond between the β-carbon and the oxo-group in the deprotonated 3-oxo-decanoic acid ion.
Discussion
Through generating a series of targeted gene knockdown mutants, we identified the genes encoding the hypothetical biosynthesis pathway for the red pigment in P. marneffei (Fig. 5). The pathway involves a gene cluster of five genes, including pks3, rp1, rp2, rp3 and rp4. The red pigment of P. marneffei is composed of at least 16 compounds when cultured in RPMI 1640 medium, including monascorubrin and its arginine, lysine, asparagine, serine, glutamic acid, tyrosine, glycine, aspartic acid, tryptophan, valine, methionine, leucine, isoleucine and phenylalaine amino acid conjugates and rubropunctatin and its arginine and phenylalaine amino acid conjugates that we detected in the present study. These results and speculations are in line with the findings of experiments that showed a perfect match between the colors of the medium culturing P. marneffei with the presence of only one amino acid and those of monascorubrin and rubropunctatin conjugated with the same amino acid (Fig. 3). We infer that the amino acids were added to monascorubrin and rubropunctatin, resulting in Schiff base formation, after generation of monascorubrin and rubropunctatin. This is in line with experiments that showed that amino acids can be conjugated to monascorubrin and rubropunctatin under specific conditions without enzymatic catalysis20. The conclusions drawn on the identity of the red pigment in the present study is in contrast to that of a previous article by Bhardwaj et al., which claimed that the red pigment of P. marneffei structually resembled herquinone and appeared as a dimer through disulfide bond21. In the present study, such a compound was not observed in wild type P. marneffei and the knockdown mutants.
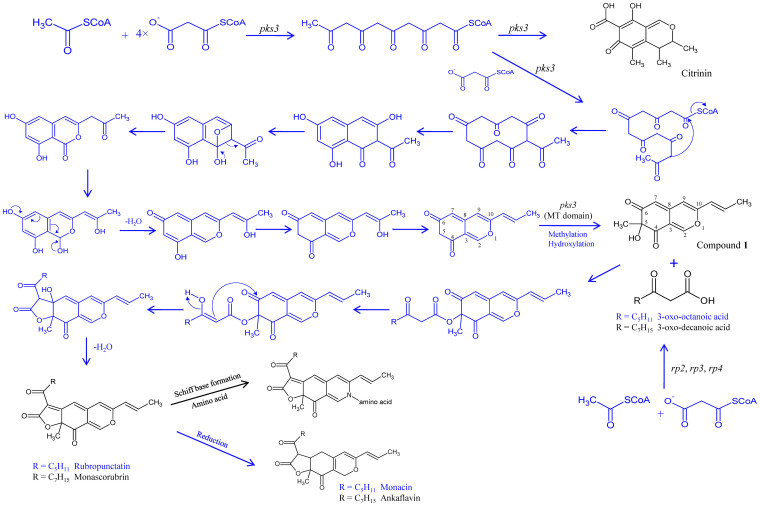
Figure 5. Hypothetical pathway for monascorubrin, ankaflavin and citrinin biosynthesis in P. marneffei.
Compounds and the five genes of the red pigment biosynthesis cluster identified in this study are in black. Hypothetical compounds, intermediates and pathways are depicted in blue.
PKS3 is responsible for catalyzing multiple steps on the biosynthesis pathway, whereas RP2, RP3 and RP4 catalyze the formation of 3-oxo-decanoic acid, and RP1 is the transcriptional activator of PKS3 (Fig. 5). Since RP1 is a predicted transcriptional activator and the metabolomic profile of rp1 knockdown mutant was identical to that of pks3 knockdown mutant, RP1 should be the transcriptional activator of pks3. Since rp2, rp3 and rp4 knockdown mutants shared the same metabolomic profile, their gene products should catalyze the same part of the red pigment biosynthetic pathway. In fact, RP2 and RP3 are predicted subunits of fatty acid synthase with very high amino acid identities to the subunits of fatty acid synthase of Talaromyces stipitatus, the teleomorph of Penicillium emmonsii. Therefore, RP2 and RP3, together with RP4 should catalyze the synthesis of 3-oxo-decanoic acid, which was observed in pks3 knockdown mutant because compound 1, the other substrate for 3-oxo-decanoic acid to react with and form monascorubrin, was not available. When rp2, rp3 or rp4 was knocked down, compound 1 was accumulated to a level much higher than that of wild type PM1. On the other hand, compound 1 was not detected in pks3 and rp1 knockdown mutants. These indicate that PKS3 catalyzes the synthesis of compound 1 and the utilization of compound 1 is catalyzed by RP2, RP3 and RP4. Specifically, the addition of a methyl group at C5 of the precursor of compound 1 should be catalyzed by the methyltransferase domain of PKS3. Both 3-oxo-decanoic acid and a polyketide chromophore were postulated intermediates of monascorubrine biosynthesis in M. purpureus, although the polyketide chromophore they postulated lacked a keto group at C4 as compared to compound 122.
The red pigment biosynthetic pathway in P. marneffei is also responsible for biosynthesis of ankaflavin and citrinin. It has been speculated that monascorubrin and ankaflavin are synthesized by the same biochemical pathway in M. purpureus due to their structural resemblance23. In this study, we showed that both monascorubrin and ankaflavin were not detectable in all the pks3, rp1, rp2, rp3 and rp4 knockdown mutants. Therefore, it is logical to deduce that monascorubrin and ankaflavin are both end products of this biochemical pathway, where ankaflavin is the reduced product of monascorubrin. As for citrinin, it was not detectable in pks3 and rp1 knockdown mutants, but detectable in rp2, rp3 and rp4 knockdown mutants. We infer that citrinin is an early side product of the pathway (Fig. 5). This is in contrast to M. purpureus, where it has been shown that monascorubrin and citrinin are synthesized by two separate pathways because when the PKS gene responsible for synthesis of citrinin was disrupted, red pigment production from the fungus was not affected4. We speculate that the yellow pigment observed in rp2, rp3 and rp4 knockdown mutants is a mixture of citrinin and compound 1.
The present study on elucidation of the biosynthetic pathway of monascorubrin is a proof-of-the-concept study that serves as a cornerstone for future studies on monascorubrin biosynthesis pathway dissection in Monascus species. Carbon-13 labeling experiment in M. purpureus has shown that monascorubrin was synthesized from one acetate and five malonate molecules24. The only genetic information on red pigment biosynthesis in M. purpureus includes a gene and its possible transcriptional activator, named MpPKS5 and mppR125. Deletion of mppR1 resulted in a loss of red pigment and monascorubrin25. Since MpPKS5 and mppR1 possess 65% and 49% amino acid identities to PKS3 and RP1 of the red pigment biosynthesis cluster in P. marneffei, it is logical to deduce that a similar cluster should also be present in M. purpureus and other Monascus species. Further studies would shed light on ex vivo manufacturing of these ancient pigments.
Methods
Strain and culture conditions
P. marneffei strain PM1 was isolated from an HIV-negative patient suffering from culture-documented penicilliosis in Hong Kong14. The yeast form of PM1 was used for DNA extraction for knockdown of the PKS genes and genes in the neighborhood of pks3. A single colony of the fungus grown on Sabouraud dextrose agar at 37°C was inoculated into yeast peptone broth and incubated in a shaker at 37°C for 10 days.
Knockdown of PKS genes and genes in the neighborhood of pks3
DNA extraction was performed using the DNeasy Plant Mini Kit according to manufacturer's instructions (Qiagen, Hilden, Germany). The extracted DNA was eluted in 50 μl of AE buffer and the resultant mixture was diluted 10× and 1 μl of the diluted extract was used for PCR.
Plasmid construction was performed according to our previous publication17. Knockdown of pks1 to pks25 was reported previously18. For knockdown of rp1, plasmid pSilent-126, obtained from the Fungal Genetics Stock Center, was used to construct the pPW2422 plasmid. First, the internal rp1 fragment (sense) was amplified using primers LPW18206 5′-CCGCTCGAGCTGCTGGCGATACCGAGTTC-3′ and LPW18207 5′-CCCAAGCTTGGGGCAAGGCATCAGCTCAATGA-3′ (Invitrogen, USA) (Supplementary Table S4). The PCR mixture (25 μl) contained P. marneffei DNA, PCR buffer (10 mM Tris-HCl pH 8.3, 50 mM KCl, 2 mM MgCl2 and 0.01% gelatin), 200 μM of each deoxynucleoside triphosphates and 1.0 U Taq polymerase (Applied Biosystem, Foster City, CA, USA). The mixtures were amplified in 32 cycles of 95°C for 30 s, 56°C for 30 s and 72°C for 40 s, and a final extension at 72°C for 10 min in an automated thermal cycler (Applied Biosystem, Foster City, CA, USA). The PCR product was purified using the QIAquick Gel Extraction kit (QIAgen, Hilden, Germany), digested with XhoI and HindIII, and cloned into the XhoI-HindIII site of the pSilent-1 plasmid, resulting in pPW2422-1. Second, the internal rp1 fragment (antisense) was amplified with primers LPW18208 5′-GGGGTACCCTGCTGGCGATACCGAGTTC-3′ and LPW18209 5′-GAAGATCTGCAAGGCATCAGCTCAATGA-3′ (Invitrogen, USA), using the PCR conditions described above. This amplified fragment was purified as described above, digested with BglII and KpnI, and cloned into the BglII-KpnI site of the pPW2422-1, resulting in pPW1459. The wild type P. marneffei strain PM1 was transformed with linearized pPW2422, using 200 μg/ml hygromycin for selection. For the other genes in the neighborhood of pks3, they were knocked down using the protocol described above with primers listed in Supplementary Table S4.
RT-PCR and real-time quantitative RT-PCR
Total RNA was extracted using RiboPure-Yeast (Ambion, USA). The RNA was eluted in 70 μl of RNase-free water and was used as the template for RT-PCR and real-time quantitative RT-PCR. Reverse transcription was performed using the SuperScript III kit (Invitrogen, USA). PCR and agarose gel electrophoresis were performed according to our previous publication27. Real-time RT-PCR assays was performed as described previously17,18, for rp1 fragment with primers LPW18206 and LPW18207 (Supplementary Table S4), using actin with primers LPW8614 5′-CAYACYTTCTACAAYGARCTCC-3′ and LPW8615 5′-KGCVARRATRGAACCACC-3′ for normalization. cDNA was amplified in a LightCycler 2.0 (Roche, Switzerland) with 20 μl reaction mixtures containing FastStart DNA Master SYBR Green I Mix reagent kit (Roche, Switzerland), 2 μl cDNA, 2 mM MgCl2 and 0.5 mM primers at 95°C for 10 min followed by 50 cycles of 95°C for 10 s, 57°C (55°C for actin gene) for 5 s and 72°C for 23 s (36 s for actin gene). For the other genes, real-time RT-PCR was performed using the protocol described above with primers listed in Supplementary Table S4.
Sequence analysis of PKS3 and genes in the neighborhood for red pigment biosynthesis
Introns were predicted by performing pairwise alignment with the annotated Talaromyces stipitatus (teleomorph of Penicillium emmonsii) complete genome sequence. Domains of PKS3 were predicted using the Conserved Domains Database of NCBI and PFAM (http://pfam.sanger.ac.uk/search?tab=searchSequenceBlock) and manual inspection of multiple alignments of PKS3 and its homologous sequences.
Fermentation and extraction of red pigment
Ten-day-old yeast cultures of wild type, rp1, rp2, rp3, rp4 and pks3 knockdown mutant strains of P. marneffei were washed twice with Milli-Q water. Four hundred microliters of each culture at a turbidity of McFaland 1, and 400 μl of Milli-Q water as the negative control, were grown in 10 ml RPMI 1640 medium (Gibco) supplemented with 5% glucose at 25°C with shaking at 250 rpm for 100 h. After incubation for 100 h, 5 μl of culture medium from each strain and control were filtered with a 0.22 μm filter. Metabolic activities in the culture medium were quenched by incubating the filtrates in liquid nitrogen for 10 min. The filtrates were lyophilized for 48 h. The lyophilized samples were reconstituted in 800 μl solution of water, methanol and acetonitrile (2:4:4) mixture and vortexed for 5 min. The samples were undergone ultrasonic extraction for 10 min, followed by centrifugation at 12000 rpm for 5 min. Supernatants were used for UV-Vis spectroscopic examination and UHPLC-DAD/ESI-Q-TOF-MS analysis.
UV-Vis spectroscopic analysis
UV-Vis spectroscopic analysis was performed according to our previous publication18. The maximum absorbance of the extracts was examined by a UV-Vis spectrometer (NanoDrop 1000 spectrophotometer, Thermo scientific, USA) from 300 to 750 nm. The absorbance was measured by using 0.2 mm path.
UHPLC-DAD/ESI-TOF-MS analysis
UHPLC-DAD/ESI-TOF-MS analysis was performed according to our previous publication18. Separations were performed using Agilent 1290 UHPLC (Agilent Technologies, USA) and Agilent Eclipse Plus RRHD C18 (2.1 × 100 mm, 1.8 μm) column with Agilent SB-C8 (2.1 × 30 mm, 3.5 um) guard column, in both positive and negative ionization modes. The injection volume was 8 μl of each sample. The column and the autosampler temperature were maintained at 50°C and 10°C respectively. For the positive mode, the separation was performed at a flow rate of 0.4 ml/min under a gradient program. Mobile phase A was 0.1% acetic acid (v/v) in water. Mobile phase B was pure methanol. The gradient program was applied as follows: t = 0 min, 2% B; t = 1 min, 2% B; t = 10 min, 45% B; t = 24 min, 99% B; t = 27 min, 99% B; t = 27.01 min, 2% B; t = 30 min, 2% B. The capillary voltage was kept at +3800 V with nozzle voltage at +0 V. For the negative mode, the separation was performed at a flow rate of 0.4 ml/min under a gradient program. Mobile phase A was 5 mM ammonium acetate with 0.1% acetic acid (v/v) in water. Mobile phase B was pure methanol. The gradient program was applied as follows: t = 0 min, 25% B; t = 1 min, 25% B; t = 15 min, 99% B; t = 20 min, 99% B; t = 20.1 min, 25% B; t = 25.0 min, 25% B. The capillary voltage was kept at −3800 V with nozzle voltage at −0 V. Mass spectrometer was operated in both positive and negative ESI mode using Agilent 6540 A Q-TOF mass spectrometer (Agilent Technologies, USA) with Agilent Jet Stream ESI source. The gas temperature was kept at 300°C. The drying gas (nitrogen) was set at 7 L/min and the pressure of the nebulizer gas (nitrogen) was maintained at 45 psi. The sheath gas was kept at a flow rate of 10 L/min and was maintained at a temperature of 350°C. The voltages were kept at 130 V, 50 V and 500 V of the fragmentor, skimmer 1, and octopoleRFPeak, respectively. The full scan mass range was adjusted to 50–1700 m/z at the acquisition rate of 2 spectra/s. Data were acquired and processed using Agilent MassHunter Qualitative Analysis software (version B.03.01, Agilent Technologies, USA). Rubropunctatin (ReseaChem GmbH, Switzerland), monascorubrin (ReseaChem GmbH), octanoic acid (Sigma Aldrich), 7-oxo-octanoic acid (Sigma Aldrich) and citrinin (Sigma Aldrich) were used as authentic standards in both MS and MS/MS analysis.
Culture of P. marneffei in Aspergillus minimal medium with different amino acids
Ten-day-old yeast cultures of wild type and pks3 knockdown mutant strains of P. marneffei were washed twice with Milli-Q water. Four hundred microliters of each culture at a turbidity of McFaland 1 were grown in 10 ml Aspergillus minimal medium28 supplemented with one of the 20 essential amino acids at a concentration of 25 mM with shaking at 25°C for 72 h at 250 rpm and observed for pigment production. The colors of the cultures were compared to monascorubrin and rubropunctatin standards conjugated with the corresponding amino acids.
Author Contributions
P.C.Y.W., C.-W.L., S.K.P.L. and K.Y.Y. conceived and designed the experiments. E.W.T.T., K.-C.L., K.K.Y.Y., C.K.F.L. and K.-H.S. performed the experiments. P.C.Y.W., C.-W.L. and E.W.T.T. wrote the paper.
Supplementary Material
Supplementary information
Acknowledgments
This work was partly supported by the Research Fund for the Control of Infectious Diseases (Commissioned study) of the Health, Welfare and Food Bureau of the Hong Kong SAR Government; the Strategic Research Theme Fund and University Development Fund of The University of Hong Kong; Research Grant Council Grant; Providence Foundation Limited in memory of the late Dr. Lui Hac Minh, HKU Award for CAE Membership, HKU Medical Faculty Award for CAE Membership, Dr Hector T.G. Ma.
References
- Feng Y., Shao Y. & Chen F. Monascus pigments. Appl Microbiol Biotechnol 96, 1421–1440 (2012). [DOI] [PubMed] [Google Scholar]
- Huang H. T. Fermentations and Food Science. in Science and Civilisation in China (ed. Needham, J.) 741 (University Press, Cambridge, 2000). [Google Scholar]
- Li S. Z. Pen Ts'ao Kang Mu (Compendium of Materia Meica, Systematic Pharmacopoeia), (China, 1596). [Google Scholar]
- Fu G., Xu Y., Li Y. & Tan W. Construction of a replacement vector to disrupt pksCT gene for the mycotoxin citrinin biosynthesis in Monascus aurantiacus and maintain food red pigment production. Asia Pac J Clin Nutr 16 Suppl 1, 137–142 (2007). [PubMed] [Google Scholar]
- Dufossé L. et al. Microorganisms and microalgae as sources of pigments for food use: a scientific oddity or an industrial reality? Trends Food Sci. Technol. 16, 389–406 (2005). [Google Scholar]
- Mapari S. A., Meyer A. S., Thrane U. & Frisvad J. C. Identification of potentially safe promising fungal cell factories for the production of polyketide natural food colorants using chemotaxonomic rationale. Microb. Cell Fact. 8, 24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T. et al. Polyketide synthase gene responsible for citrinin biosynthesis in Monascus purpureus. Appl Environ Microbiol 71, 3453–3457 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen K. Y., Wong S. S., Tsang D. N. & Chau P. Y. Serodiagnosis of Penicillium marneffei infection. Lancet 344, 444–445 (1994). [DOI] [PubMed] [Google Scholar]
- Wong S. S., Siau H. & Yuen K. Y. Penicilliosis marneffei--West meets East. J Med Microbiol 48, 973–975 (1999). [DOI] [PubMed] [Google Scholar]
- Woo P. C. et al. MP1 homologue-based multilocus sequence system for typing the pathogenic fungus Penicillium marneffei: a novel approach using lineage-specific genes. J Clin Microbiol 45, 3647–3654 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. et al. Penicillium marneffei fungaemia in an allogeneic bone marrow transplant recipient. Bone Marrow Transplant 35, 831–833 (2005). [DOI] [PubMed] [Google Scholar]
- Hsueh P. R. et al. Molecular evidence for strain dissemination of Penicillium marneffei: an emerging pathogen in Taiwan. J Infect Dis 181, 1706–1712 (2000). [DOI] [PubMed] [Google Scholar]
- Supparatpinyo K., Khamwan C., Baosoung V., Nelson K. E. & Sirisanthana T. Disseminated Penicillium marneffei infection in southeast Asia. Lancet 344, 110–113 (1994). [DOI] [PubMed] [Google Scholar]
- Woo P. C. et al. Draft genome sequence of Penicillium marneffei strain PM1. Eukaryot Cell 10, 1740–1741 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P. C. et al. Genomic and experimental evidence for a potential sexual cycle in the pathogenic thermal dimorphic fungus Penicillium marneffei. FEBS Lett 580, 3409–3416 (2006). [DOI] [PubMed] [Google Scholar]
- Woo P. C. et al. The mitochondrial genome of the thermal dimorphic fungus Penicillium marneffei is more closely related to those of molds than yeasts. FEBS Lett 555, 469–477 (2003). [DOI] [PubMed] [Google Scholar]
- Woo P. C. et al. High diversity of polyketide synthase genes and the melanin biosynthesis gene cluster in Penicillium marneffei. FEBS J 277, 3750–3758 (2010). [DOI] [PubMed] [Google Scholar]
- Woo P. C. et al. First discovery of two polyketide synthase genes for mitorubrinic acid and mitorubrinol yellow pigment biosynthesis and implications in virulence of Penicillium marneffei. PLoS Negl Trop Dis 6, e1871 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroken S., Glass N. L., Taylor J. W., Yoder O. C. & Turgeon B. G. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A 100, 15670–15675 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. F., Yakushijin K., Büchi G. H. & Demain A. L. Formation of water-solubleMonascus red pigments by biological and semi-synthetic processes. J Ind Microbiol 9, 173–179 (1992). [Google Scholar]
- Bhardwaj S. et al. Putative structure and characteristics of a red water-soluble pigment secreted by Penicillium marneffei. Med Mycol 45, 419–427 (2007). [DOI] [PubMed] [Google Scholar]
- Hajjaj H. et al. Medium-chain fatty acids affect citrinin production in the filamentous fungus Monascus ruber. Appl Environ Microbiol 66, 1120–1125 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jůzlová P., Martínková L. & Křen V. Secondary metabolites of the fungus Monascus: A review. J. Ind. Microbiol. Biotechnol. 16, 163–170 (1996). [Google Scholar]
- Hajjaj H. et al. Biosynthetic pathway of citrinin in the filamentous fungus Monascus ruber as revealed by 13C nuclear magnetic resonance. Appl Environ Microbiol 65, 311–314 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan B. et al. Genetic localization and in vivo characterization of a Monascus azaphilone pigment biosynthetic gene cluster. Appl Microbiol Biotechnol 97, 6337–6345 (2013). [DOI] [PubMed] [Google Scholar]
- Nakayashiki H. et al. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol 42, 275–283 (2005). [DOI] [PubMed] [Google Scholar]
- Tam E. W. et al. Misidentification of Aspergillus nomius and Aspergillus tamarii as Aspergillus flavus: characterization by internal transcribed spacer, beta-Tubulin, and calmodulin gene sequencing, metabolic fingerprinting, and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 52, 1153–1160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. P. et al. Aspergillus vadensis, a new species of the group of black Aspergilli. Antonie Van Leeuwenhoek 87, 195–203 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information