Abstract
Disruptions in mitochondrial dynamics may contribute to the selective degeneration of dopamine (DA) neurons in Parkinson's disease (PD). However, little is known about the normal functions of mitochondrial dynamics in these neurons, especially in axons where degeneration begins, and this makes it difficult to understand the disease process. To study one aspect of mitochondrial dynamics—mitochondrial fission—in mouse DA neurons, we deleted the central fission protein dynamin-related protein 1 (Drp1). Drp1 loss rapidly eliminates the DA terminals in the caudate–putamen and causes cell bodies in the midbrain to degenerate and lose α-synuclein. Without Drp1, mitochondrial mass dramatically decreases, especially in axons, where the mitochondrial movement becomes uncoordinated. However, in the ventral tegmental area (VTA), a subset of midbrain DA neurons characterized by small hyperpolarization-activated cation currents (Ih) is spared, despite near complete loss of their axonal mitochondria. Drp1 is thus critical for targeting mitochondria to the nerve terminal, and a disruption in mitochondrial fission can contribute to the preferential death of nigrostriatal DA neurons.
Keywords: axon, Drp1, mitochondria, neurodegeneration, Parkinson's disease
Introduction
Disruption of mitochondrial dynamics has been implicated in the pathogenesis of several neurodegenerative diseases, especially PD (Itoh et al., 2013). A number of PD-associated proteins, including α-synuclein, PINK1, parkin, DJ-1, and LRRK2, regulate the relative rates of mitochondrial fission and fusion (Poole et al., 2008; Yang et al., 2008; Irrcher et al., 2010; Kamp et al., 2010; Tanaka et al., 2010; Nakamura et al., 2011; X. Wang et al., 2012a,b), raising the possibility that changes in mitochondrial dynamics contribute to neuronal degeneration. However, to understand the role of mitochondrial dynamics in PD, it is necessary to study dynamics specifically in the nigrostriatal DA neurons that degenerate. Indeed, disrupted proteins that regulate mitochondrial dynamics produce selective neuronal degeneration despite being ubiquitously expressed (Alexander et al., 2000; Züchner et al., 2004; Niemann et al., 2005). Furthermore, mitochondrial dynamics have been studied almost exclusively in the cell body of neurons in the CNS, even though the vast majority of mitochondria in many neuron types probably reside in axons (Grafstein and Forman, 1980), and axonal mitochondria exist in unique micro-environments and have different properties from those at the cell body (Pathak et al., 2013). Understanding mitochondrial biology in this compartment is critical, as axons are consistently lost before the cell body in neurodegenerative diseases that are thought to involve energy depletion (Li et al., 2001; Scheff et al., 2007; Cheng et al., 2010).
In mammals, most fission pathways depend on dynamin-related protein 1 (Drp1), a dynamin-like GTPase recruited to fission sites on the outer mitochondrial membrane (Itoh et al., 2013). Mitochondrial fission can help sequester poorly functioning (depolarized) mitochondria for degradation, and loss of mitochondrial fission prevents depolarized mitochondria from being degraded (Tanaka et al., 2010). However, the functional consequences of any changes in mitochondrial turnover are unclear, and immortalized mouse embryonic fibroblasts that lack Drp1 have normal respiration (Ishihara et al., 2009; Wakabayashi et al., 2009). Drosophila with impaired mitochondrial fission also have fewer mitochondria at the neuromuscular junction (Verstreken et al., 2005), although the mechanism underlying this depletion is not understood.
To determine how mitochondrial fission affects mitochondrial behavior in axons and how this, in turn, affects axonal integrity and neuronal survival, we selectively deleted Drp1 in DA neurons. Loss of Drp1 produced a surprising decrease in mitochondrial mass and greatly impaired the coordination of mitochondrial movements, leading to a decrease in mitochondria at the nerve terminal, degeneration of synaptic terminals, and cell loss. However, we also identified a population of resistant DA neurons with specific electrophysiological properties that were scattered throughout the midbrain but enriched in the medial VTA. These neurons survived despite showing similar changes in mitochondrial morphology at the cell body and losing the mitochondria in their axons.
Materials and Methods
Molecular biology.
Mitochondria-targeted GFP (mitoGFP) and mCherry fused to the N terminus of rat synaptophysin (Hua et al., 2011; Nakamura et al., 2011) were subcloned into pAAV-EF1a-DIO-hChR2(H134R)-EYFP-WPRE (Addgene), and recombinant adeno-associated virus (AAV) 1 was made by the Vector Core at the University of North Carolina. mitoBFP was made by fusing the mitochondria-targeting sequence, cytochrome c oxidase subunit VIII, to mTagBFP (a kind gift from Vladislav Verkhusha, Albert Einstein College of Medicine, New York; Subach et al., 2008).
Animals.
Floxed Drp1KO mice (Wakabayashi et al., 2009) have been described. DATcre mice (Bäckman et al., 2006) and floxed tdTomato mice (Madisen et al., 2010) were obtained from The Jackson Laboratory. Mice were group housed in a colony maintained with a standard 12 h light/dark cycle and given food and water ad libitum. All mice received food on the cage floor. Experiments were performed on age-matched mice of either sex, with the specific number of males and females matched between groups in each experiment (typically divided ≈50:50 between males and females). No differences between genders were noted in any of the experiments. Experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals, as adopted by the National Institutes of Health, and with approval of the University of California, San Francisco Institutional Animal Care and Use Committee.
Behavioral testing.
Spontaneous locomotor activity in an open field was measured in an automated Flex-Field/Open Field Photobeam Activity System (San Diego Instruments), with the examiner blind to the genotype. Before testing, mice were acclimated to the testing environment for at least 1 h. Activity was then assessed in a clear plastic chamber (41 × 41 × 30 cm) for 15 min, with two 16 × 16 photobeam arrays detecting horizontal and vertical movements. These assessments were used to quantify total ambulatory movements and rearing. For experiments with levodopa, after we assessed baseline activity, we returned mice to their home cages and injected them intraperitoneally with a mixture of l-3,4-dihydroxyphenylalanine methyl ester (3 mg/kg, Sigma) and the DOPA decarboxylase inhibitor benserazide hydrochloride (12 mg/kg; Sigma). Activity was retested 45 min after the injections.
Stereotaxic recombinant AAV and injection.
For stereotaxic injection of AAV1 expressing mitoGFP and mCherrySynaptophysin, 3-week-old Daticre/icre, Drp1w/w and Daticre/wt, and Drp1lox/lox mice were used. For stereotaxic injection of AAV2/1.CMV.PI.Cre.RBG, purchased from Penn Vector Core, 2- to 3-month-old Drp1w/w, tdTomatolox/lox and Drp1lox/lox, tdTomatolox/lox mice were used. Mice were anesthetized with ketamine/xylazine, and their skulls were immobilized with a stereotaxic frame (Kopf). A 0.5 μl volume of AAV1-EF1α-DIO-mitoGFP [8 × 1012 vector genomes (VG)/ml] and 0.5 μl of AAV1-EF1α-DIO-Cherry-synaptophysin (3 × 1012 VG/ml) were coinjected, and 0.75 μl of AAVcre (2.16 × 1013 genome copies /ml) was injected using a Hamilton syringe and a cannula (33 gauge) unilaterally into the substantia nigra pars compacta (SNc)/VTA (anteroposterior: –3.0 mm from bregma; mediolateral: 1.1 mm; dorsoventral: 4.3 mm), at a rate of 0.2 μl/min (Shin et al., 2011). Animals were killed 4 weeks after mitoGFP and mCherrySynaptophysin coinjection and 2 months after AAVcre injection.
Brain monoamine levels.
To measure tissue monoamine content, mice were killed with CO2, their brains rapidly removed, and immediately frozen in superchilled isopentane. Coronal sections of 300 μm thickness were cut on a cryostat, and punches were made from the dorsal striatum (using a pipette tip cut to 1 mm in diameter) and stored at −80°C. Dopamine levels were measured by the Vanderbilt Neurochemistry Core by HPLC coupled to an electrochemical detector (Hnasko et al., 2010).
Slice preparation and electrophysiology.
Mice were anesthetized with isoflurane and decapitated, and the brains were removed. Horizontal brain slices (150 μm thick) containing the SNc/VTA were prepared using a vibratome (Leica Instruments). The landmarks for VTA-containing slices were the interpeduncular fossa and the medial terminal nucleus of the accessory optic tract (MT). Slices were submerged in aCSF containing the following (in mm): 119 NaCl, 2.5 KCl, 1.3 MgSO4, 1.0 NaH2PO4, 2.5 CaCl2, 26.2 NaHCO3, and 11 glucose, saturated with 95% O2–5% CO2 and allowed to equilibrate at 33°C for at least 1 h.
Individual slices were visualized under a Zeiss Axioskop with differential interference contrast optics and infrared illumination with a Zeiss AxioCam MRm and Microlucida software (MicroBrightField). The examiner was unblinded to the genotype while performing the recordings, but was blinded during the subsequent analysis (no cells were excluded from the analysis while unblinded). Whole-cell patch-clamp recordings were made at 31°C using 2.5–5 MΩ pipettes containing the following (in mm): 123 potassium gluconate, 10 HEPES, 0.2 EGTA, 8 NaCl, 2 MgATP, and 0.3 Na3GTP, pH 7.2, osmolarity adjusted to 275. Biocytin (0.1%) was added to the internal solution to mark the recorded neuron for later cytochemical characterization. Recordings were made using an Axopatch 1D (Molecular Devices) filtered at 5 kHz and collected at 20 kHz using IGOR Pro (WaveMetrics, RRID:nlx_156887). Liquid junction potentials were not corrected during current-clamp or voltage-clamp recordings. Ih was measured by voltage clamping cells and stepping from −60 to −40, −50, −70, −80, −90, −100, −110, and −120 mV. Input resistance was monitored with hyperpolarizing pulses while holding the cell in current-clamp mode (current set to 0 pA).
Cell sizes were measured using Neurolucida (MicroBrightField, RRID:nif-0000-10294). The Ih magnitude was measured as the difference between the initial capacitative response to a voltage step from −60 to −120 mV and the final current during the same 200 ms step. Neurons were considered Ih(−) if the slope of the I–V curve for hyperpolarizing steps from −60 to −90, −100, −110, and −120 mV was 0. For all action potential (AP) duration measurements, at least 10 APs were averaged together to reduce the influence of noise on the measurements. AP duration was measured during the first 2 min after establishing whole-cell access. The intracellular AP duration was measured from the threshold (when the rate of rise exceeded 10 V/s) to the time at which the descending voltage recrossed the threshold after the AP peak. The reported firing rates are averages of the instantaneous firing rate over at least 2 min and up to 10 min, usually at the beginning of the experiment. The interspike interval (ISI) coefficient of variation (CV) was calculated as the SD divided by the mean for 50 ISIs at the beginning of each experiment, excluding delays due to brief hyperpolarizations to monitor input resistance. Firing rates and ISI CVs were only included in the analysis if the firing appeared stable over the duration of the measurement.
Histology.
Mice were anesthetized and perfused with PBS and then 4% PFA. Brains were removed, postfixed, cryoprotected in 30% sucrose, and frozen in superchilled isopentane. Coronal sections of 40 μm thickness were cut on a cryostat throughout the brain, including the SNc/VTA, and every sixth section was analyzed. Cells were fixed for 15 min in medium with 4% PFA. For electrophysiology, slices were fixed immediately after recording in 4% PFA for 2 h.
For immunofluorescence, brain section and cells were rinsed with PBS and transferred in blocking solution containing 0.2% Triton X-100 with either 3% bovine serum albumin and 10% goat serum for 1 h. Tissue sections of 150 mm used for electrophysiological recordings were blocked 2 h with 0.2% bovine serum albumin and 5% normal goat serum in 0.3% Tween 20. Samples were then incubated overnight at room temperature, or for 48 h at 4°C, with the appropriate primary antibody in blocking solution.
The following primary antibodies were used: mouse anti-Drp1 (1:200; clone 8; BD Biosciences), rabbit anti-Tom20 (1:1000; Santa Cruz Biotechnology, catalog #SC-11415, RRID:AB_2207533), anti-TH mouse (1:20,000; EMD Millipore, catalog #MAB318, RRID:AB_2201528) or rabbit (1:1000; Millipore, catalog #AB152, RRID:AB_390204), rabbit anti-DsRed (1:1000; Clontech, catalog #632496, RRID:AB_10015246), mouse anti-COX1 (1:200; Abcam, catalog #ab14705, RRID:AB_2084810), mouse pyruvate dehydrogenase (PDH; 1:200; Abcam, catalog #ab110333, RRID:AB_10862029; Kageyama et al., 2012), and mouse anti-α-synuclein (1:400; BD Biosciences, catalog #610787, RRID:AB_398108). Sections were rinsed and incubated for 2 h with the appropriate secondary antibodies: Alexa Fluor 488, 594, or 647 anti-mouse or rabbit IgG (1:400; Invitrogen) or FITC goat anti-rabbit (1:100; catalog #111-095-003) and Cy5-streptavidin (1:200; Jackson ImmunoResearch, catalog #016-170-084). Samples were viewed using a laser-scanning confocal microscope (LSM510-Meta; Carl Zeiss) equipped with a 63× (1.4 NA), and 100× (1.3 NA) PlanApo objectives, or, for slices, a Zeiss Axioskop running Neurolucida. Calbindin and tdTomato/TH immunofluorescence experiments were imaged with the examiner blind to the genotype with 10× (0.3 NA) and 100× oil-immersion (1.4 NA) objectives, respectively, on a Nikon Ti-E inverted microscope with an Andor iXon EMCCD camera. For peroxidase experiments, sections were incubated with rabbit polyclonal anti-TH and visualized with biotinylated goat anti-rabbit IgG (1:300; Vector Laboratories, BA-1000, RRID:AB_2313606), followed by streptavidin-conjugated HRP (1:300; Vectastain ABC kit; Vector Laboratories). Positive immunostaining was visualized with DAB (Sigma) after reaction with hydrogen peroxide.
Stereology.
Total numbers of TH-positive neurons in the SNc and VTA were quantified with the examiner blind to the genotype using a computer-assisted image analysis system, consisting of an Olympus BX-51 microscope equipped with an XYZ computer-controlled motorized stage and an SIA-L9C Digital Camera (Scientific Instruments and Applications) and counted using the Optical Fractionator probe of Stereo Investigator software (MicroBrightField, RRID:nif-0000-00110), according to described and validated procedures (J. Zhang et al., 2007). TH-stained neurons were counted in the SNc or VTA of every fourth section for P14 mice, or every sixth section for mice 1 month and older, throughout the entire SNc or VTA. Each section was viewed at lower power and outlined. The numbers of TH-stained cells were counted at high power (100× oil; NA 1.4) using a 75 × 75 mm counting frame. A 15 μm dissector was placed 1.5 μm below the surface of the section (J. Zhang et al., 2007).
Electron microscopy.
For immuno-EM studies, TH was detected using mouse anti-TH using validated procedures (Dumartin et al., 1998; Berthet et al., 2009). Mice were perfused with 2% PFA/0.2% glutaraldehyde in 0.1 M phosphate buffer at pH 7.4. Brains were quickly removed, left overnight in 2% PFA at 4°C, and cut into 60-μm-thick frontal sections with a vibratome. To enhance the penetration of the immunoreagents, the sections were equilibrated in a cryoprotectant solution, freeze thawed, and stored in PBS with 0.03% sodium azide. To visualize mitochondria in terminals, TH was detected by immunoperoxidase with DAB-nickel. To visualize mitochondria in the cell body, TH was detected by the pre-embedding immunogold technique, as described previously (Dumartin et al., 1998; Berthet et al., 2009). After immunodetection, the sections were postfixed, dehydrated, and included in resin (Durcupan ACM; Fluka). Serial ultrathin sections were cut with a Reichert Ultracut S, contrasted with lead citrate, and analyzed with the examiner blind to the genotype using a Phillips Tecnai10 transmission EM with FEI software.
Quantitative analysis of electron microscopy experiments and calbindin levels.
The analysis was performed on digital images obtained with a computer linked directly to a CCD camera on the EM at a final magnification of 11500. The quantification of mitochondria size was performed with the examiner blind to the genotype using the MetaMorph software (version 7.7.3.0; Universal Imaging, RRID:SciRes_000136). Measurements of mitochondrial size were performed on 10–15 perikarya per animal with three animals per group and expressed TH. Calbindin levels were quantified in DA cells, whose boundaries were defined using a binary mask created with the “count nuclei” function on TH-immunostained sections.
Live imaging of mitochondrial motility and mitochondrial membrane potential.
Primary hippocampal cultures were prepared from early postnatal (P0 to P1) mouse Drp1lox/lox pups, cotransfected by electroporation (Amaxa) with either mitoGFP or mitoBFP to visualize mitochondria, mCherry, or GFP fused to synaptophysin (mCherrySynaptophysin or GFPSynaptophysin) to visualize axons (Hua et al., 2011; Nakamura et al., 2011), and either Cre or a vector control to delete Drp1. At D9–D10, cells were imaged live in Tyrode's medium containing the following (in mm): 127 NaCl, 10 HEPES-NaOH, pH 7.4, 30 glucose, 2.5 KCl, 2 CaCl2, and 2 MgCl2 with either a 40× air objective (0.95 NA) or a 60× water-immersion objective (1.2 NA) on a Nikon Ti-E inverted microscope at 25°C. For experiments with calcimycin, cells were pretreated for 5 min with 4-bromo-calcium ionophore A23187 (Sigma) before imaging.
For mitochondrial motility experiments, images were captured every 2 s for a total of 200 s with an Andor iXon EMCCD camera, and kymographs were generated from each live-imaging movie with MetaMorph software. The examiner was unblinded to genotype, but no runs were excluded from analysis. Mitochondria were considered moving if they traveled >0.67 mm during the 200 s imaging. For mitochondrial membrane potential experiments, cells were pretreated for 60 min with tetramethylrhodamine methyl ester (TMRM; 20 nm) and then also imaged live in the TMRM (Nakamura et al., 2011), in the presence and absence of oligomycin (5 μm) and carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP; 10 μm). TMRM fluorescence was calculated by an examiner blind to the genotype, as the mean background-subtracted fluorescence over the cytoplasm (area of the cell body–area of the nucleus). The intensity of TMRM fluorescence of the mitochondria themselves (an estimate of the extent of polarization of these mitochondria) was calculated as the mean background-subtracted fluorescence of the TMRM-positive mitochondria, identified by thresholding all TMRM images to the same value, and then creating a mask. The percentage of the cytoplasm covered by these polarized mitochondria was then calculated by dividing the area of the mask/area of the cytoplasm.
Results
Most midbrain DA neurons are susceptible to the toxicity of Drp1 loss
To selectively delete Drp1 from DA neurons, mice carrying a floxed Drp1 gene (Wakabayashi et al., 2009) were bred with mice that express Cre recombinase selectively in Slc6a3 (dopamine transporter, DAT)-expressing DA neurons (Bäckman et al., 2006). The homozygous progeny of floxed Drp1 mice (Drp1lox/lox;DATw/w) and floxed Drp1 and DATcre heterozygous mice (Drp1wt/lox;DATwt/cre) are referred to as Drp1KO-DATcre (Drp1lox/lox;DATwt/cre). Heterozygotes are Drp1HET-DATcre (Drp1wt/lox;DATwt/cre) and controls are Drp1WT (Drp1wt/lox;DATw/w and Drp1lox/lox;DATw/w). Drp1KO-DATcre mice were born in normal Mendelian proportions (control 49.8%, het 25.0%, and KO 25.2%, n = 631) and had similar body weights to controls through age P21. However, they gained weight much more slowly, weighing only ∼50% as much as control mice by 2 months (Fig. 1A). Drp1KO-DATcre mice also developed parkinsonism by 1 month with less activity and rearing (Fig. 1C), with head tremor (data not shown), suggesting the nigrostriatal system was significantly dysfunctional. After they were given low-dose levodopa, Drp1KO-DATcre mice were much more active. These findings are consistent with severe dopaminergic denervation of the caudate–putamen (CPu) with secondary sensitization of DA receptors (Kim et al., 2000). Nonetheless, ∼50% of the mice survived for at least 12 months without receiving levodopa (Fig. 1B), indicating that the remaining nigral and/or VTA DA neurons had some function: animals without functional midbrain DA neurons stop gaining weight by P17 and die by 1 month if they do not receive levodopa (Zhou and Palmiter, 1995; Hnasko et al., 2006).
Figure 1.
Loss of Drp1 in DA neurons leads to levodopa-responsive parkinsonism. A, Plot showing monthly weight measurements. Each point represents the average weight ± SEM (n = 4–101) for each genotype with the genders combined. B, Kaplan–Meier survival curve of control (n = 26), heterozygous (het; n = 18), and Drp1KO mice (n = 36). Drp1KO mice were significantly more likely to die than controls [HR 7.50, 95% CI: 2.82–19.9, p < 0.0001 by log-rank (Mantel–Cox) test]. C, Total ambulatory movements and rearing over the first 15 min after mice were placed in an open field cage was lower in Drp1KO (Daticre/wt, Drp1lox/lox; light gray column) than in Drp1 heterozygotes (Daticre/wt, Drp1lox/wt; dark gray) or controls (Datw/w, Drp1lox/lox or lox/wt; black; ***p < 0.001 vs controls at both time points). After mice were given levodopa (3 mg/kg) and retested 45 min later, the ambulatory movement of Drp1KO mice significantly increased (*p < 0.05; NS, not significant). The movement of controls probably decreased as a result of habituation. Data show mean ± SEM, n = 7–13 mice per group.
We next sought to determine how losing Drp1 affects the nigrostriatal and mesolimbic pathways. We assessed the integrity of surviving DA terminals by measuring the optical density of TH immunostaining (Fig. 2A,B). In Drp1KO-DATcre mice, the TH signal in the CPu was almost completely lost by P14, and DA levels were undetectable by HPLC in the CPu of 1-month-old Drp1KO-DATcre mice (Fig. 2C). The phenotype appeared to be degenerative, given that in Drp1KO-DATcre pups, TH optical density of the CPu decreased by only ∼35% at P7 and was indistinguishable from controls at P1 (data not shown). Remarkably, despite that almost no fibers projected into the CPu in Drp1KO-DATcre mice by P14, TH-positive nerve terminals were completely preserved in the nucleus accumbens (NAc) shell through 2.5 months, while those in the NAc core and olfactory tubercle (OT) were partially preserved (Fig. 2B).
Figure 2.

Loss of Drp1 promotes preferential death of nigrostriatal DA neurons, beginning at the nerve terminal. A, TH staining of brain sections from control and Drp1KO mice. Left, Striatal terminals from 1-month-old mice are almost completely lost in the CPu, but fibers projecting to the NAc are well preserved, and fibers in the OT are partially preserved. Right, DA neuron cell bodies from 2.5-month-old mice in the SNc and VTA are lost in both areas, but a subset of VTA neurons enriched in the ventromedial VTA are preserved. Scale bar, 500 μm. B, Quantitation of TH fiber loss by optical density reveals the early loss of fibers projecting to the CPu from age P7 with near complete denervation by P14. Fibers projecting to the NAc shell are largely preserved, and fibers to the NAc core and OT are partially preserved through 2.5 months. Data show mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001 versus respective control group by two-way ANOVA and Bonferroni post hoc test; n = 3–4 mice with 6–19 fields per mouse. C, HPLC of CPu dissected from fresh-frozen brain tissue from 1-month-old mice reveals complete loss of DA in Drp1KO mice, ***p < 0.001 by one-way ANOVA with Tukey post hoc test, n = 4 mice per group. D, Stereology shows severe loss of DA neurons in both the SNc and VTA. Data show mean ± SEM; **p < 0.01, ***p < 0.001 versus respective control group, n = 3–4 mice per group by two-way ANOVA and Bonferroni post hoc test. E, Fluorogold was injected into the NAc of 4-month-old control and Drp1KO mice 4 d before perfusion. Double staining against TH and Fluorogold shows that surviving NA fibers originate from DA neurons in the ventromedial VTA. Scale bars: 200 μm; inset, 10 μm.
To determine whether losing terminals in the CPu was accompanied by neuronal death, we used stereology to quantify the survival of DA neurons in the SNc (Fig. 2D). At P14, when terminals in the CPu were absent, the number of TH-positive DA neurons in the SNc dropped by ∼35%, indicating that synaptic terminals degenerate before cell bodies do. By 1 month, ∼85% of SNc DA neurons was absent. The majority of DA neurons in the VTA were also lost, although we consistently observed a subpopulation of DA neurons enriched in the ventromedial VTA that were preserved (Fig. 2A). To confirm that surviving terminals in the NAc originated from this subset of surviving DA neurons in the VTA, we injected Fluorogold, a retrograde axonal tracer, into the NAc. As expected, the Fluorogold accumulated within the surviving cell bodies in the VTA, many of which were in the ventromedial VTA, indicating that axonal integrity is preserved in the surviving DA neurons (Fig. 2E).
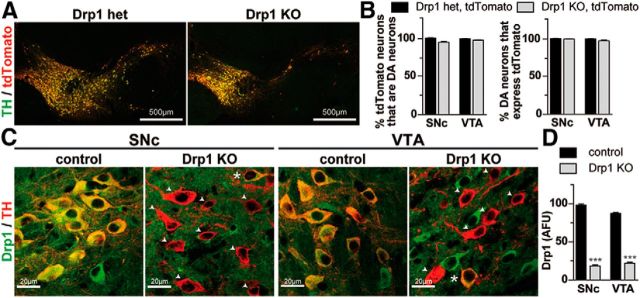
One reason this subpopulation of DA neurons survived might be because they did not express Cre. However, when mice were crossed with floxed tdTomato reporter mice (Madisen et al., 2010), the vast majority (≥98%) of neurons that showed tyrosine hydroxylase (TH+ in the SNc and VTA of the tdTomato-DATcre control (tdTomatolox/lox;DAT+/cre), and Drp1KO-tdTomato-DATcre (Drp1lox/lox;tdTomatolox/lox;DAT+/cre) mice also showed tdTomato fluorescence, indicating that they expressed Cre (Fig. 3A,B). As expected, the majority of tdTomato+ neurons in Drp1KO-tdTomato-DATcre mice also lost Drp1 expression in the SNc and VTA at P14 (Fig. 3C,D). Importantly, the vast majority (≥95%) of tdTomato+ neurons in the tdTomato control and Drp1KO mice also had detectable levels of TH, indicating that Cre is expressed specifically in midbrain DA neurons in control and Drp1KO mice (Fig. 3A,B). Rare tdTomato+ TH-negative neurons (neurons that expressed Cre but did not produce detectable TH) in the midbrain might have resulted from ectopic expression of Cre or severe downregulation of TH in DA neurons (Levinson et al., 1998). We also confirmed tdTomato expression in the control and surviving Drp1KO fibers projecting to the NAc and OT (data not shown). The loss of both tdTomato-positive cell bodies in the SNc and VTA and tdTomato-positive fibers in the CPu provides confirmation that the loss of TH immunoreactivity reflected an actual loss of cell bodies and processes rather than simply a downregulation of TH levels.
Figure 3.
Midbrain DA neurons in Drp1KO animals express Cre and lose Drp1. A, One-month-old Drp1KO-tdTomato-DATcre (Drp1lox/lox;tdTomatolox/wt;DATcre/wt) and tdTomato-DATcre control (tdTomatolox/wt;DATcre/wt) brains were processed for immunostaining against TH (to identify DA neurons) and tdTomato (as a surrogate for cre expression). Scale bar, 500 μm. B, Quantitation reveals that the vast majority of TH+ cell bodies are tdTomato+, indicating that essentially all DA neurons in the SN and VTA express Cre. Similarly, almost all tdTomato+ neurons are TH+, indicating that there is essentially no ectopic expression of Cre in non-DA neurons. Data show mean ± SEM, N = 3–4 mice per group, 84–590 cells per group. C, Midbrain sections from P14 Drp1KO-tdTomato-DATcre and tdTomato-DATcre mice were immunostained against Drp1 and tdTomato, and the intensity of Drp1 staining quantified on a cell-by-cell basis. Drp1 fluorescence was markedly decreased in DA neurons in both the SNc and VTA (arrowheads), although occasional neurons in both regions (stars) showed preservation of Drp1. Scale bar, 20 μm. D, Quantitation of mean Drp1 staining intensity in Drp1KO versus control cell bodies; ***p < 0.001 versus respective control region by unpaired two-tailed t test. Data show mean ± SEM, n = 4 mice per group, 56–126 cells per mouse.
The progressive loss of synaptic terminals from P7 and cell bodies from P14 suggests that cell loss results from degeneration rather than developmental changes. However, because Cre expression is detected in mouse embryos by age E17 in DATcre mice (Bäckman et al., 2006), developmental factors may contribute somewhat to these losses. To definitively determine whether Drp1 is required by nigrostriatal DA neurons to survive in adult animals, we also delivered an AAV-expressing Cre to the SNc/VTA of 3-month-old tdTomatolox/lox;Drp1lox/lox and tdTomatolox/lox mice. Mice were killed 2.5 months after injection, and we assessed the extent of striatal denervation and neuronal death in the SNc. Again, DA nerve terminals disappeared from Drp1lox/lox;tdTomatolox/lox mice but not tdTomatolox/lox control mice (Fig. 4), proving that Drp1 is required for adult nigral DA neurons to survive. We also saw that nerve terminals to the NAc were relatively preserved in all Drp1lox/lox;tdTomatolox/lox mice versus control animals, although not as much as in the DATcre mice.
Figure 4.
Adult DA neurons require Drp1 for survival. AAV-cre was delivered to the SNc of 3-month-old tdTomatolox/lox and Drp1lox/lox, tdTomatolox/lox mice. A, At 2.5 months later, TH immunohistochemistry revealed severe loss of midbrain DA neurons in the SNc and VTA of Drp1KO mice. Drp1KO also produced near complete loss of terminals in the CPu, although some fibers projecting to the NAc were again spared. Scale bar, 500 μm. B, tdTomato immunohistochemistry confirms Cre expression in residual TH+ fibers in the NAc. C, Quantitation of TH optical density in the striatum. Mice receiving AAV-cre had markedly decreased TH density in all regions examined (p < 0.0001 vs respective control region by one-way ANOVA with Bonferroni post hoc test). However, the residual optical density was significantly higher in the NAc shell than in the CPu (**p < 0.01), indicating that fibers projecting to the NAc shell were relatively spared. NAc core and OT also had a trend to greater residual optical density, but this did not reach significance versus CPu. Data show mean ± SEM, n = 4–7 mice per group.
Drp1 loss depletes mitochondria from axons
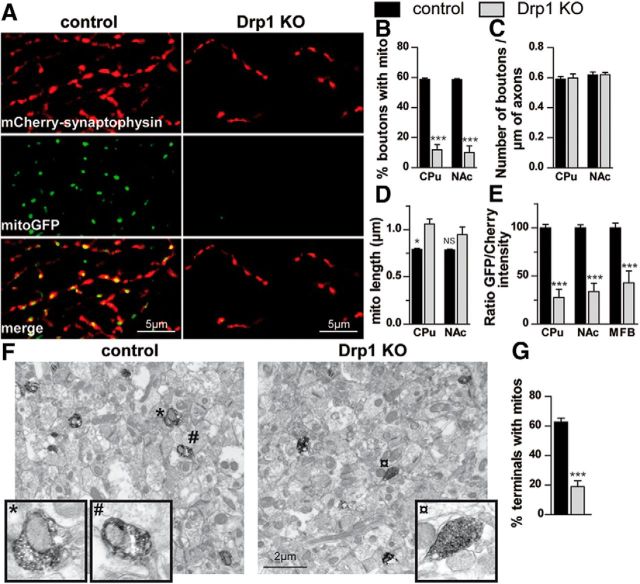
To begin to understand why Drp1KO is toxic to nigrostriatal DA neurons, we looked at the terminals where degeneration begins. Because axonal mitochondria are small and easily obscured by larger and more abundant mitochondria in other cellular compartments and in glia, it is nearly impossible to study axonal mitochondria by immunohistochemistry. We therefore engineered AAV-based viral reporters in the cre-dependent DIO construct (Sohal et al., 2009) that express mitochondria-targeted GFP (mitoGFP) to visualize mitochondria and mCherry-synaptophysin to visualize the surrounding synapses. When these constructs were delivered to DATcre mice, they expressed selectively in DA neurons. When we observed individual axons in the control DA neurons from DATcre mice, we found that ∼60% of mCherry-synaptophysin-positive axonal boutons in the CPu or NAc normally contain mitochondria. In contrast, only ∼10% of Drp1KO axonal boutons contained mitochondria (Fig. 5A,B), and these axonal mitochondria were slightly longer than those in the control DA neurons (Fig. 5D). When we calculated the ratio of total mitochondria to axonal boutons (i.e., total fluorescence of mitoGFP/total fluorescence of mCherry-synaptophysin) in the CPu and NAc, we also found that synaptic mitochondria were severely depleted (Fig. 5E). To further confirm this, we performed immuno-EM against TH to identify DA axons in the NAc. Consistent with our fluorescent reporters, Drp1KO mice showed a much lower percentage of TH+ axonal boutons that contained mitochondria (Fig. 5F,G). To determine whether the extent of axonal mitochondrial loss depended on the distance from the cell body, we also looked in the medial forebrain bundle (MFB), a more proximal location in the nigrostriatal and mesoaccumbens pathways. However, the extent of mitochondrial depletion was similar, indicating that even proximal axons had markedly fewer mitochondria (Fig. 5E). Somewhat surprisingly, despite that almost all mitochondria had disappeared from axons, Drp1KO did not decrease the density of axonal boutons within surviving axons in the NAc (Fig. 5C), although we do not yet know if these surviving axons function normally.
Figure 5.
Loss of Drp1 depletes mitochondria from axons. A–E, AAVs expressing mitochondria-targeted GFP (mitoGFP; green, to visualize mitochondria) and mCherrySynaptophysin (red, to visualize synaptic boutons) in DIO constructs (Sohal et al., 2009) that express only in Cre-expressing neurons were coinjected into the SNc of 21-d-old DATcre control and Drp1KO-DATcre mice. One month later, ∼60% of control synaptic boutons show mitochondria in the CPu and NAc. In contrast, remaining Drp1KO synapses in the CPu and NAc contain very few mitochondria, despite having normal numbers of boutons per length of axon. Scale bars, 5 μm. Data show mean ± SEM; *p < 0.05, ***p < 0.001 versus respective control group by one-way ANOVA and Tukey post hoc test; n = 4 mice per group, where each value is the mean of 8–20 fields. E, Images from regions of interest—the CPu, NAc, and the MFB—were also quantified based on the ratio of total GFP/mCherry fluorescence to approximate the ratio of total mitochondria/total axons. ***p < 0.001 versus indicated group; n = 5 mice per group, where each value is the mean of 2–8 fields. F, G, Immuno-EM of NAc, stained against TH to identify DA terminals show that 60% TH-positive boutons contain mitochondria in the NAc of control mice and only 20% of those in Drp1KO contain mitochondria. *, #, and ¤ mark boutons in inset. ***p < 0.001 by unpaired two-tailed t test, n = 4 mice per group, 17–41 terminals per mouse. Scale bar, 2 μm.
Decreased mitochondrial mass at the synapse might occur if there is a general decrease in mitochondrial mass throughout the neuron and/or if mitochondria are not being properly transported to the synapse. The first possibility seemed unlikely because Drp1 is required for parkin-induced removal of damaged mitochondria (mitophagy; Tanaka et al., 2010), and therefore, Drp1 loss might cause mitochondria to accumulate. In vivo, individual mitochondria in the SNc and VTA from 1-month-old Drp1KO mice were often swollen and rounded (Fig. 6A,B,G,H), as in cerebellar Purkinje cells (Kageyama et al., 2012). However, although individual mitochondria were larger, the total mass of mitochondria per cell area was markedly lower in DA neurons lacking Drp1 in both the SNc and VTA, as assessed by mitoGFP fluorescence (Fig. 6D) and by immunofluorescence for Tom20 (translocase of the outer mitochondrial membrane, a protein in the outer mitochondrial membrane; Fig. 6C), PDH (pyruvate dehydrogenase, matrix protein; Fig. 6E), and COX1 (mitochondrial cytochrome c oxidase subunit I, inner membrane; Fig. 6F). This loss appeared to reflect a true decrease in mitochondrial mass rather than an increase in density leading to less volume occupied, given that the density of Drp1KO mitochondria, assessed by immuno-EM against TH, was similar to that of controls (Fig. 6G). This mitochondrial mass loss may limit bioenergetic and other mitochondrial functions in surviving DA neurons, especially because the function of residual mitochondria may also be compromised (Kageyama et al., 2012). Indeed, although the cristae structure was normal in most mitochondria, it tended to be disrupted in the largest Drp1KO mitochondria, suggesting that at least some of the largest mitochondria may not function normally. Although these findings might indicate that only a subpopulation of neurons with lower mitochondrial masses survive, a scatter plot of integrated Tom20 fluorescence per cell body revealed a clear downward shift in the mitochondrial mass of Drp1KO DA neurons (data not shown), suggesting that mitochondrial mass truly drops when Drp1 is lost, especially in neurons without swollen mitochondria.
Figure 6.

Loss of Drp1 disrupts mitochondrial morphology and decreases mitochondrial mass in DA neurons. A–F, Mitochondrial morphology and mass in midbrain DA neurons from 1-month-old Drp1KO and control mice. A, Loss of Drp1 causes mitochondria to become visibly swollen in most midbrain DA neurons (identified by TH staining, red). B, Mitochondrial mass decreased upon determining the mean fluorescence (normalized to cell area) of neurons stained with Tom20 (C), AAVmitoGFP (AFU, arbitrary fluorescent units) (D), PDH (E), and cytochrome c oxidase I (Cox I; F). Scale bars: 5 μm. Data show mean ± SEM, *p < 0.05, **p < 0.001, ***p < 0.001 by unpaired two-tailed t test, n = 3–4 mice per group, 36–76 cells quantified per mouse. G, H, Ultrastructural analysis at the cell body by immunogold staining against TH revealed larger mitochondria in Drp1KO mice than in controls. Scale bars: 1 μm. Data show mean ± SEM, n = 3 mice per group, 11–16 cell bodies quantified per mouse.
Drp1 knock-out affects mitochondrial motility
In Drp1KO DA neurons, mitochondrial mass decreases more in the axons than in the cell bodies, indicating that additional axon-specific factors help to deplete axonal mitochondria. One possibility is that axonal mitochondria are too big to fit down narrow distal axons. However, the diameters of mitochondria in Drp1KO axons and control axons were similar (Fig. 7A), suggesting that mitochondrial size does not fully explain the misdistribution. We, therefore, determined whether axonal transport of mitochondria is impaired in Drp1KO axons. We examined hippocampal neurons, which are easier to image live than DA neurons. Postnatal hippocampal neurons from Drp1lox/lox mice were cotransfected with either Cre (to delete Drp1, referred to as Drp1KO) or vector control, in addition to mitochondria-targeted mito-GFP (to visualize mitochondria) and mCherry-synaptophysin (to identify axons).
Figure 7.
Loss of Drp1 inhibits mitochondrial mobility in axons. A, B, Depletion of Drp1 in hippocampal neurons produces long mitochondria with normal widths (mitoGFP) in axons (mCherrySynatophysin). Loss of Drp1 markedly decreases the percentage of moving mitochondria. There is also a trend for decreased mean velocity of movement, as depicted on kymographs. Scale bar, 10 μm. C, There was no difference in the length of moving and stationary Drp1KO mitochondria. D, In contrast to controls, Drp1KO mitochondria often showed extension–retraction movements, where one or both sides moved without any net movement of the mitochondria [uncoordinated (unc) movements]). E, These were more frequent on the retrograde side. F, G, Treatment with the calcium ionophore calcimycin (1 μm) decreases net movement of control mitochondria, but does not block the uncoordinated movements of Drp1KO mitochondria. Data show mean ± SEM, *p < 0.05, **p < 0.001, ***p < 0.001 by unpaired two-tailed t test (A–D, F, G) or one-way ANOVA and Tukey post hoc test (E), n = 16–48 fields per group, from 2—4 experiments.
Deleting Drp1 from cultured hippocampal neurons produced rounded, enlarged mitochondria at the cell body, similar to what we observed in DA neurons in vivo. Axonal mitochondria were also longer—significantly longer than the elongated residual axonal mitochondria we found in vivo—and separated by large gaps (Figs. 5, 7). A much smaller percentage of mitochondria was moving in Drp1KO axons, independent of mitochondrial length. There was also a trend for the velocity of the moving mitochondria to be slower than in controls (Fig. 7A,B). In addition, we imaged parts of axons that were relatively straight (Fig. 7A), so this impaired motility did not appear to arise because mitochondria had trouble turning corners.
To better understand why many Drp1KO mitochondria could not move, we focused on the stationary mitochondria. Interestingly, in contrast to controls, Drp1KO mitochondria often showed asymmetric extension–retraction movements, whereby only one side of a mitochondrion moved (Fig. 7D,E). The proportion of Drp1KO mitochondria with these asymmetric movements correlated well with the drop in mitochondrial movement between control and Drp1KO axons. Therefore, poor coordination may impair mitochondrial movement in Drp1KO axons, which would contribute to the paucity of mitochondria at the nerve terminal. Interestingly, these asymmetric movements occurred more frequently on the cell body (retrograde) side rather than the distal (anterograde) side (Fig. 7E), suggesting that mitochondria perhaps have particular difficulty coordinating their movements in this direction. Surprisingly, these asymmetric retraction–extension movements did not respond to calcium; calcimycin (1 μm or 10 μm) failed to block the retraction–extension movements of Drp1KO mitochondria, despite decreasing the movement of control mitochondria (Fig. 7F,G). More Drp1KO mitochondria rapidly fragmented at higher calcimycin doses (∼40 and 65% at 1 and 10 μm, respectively), and we excluded these mitochondria from further analysis. These data suggest that poorly coordinated mitochondrial movements and an overall decrease in mitochondrial mass cause mitochondrial content at the synapse to decrease, followed by subsequent degeneration.
Drp1 knock-out decreases total mitochondrial membrane potential per cell
To gain insight into whether Drp1KO compromises the respiratory function of mitochondria, we measured mitochondrial membrane potential with the dye TMRM (20 nm). Drp1KO neurons had a lower baseline mitochondrial membrane potential than controls (i.e., a lower mean TMRM fluorescence over the cell body, excluding the nucleus), suggesting that Drp1KO neurons have less capacity for respiratory function (Fig. 8A,B). Interestingly, this resulted because the ratio of the (area of TMRM-labeled mitochondria)/(area of the cytoplasm) was decreased in Drp1KO neurons (Fig. 8C). This decrease in the content of polarized mitochondria reflects a decrease in the total content of mitochondria, rather than in the percentage of mitochondria that are polarized, as the area of total mitochondria per cell assessed by Tom20 staining was also decreased (Fig. 8D). Indeed, those mitochondria that are present have normal mitochondrial membrane potential (Fig. 8E), suggesting that they have normal respiratory function. Consistent with this, Drp1KO mitochondria maintain their membrane potential in the presence of oligomycin (Figs. 8B), indicating that they are sufficiently functional to maintain their polarization by respiration, rather than by hydrolysis of ATP by complex V with reverse pumping of protons out of the mitochondrial matrix (Gandhi et al., 2009).These data suggest that Drp1KO neurons have less capacity to produce ATP due to decreased mitochondrial mass, with the effects more prominent at the nerve terminal where mitochondrial mass is decreased to a greater degree.
Figure 8.

Loss of Drp1 decreases mitochondrial membrane potential at the cell body. A, B, Drp1KO and control hippocampal neurons expressing mitoBFP were loaded with the membrane potential-sensitive dye TMRM (20 nm) for 1 h before imaging live. Selected on the basis of BFP, TMRM fluorescence was quantified in individual cells. At baseline, Drp1KO neurons had a lower mean TMRM fluorescence (normalized to the cytoplasmic area) that was not affected by the ATP synthase inhibitor oligomycin (5 μm), but markedly decreased by the proton ionophore FCCP (10 μm). Scale bar, 5 μm. Data show mean ± SEM, *p < 0.05 by two-way ANOVA, n = 5–9 fields (9–12 cells) per group, from two experiments. The experiment was repeated two additional times using a lower oligomycin dose (2.5 μm) with similar results. C, The percentage of the cytoplasmic area covered by TMRM+ mitochondria was decreased in Drp1KO neurons. D, In addition, the area of total mitochondria per cell assessed by Tom20 staining was also decreased. E, Drp1KO did not affect the mean fluorescence of the TMRM-labeled mitochondria themselves, indicating that the mitochondria in Drp1KO neurons had normal polarization. *p < 0.05, ***p < 0.001 by unpaired two-tailed t test, n = 22–26 cells per group (15–20 fields per group).
Intrinsic properties of susceptible versus resistant DA neurons
The subpopulation of resistant Drp1KO-DATcre DA neurons are of particular interest. The mechanisms of their resistance may underlie the resistance of midbrain DA neurons in other mitochondrial models of PD and in PD itself. Indeed, although the resistant DA neurons are enriched in the ventromedial VTA, they are also scattered through other parts of the VTA and even occasionally in the SNc and, hence, share the same vasculature and local environment as the susceptible DA neurons. So, why are they resistant? One possibility is that resistant DA neurons have parallel fission pathways that can compensate for losing Drp1. To examine this, we looked at levels of α-synuclein, which accumulates in the brains of essentially all patients with sporadic PD (Spillantini et al., 1998). α-Synuclein interacts with and fragments mitochondria in cultured cells in a pathway parallel to Drp1's pathway (Kamp et al., 2010; Nakamura et al., 2011). Therefore, it might be upregulated in Drp1KO DA neurons to compensate for their loss of Drp1. However, in 2-week-old Drp1KO mice—at an age where mice have lost their projections of DA terminals into the CPu but most cell bodies are intact (Fig. 2)—α-synuclein levels were markedly decreased in SNc DA neurons. Therefore, α-synuclein was downregulated rather than upregulated to compensate for Drp1 loss. While, α-synuclein levels were the same in the resistant DA neurons in the ventromedial VTA of 2-week-old Drp1KO and control mice (Fig. 9) by 9 months, the α-synuclein levels even in these surviving Drp1KO DA neurons dropped. Therefore, α-synuclein levels decrease to compensate for Drp1 loss in both susceptible and resistant neurons. This is particularly surprising as mitochondrial and other toxins consistently increase synuclein levels and/or aggregation (Vila et al., 2000; Lee et al., 2002; Manning-Bog et al., 2002; Sherer et al., 2002, 2003). These findings do not represent a general decrease in protein synthesis, as the levels of other proteins, such as calbindin and the tdTomato reporter, remained unchanged in Drp1KO mice (Fig. 10B,C). Instead, the lower levels of α-synuclein may protect against the toxicity associated with decreased fission, as in other cases of compromised mitochondrial function (Klivenyi et al., 2000; Dauer et al., 2002).
Figure 9.
α-Synuclein levels are decreased in Drp1KO DA neurons. A, B, Two-week-old Drp1KO-tdTomato-DATcre and tdTomato-DATcre mice were perfused and then immunostained against α-synuclein and tdTomato, and the mean intensity of α-synuclein staining (normalized per cell) was quantified on a cell-by-cell basis. At 2 weeks, when DA terminals have degenerated in the CPu but most DA cell bodies remain in the midbrain, α-synuclein levels were markedly decreased in SNc but not in ventromedial VTA (VTAVM) DA neurons. Data show mean ± SEM, *p < 0.05 by unpaired two-tailed t test, n = 3 mice per group, 33–67 cell bodies quantified per mouse. Scale bar, 5 μm. C, By 9 months, α-synuclein levels had also decreased in surviving Drp1KO versus Drp1Het DA neurons in the VTAVM. Data show mean ± SEM, n = 2–3 mice per group, 25–50 cell bodies quantified per mouse.
Figure 10.
Increased calbindin does not identify resistant Drp1KO DA neurons. A–C, Midbrain sections from 1-month-old Drp1KO-tdTomato-DATcre and tdTomato-DATcre mice were immunostained against calbindin and tdTomato, and the intensity of calbindin staining was quantified on a cell-by-cell basis. B, VTA DA neurons had higher mean calbindin than SNc DA neurons (AFU, arbitrary fluorescence units). However, surviving Drp1KO DA neurons had calbindin levels similar to controls in the VTA, and only slightly elevated in the SNc. Scatter graphs of calbindin levels in these same individual DA neurons reveal that many of the surviving Drp1KO DA neurons have low calbindin. C, tdTomato expression is similar between controls and Drp1KO, suggesting a similar capacity to synthesize protein. Data show mean ± SEM, *p < 0.05, ***p < 0.001 by one-way ANOVA and Tukey post hoc test, n = 4 mice per group, 6–12 fields, 374–2515 cell bodies quantified per mouse. Scale bars: A, 100 μm; inset, 10 μm.
An as yet undefined fission mechanism might compensate for Drp1 loss in resistant DA neurons. However, this seems unlikely: Drp1 loss caused mitochondria to swell in both the resistant and susceptible midbrain DA neurons, and mitochondria were also depleted from the axons of resistant DA neurons. Drp1 thus appears to regulate mitochondrial fission in both resistant and susceptible DA neurons. Therefore, the resistant DA neurons must have core properties that make them intrinsically resistant to Drp1 loss or other differences (e.g., in activity level) obtained through the basal ganglia circuitry.
To better understand these differences in susceptibility, we must identify the populations of resistant and susceptible neurons prospectively, before the cells die. This would first allow us to understand if the surviving DA neurons are essentially the same as those that are lost, but they survive simply because they are a part of a normal standard distribution. Conversely, we may learn that they actually represent a distinct subpopulation of DA neurons that does not require mitochondrial fission. To address this, we examined calbindin levels, because DA neurons that express calbindin tend to be more resistant on average to the neurotoxin MPTP (Liang et al., 1996). Consistent with prior data, we found that, in Drp1KO DA neurons, mean calbindin levels were higher in the VTA than in the SN (Fig. 10). In the SNc, on average, surviving Drp1KO SNc DA neurons also had slightly higher calbindin than controls. However, calbindin did not distinguish resistant from susceptible VTA DA neurons, and many surviving Drp1KO DA neurons in the SNc and VTA lacked calbindin (Fig. 10A,B). Therefore, high calbindin level is only a weak predictor of resistance to Drp1 loss.
We next considered if other known DA neuron subtypes are particularly resistant. Because electrophysiologically distinct subsets of midbrain DA neurons exist (Lammel et al., 2008, 2011), we aimed to determine whether the surviving Drp1KO DA neurons represent one of these subsets by using whole-cell recordings in horizontal brain slices. We identified surviving neurons scattered throughout the VTA and SNc as DA or non-DA during recording by visually detecting tdTomato expression. Their identities were later confirmed by post hoc immunocytochemical localization of TH in the biocytin-filled recorded neurons (Fig. 11). As expected, TH-negative (non-DA) neurons from both control and Drp1KO-DATcre mice had small to undetectable hyperpolarization-activated current (Ih) magnitudes (Fig. 11G,H), while DA neurons in wild-type mice showed a range of Ih magnitudes in the VTA and SN. In contrast, in Drp1KO-DATcre mice, all recorded surviving DA neurons in the VTA expressed a small or absent Ih (Fig. 11C). Most Drp1KO DA neurons in the SNc died, but among the 11 that we detected for recording, 10 of these also expressed unusually small Ih magnitudes for the SNc. Therefore, resistant DA neurons have small Ih, consistent with prior reports that NA-projecting VTA neurons have small Ih, (Margolis et al., 2006; T. A. Zhang et al., 2010; Lammel et al., 2011) and our observation that NA-projecting neurons tend to survive in the Drp1KO mice.
Figure 11.

Electrophysiological properties of the subpopulation(s) of resistant DA neurons. Whole-cell recordings were made in horizontal brain slices. DA and non-DA neurons were identified live based on the presence or absence of the tdTomato reporter, respectively, and confirmed post hoc by TH immunostaining. A, Anatomic locations of the recorded and recovered wild-type (blue) and knock-out (red) TH-positive cells (A, anterior; P, posterior; M, medial; L, lateral), which were scattered throughout the VTA, and occasionally in the SNc. SNr, substantia nigra pars reticulata; MT, medial terminal nucleus of the accessory optic tract; IPF, interpeduncular fossa. B, Firing rate of surviving Drp1KO DA neurons is similar to controls in the VTA and SN overall, although was markedly increased in three of the SNc neurons sampled. C, While all DA neurons in wild-type mice exhibited a detectable Ih (a hyperpolarization-activated nonselective cation current; closed circles, detectable; open, not detectable), almost all surviving DA neurons had very small or absent Ih. D, Surviving Drp1KO DA neurons also had smaller input resistance (Ri), consistent with their smaller size. E, AP durations were similar between DA neurons from wild-type (wt) and Drp1KO mice (ko). F, VTA DA neurons from Drp1KO mice had higher coefficients of variation of the CV-ISI than controls. G, Anatomic locations of the recorded and recovered TH-negative neurons, which were scattered throughout the VTA, with occasional cells in the SN. H, Non-DA neurons from both control and Drp1KO mice had very small Ih values. Some of the non-DA SNc neurons expressed larger than expected Ihs. I, J, Input resistances (Ri) and AP durations in both brain regions were overlapping between wild-type and Drp1KO samples. K, No differences were observed between the coefficient of variation of the CV-ISI of non-DA neurons in Drp1KO animals. *p < 0.05, **p < 0.01,***p < 0.001 by unpaired two-tailed t test. Horizontal lines show mean value for each group. Recordings were made in four Drp1KO mice and four wild-type mice. Scale bars: horizontal, 200 ms; vertical, 20 mV.
To further characterize these resistant DA neurons, we checked their input resistance (Ri)—a value related to the number of open channels on the plasma membrane—which is generally proportional to cell size. As expected, surviving VTA and SN DA neurons had higher Ri values, consistent with their smaller somatic size (cross-sectional areas of recorded neurons: VTA wild-type: 245 ± 16 μm2, n = 10; Drp1KO-DATcre: 103 ± 17 μm2, n = 7; p < 0.0001; SN wild-type: 237 ± 13 μm2, n = 16; Drp1KO-DATcre: 150 ± 16 μm2, n = 11; p < 0.001; Fig. 11). Because recordings tend to be biased toward larger neurons, these data suggest that larger, low Ri, and large Ih neurons were absent in Drp1KO-DATcre mice. To further investigate the size discrepancy, we analyzed a larger subset of cells and confirmed that surviving Drp1KO DA neurons in the VTA were slightly smaller than controls (104 ± 8.6 vs 137 ± 16.7 μm2, n = 3 mice per group, 37–76 cells per mouse) while Drp1KO DA neurons in the SNc were markedly smaller (102 ± 5.3 vs 175 ± 10.9 μm2, n = 3 mice per group, 38–55 cells per mouse). This size discrepancy may mean that these DA neurons shrunk after Drp1 loss (e.g., due to insufficient energy), or that surviving DA neurons were smaller to begin with (or perhaps both).
While knocking out Drp1 produced differences in Ih and Ri between surviving Drp1KO DA neurons and control DA neurons, the Drp1KO did not affect action potential duration (Fig. 11E). In addition, there was no overall difference in the firing rate of VTA control versus Drp1KO-DATcre DA neurons (Fig. 11B), which use a persistent Na+ current to maintain pacemaker firing (Khaliq and Bean, 2010). In contrast, some SNc DA neurons maintain pacemaker firing by Ih activity (Neuhoff et al., 2002; Chan et al., 2007); however, three SNc Drp1KO-DATcre DA neurons actually displayed unusually elevated firing rates for ex vivo recordings. Even though firing rates appeared to be unchanged in the Drp1KO-DATcre mice, the firing pattern was somewhat disrupted: the coefficient of variation of the ISI-CV was significantly higher in VTA DA neurons and there was a trend (p = 0.08) toward higher ISI-CV in SNc DA neurons from Drp1KO-DATcre mice. This indicates that pacemaker firing was less regular in these neurons even though the firing rates were unchanged. It is unclear how this might affect neural firing in the intact circuit in vivo.
Discussion
We show that nigrostriatal DA neurons require Drp1 to maintain their axons and survive. In vivo, losing Drp1 depletes the mitochondria in axons by impairing the coordination of mitochondrial movements and decreasing the overall mitochondrial mass. However, we also show that the midbrain DA neurons resistant to disruptions in mitochondrial fission have small Ih values. This population of midbrain DA neurons survives despite also losing most mitochondria from its processes.
In mammalian neurons, Drp1 is required for normal mitochondrial mass at the synapses in vivo
We used viral reporters to visualize mitochondria in individual axons of DA neurons, and we found that Drp1KO DA neurons have markedly fewer axonal mitochondria than control neurons, thus replicating previous findings in the Drosophila neuromuscular junction (Verstreken et al., 2005) in the mammalian brain. We found that the mechanism causing axons to lose their mitochondria is defined by two key factors. (1) Mitochondrial movement is slower in Drp1KO axons than in control axons. This slower movement does not arise because mitochondria are too big to fit through axons; the width of Drp1KO axons is similar to controls, and parts of Drp1KO mitochondria easily extend and retract within axons (Fig. 7). Instead, the axonal motors on Drp1KO mitochondria are not coordinated, which disrupts normal movement and produces unusual and asymmetric extension–retraction movements that occur more commonly on the retrograde side. These movements have not been reported previously and do not occur in control neurons. Mitochondrial movement in neurons thus depends heavily on specific mitochondrial fusion and fission proteins, as losing the fusion protein mitofusin 2 (Mfn2), but not Opa1, also disrupts mobility in neurites (Misko et al., 2010; Pham et al., 2012). (2) The overall mass of mitochondria is lower when Drp1 is absent. This is surprising because mitochondrial fission is thought to help remove damaged mitochondria via mitophagy (Arnoult et al., 2005; Twig et al., 2008; Frieden et al., 2004; Park et al., 2008; Mao et al., 2013), so losing Drp1 might be expected to increase mitochondrial mass. However, we now show that in vivo, DA neurons lacking Drp1 actually have less mitochondrial mass in the cell body, and mitochondria are severely depleted from axons, where the vast majority of neuronal mitochondria probably reside (Grafstein and Forman, 1980). Therefore, the total mitochondrial mass per neuron is markedly lower in DA neurons lacking Drp1. This may be attributable to decreasing the fission–fusion ratio, as increased Mfn1 also decreases mitochondrial mass in cultured cells (Park et al., 2008). Because mitochondrial mass reflects a balance between the rate of mitochondrial synthesis and degradation, a decrease in mass reflects either less synthesis or more degradation. Further studies will determine whether Drp1KO can lead to secondary problems whereby neurons improperly synthesize mitochondria.
Mitochondrial fission can contribute to selective dopamine neuron degeneration
The PD proteins PINK1 and parkin function as mitochondrial fission proteins in Drosophila. Mutations in these proteins can decrease the fission–fusion ratio, and their toxicity can be blocked by increasing Drp1 (Poole et al., 2008; Yang et al., 2008). Conversely, increased levels and/or mutations of the autosomal-dominant PD proteins α-synuclein and LRRK2 increase the fission–fusion balance (Kamp et al., 2010; Nakamura et al., 2011; X. Wang et al., 2012b), suggesting that disrupting the fission–fusion balance in either direction might promote degeneration (Nakamura, 2013). In agreement with this theory, mutations in either the fusion protein Mfn2 or the fission protein ganglioside-induced differentiation associated-protein 1 (cause peripheral neuropathy in Charcot-Marie-Tooth disease (Züchner et al., 2004; Niemann et al., 2005). Interestingly, losing Mfn2 also promotes degeneration of SNc DA neurons beginning with axonal loss (Lee et al., 2012; Pham et al., 2012). However, the rate of degeneration is much slower than with Drp1KO, and the mechanism also appears to be different. In contrast to Drp1KO, Mfn2KO DA neurons do not appear to lose mitochondria from the remaining axons (Lee et al., 2012).
The loss of axonal mitochondria and early degeneration of Drp1KO axons strongly suggest that neurodegeneration is caused by a loss, rather than a gain, of mitochondrial function, such as accumulating ROS. This neurodegeneration likely stems from energy depletion within the axons, which depend on ATP to maintain synaptic transmission (Verstreken et al., 2005). Indeed, axons lacking mitochondria may have lower ATP levels in culture (Zala et al., 2013). Furthermore, SNc DA neurons may be particularly dependent on the mass and distribution of mitochondria in their processes: they have unusually high energy requirements because they are unmyelinated (Pickel et al., 1992; Braak and Del Tredici, 2004) and have large projection fields (Matsuda et al., 2009). Although VTA DA neurons are also unmyelinated (R. Y. Wang, 1981; Nirenberg et al., 1997), they generally have fewer collaterals (Fallon and Loughlin, 1982) and likely have smaller projection fields. That said, we do not yet know the specific anatomical properties of the resistant, small-Ih DA neurons we identified. Axonal mitochondria presumably have other important functions, including calcium buffering and lipid metabolism (Nunnari and Suomalainen, 2012), and losing these functions may also contribute to axonal degeneration. Ultimately, we do not know if axonal death alone triggers neuronal death. Other factors at the cell body might also contribute, such as insufficient energy and an accumulation of ROS (Kageyama et al., 2012).
Small Ih allows prospective identification of resistant dopamine neurons
To better understand how selective degeneration occurs, we must be able to identify prospectively—before the neurons die—the specific neuronal populations that are vulnerable and resistant. However, previously there was no way to do this. Although high levels of calbindin have been proposed as a marker for DA neurons that are resistant to certain mitochondrial stressors among adjacent neurons (Liang et al., 1996), we found that increased calbindin levels poorly predicted Drp1KO resistance. Instead, we found that midbrain DA neurons resistant to Drp1 loss all have a small Ih value. These resistant DA neurons are scattered throughout the VTA and occasionally reside in the SNc. They include the DA neurons in the ventromedial VTA that project to the NA and prefrontal cortex and were recently described to have small Ih (Lammel et al., 2011). DA neurons projecting to the NAc and OT are also less affected than those projecting to the CPu in PD and in pharmacologic and genetic models of mitochondrial dysfunction that target mitochondria (Heikkila et al., 1984; Miller et al., 1997; Betarbet et al., 2000; Ekstrand et al., 2007; Pham et al., 2012), but the properties of these surviving DA neurons have not been characterized. The Drp1KO-resistant, small-Ih DA neurons we observed may thus also be resistant to other PD-related mitochondrial insults. Therefore, having a small Ih value may be a general marker for DA neurons that are resistant to mitochondrial stressors and that might also be resistant in PD. Interestingly, recent reports also show that Ih can decrease in DA neurons in response to certain mitochondrial insults (Good et al., 2011). Although small Ih is an intrinsic property of VTA DA neurons that project to the NA (Margolis et al., 2006; T. A. Zhang et al., 2010; Lammel et al., 2011) and we now show that these small Ih neurons are resistant to Drp1KO, susceptible DA neurons may also decrease Ih as a protective response against mitochondrial insults.
Having a small Ih may be more than just a marker for resistant midbrain DA neurons. It may directly contribute to the resistance (Neuhoff et al., 2002), for instance, by directly modulating neuronal activity levels, energy requirements, and/or ROS production. Indeed, L-type calcium channels sensitize adult SNc DA neurons to mitochondrial stressors (Chan et al., 2007), and perhaps some susceptible VTA DA neurons also have these channels, or, Ih may also function similarly by priming vulnerable VTA and SNc neurons to degeneration. Better defining the molecular basis for this selective resistance is particularly important, as the resistant neurons share the same vasculature and local environment (e.g., iron levels) as susceptible neurons, so their resistance reflects differences in their intrinsic properties or connections. Understanding this resistance could help us discover why nigrostriatal DA neurons are susceptible to mitochondrial insults in PD.
Footnotes
This work was supported through the generosity of Betty Brown's Family and the Joan and David Traitel Family Trust. A.B., Jue Zhang, and K.N. were supported by a Burroughs-Wellcome Fund Award (to K.N.) and KO8NS062954 and P30NS069496 from National Institute of Neurological Disorders and Stroke; the Gladstone Behavioral core by National Institutes of Health (NIH) Grant P30NS065780; and the animal care facility by NIH/National Center for Research Resources Grant C06 RR018928. I.H., Jiasheng Zhang, and E.H. were supported by Merit Award BX001108 from the Department of Veterans Affairs and NIH Grant OD010927. E.B.M. was supported by R01DA030529 from National Institute on Drug Abuse. We thank Latrice Goss for administrative assistance and Celeste Brennecka and Gary Howard for editorial assistance.
The authors declare no competing financial interests.
References
- Alexander C, Votruba M, Pesch UE, Thiselton DL, Mayer S, Moore A, Rodriguez M, Kellner U, Leo-Kottler B, Auburger G, Bhattacharya SS, Wissinger B. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- Arnoult D, Rismanchi N, Grodet A, Roberts RG, Seeburg DP, Estaquier J, Sheng M, Blackstone C. Bax/Bak-dependent release of DDP/TIMM8a promotes Drp1-mediated mitochondrial fission and mitoptosis during programmed cell death. Curr Biol. 2005;15:2112–2118. doi: 10.1016/j.cub.2005.10.041. [DOI] [PubMed] [Google Scholar]
- Bäckman CM, Malik N, Zhang Y, Shan L, Grinberg A, Hoffer BJ, Westphal H, Tomac AC. Characterization of a mouse strain expressing Cre recombinase from the 3′ untranslated region of the dopamine transporter locus. Genesis. 2006;44:383–390. doi: 10.1002/dvg.20228. [DOI] [PubMed] [Google Scholar]
- Berthet A, Porras G, Doudnikoff E, Stark H, Cador M, Bezard E, Bloch B. Pharmacological analysis demonstrates dramatic alteration of D1 dopamine receptor neuronal distribution in the rat analog of L-DOPA-induced dyskinesia. J Neurosci. 2009;29:4829–4835. doi: 10.1523/JNEUROSCI.5884-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K. Poor and protracted myelination as a contributory factor to neurodegenerative disorders. Neurobiol Aging. 2004;25:19–23. doi: 10.1016/j.neurobiolaging.2003.04.001. [DOI] [PubMed] [Google Scholar]
- Chan CS, Guzman JN, Ilijic E, Mercer JN, Rick C, Tkatch T, Meredith GE, Surmeier DJ. ‘Rejuvenation’ protects neurons in mouse models of Parkinson's disease. Nature. 2007;447:1081–1086. doi: 10.1038/nature05865. [DOI] [PubMed] [Google Scholar]
- Cheng HC, Ulane CM, Burke RE. Clinical progression in Parkinson disease and the neurobiology of axons. Ann Neurol. 2010;67:715–725. doi: 10.1002/ana.21995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W, Kholodilov N, Vila M, Trillat AC, Goodchild R, Larsen KE, Staal R, Tieu K, Schmitz Y, Yuan CA, Rocha M, Jackson-Lewis V, Hersch S, Sulzer D, Przedborski S, Burke R, Hen R. Resistance of alpha -synuclein null mice to the parkinsonian neurotoxin MPTP. Proc Natl Acad Sci U S A. 2002;99:14524–14529. doi: 10.1073/pnas.172514599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumartin B, Caillé I, Gonon F, Bloch B. Internalization of D1 dopamine receptor in striatal neurons in vivo as evidence of activation by dopamine agonists. J Neurosci. 1998;18:1650–1661. doi: 10.1523/JNEUROSCI.18-05-01650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrand MI, Terzioglu M, Galter D, Zhu S, Hofstetter C, Lindqvist E, Thams S, Bergstrand A, Hansson FS, Trifunovic A, Hoffer B, Cullheim S, Mohammed AH, Olson L, Larsson NG. Progressive parkinsonism in mice with respiratory-chain-deficient dopamine neurons. Proc Natl Acad Sci U S A. 2007;104:1325–1330. doi: 10.1073/pnas.0605208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon JH, Loughlin SE. Monoamine innervation of the forebrain: collateralization. Brain Res Bull. 1982;9:295–307. doi: 10.1016/0361-9230(82)90143-5. [DOI] [PubMed] [Google Scholar]
- Frieden M, James D, Castelbou C, Danckaert A, Martinou JC, Demaurex N. Ca(2+) homeostasis during mitochondrial fragmentation and perinuclear clustering induced by hFis1. J Biol Chem. 2004;279:22704–22714. doi: 10.1074/jbc.M312366200. [DOI] [PubMed] [Google Scholar]
- Gandhi S, Wood-Kaczmar A, Yao Z, Plun-Favreau H, Deas E, Klupsch K, Downward J, Latchman DS, Tabrizi SJ, Wood NW, Duchen MR, Abramov AY. PINK1-associated Parkinson's disease is caused by neuronal vulnerability to calcium-induced cell death. Mol Cell. 2009;33:627–638. doi: 10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good CH, Hoffman AF, Hoffer BJ, Chefer VI, Shippenberg TS, Bäckman CM, Larsson NG, Olson L, Gellhaar S, Galter D, Lupica CR. Impaired nigrostriatal function precedes behavioral deficits in a genetic mitochondrial model of Parkinson's disease. FASEB J. 2011;25:1333–1344. doi: 10.1096/fj.10-173625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstein B, Forman DS. Intracellular transport in neurons. Physiol Rev. 1980;60:1167–1283. doi: 10.1152/physrev.1980.60.4.1167. [DOI] [PubMed] [Google Scholar]
- Heikkila RE, Hess A, Duvoisin RC. Dopaminergic neurotoxicity of 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine in mice. Science. 1984;224:1451–1453. doi: 10.1126/science.6610213. [DOI] [PubMed] [Google Scholar]
- Hnasko TS, Perez FA, Scouras AD, Stoll EA, Gale SD, Luquet S, Phillips PE, Kremer EJ, Palmiter RD. Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proc Natl Acad Sci U S A. 2006;103:8858–8863. doi: 10.1073/pnas.0603081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnasko TS, Chuhma N, Zhang H, Goh GY, Sulzer D, Palmiter RD, Rayport S, Edwards RH. Vesicular glutamate transport promotes dopamine storage and glutamate corelease in vivo. Neuron. 2010;65:643–656. doi: 10.1016/j.neuron.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Leal-Ortiz S, Foss SM, Waites CL, Garner CC, Voglmaier SM, Edwards RH. v-SNARE composition distinguishes synaptic vesicle pools. Neuron. 2011;71:474–487. doi: 10.1016/j.neuron.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrcher I, Aleyasin H, Seifert EL, Hewitt SJ, Chhabra S, Phillips M, Lutz AK, Rousseaux MW, Bevilacqua L, Jahani-Asl A, Callaghan S, MacLaurin JG, Winklhofer KF, Rizzu P, Rippstein P, Kim RH, Chen CX, Fon EA, Slack RS, Harper ME, et al. Loss of the Parkinson's disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum Mol Genet. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J Cell Biol. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp F, Exner N, Lutz AK, Wender N, Hegermann J, Brunner B, Nuscher B, Bartels T, Giese A, Beyer K, Eimer S, Winklhofer KF, Haass C. Inhibition of mitochondrial fusion by alpha-synuclein is rescued by PINK1, Parkin and DJ-1. EMBO J. 2010;29:3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaliq ZM, Bean BP. Pacemaking in dopaminergic ventral tegmental area neurons: depolarizing drive from background and voltage-dependent sodium conductances. J Neurosci. 2010;30:7401–7413. doi: 10.1523/JNEUROSCI.0143-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Szczypka MS, Palmiter RD. Dopamine-deficient mice are hypersensitive to dopamine receptor agonists. J Neurosci. 2000;20:4405–4413. doi: 10.1523/JNEUROSCI.20-12-04405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klivenyi P, Andreassen OA, Ferrante RJ, Dedeoglu A, Mueller G, Lancelot E, Bogdanov M, Andersen JK, Jiang D, Beal MF. Mice deficient in cellular glutathione peroxidase show increased vulnerability to malonate, 3-nitropropionic acid, and 1-methyl-4-phenyl-1,2,5,6-tetrahydropyridine. J Neurosci. 2000;20:1–7. doi: 10.1523/JNEUROSCI.20-01-00001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammel S, Hetzel A, Häckel O, Jones I, Liss B, Roeper J. Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron. 2008;57:760–773. doi: 10.1016/j.neuron.2008.01.022. [DOI] [PubMed] [Google Scholar]
- Lammel S, Ion DI, Roeper J, Malenka RC. Projection-specific modulation of dopamine neuron synapses by aversive and rewarding stimuli. Neuron. 2011;70:855–862. doi: 10.1016/j.neuron.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Shin SY, Choi C, Lee YH, Lee SJ. Formation and removal of alpha-synuclein aggregates in cells exposed to mitochondrial inhibitors. J Biol Chem. 2002;277:5411–5417. doi: 10.1074/jbc.M105326200. [DOI] [PubMed] [Google Scholar]
- Lee S, Sterky FH, Mourier A, Terzioglu M, Cullheim S, Olson L, Larsson NG. Mitofusin 2 is necessary for striatal axonal projections of midbrain dopamine neurons. Hum Mol Genet. 2012;21:4827–4835. doi: 10.1093/hmg/dds352. [DOI] [PubMed] [Google Scholar]
- Levinson AJ, Garside S, Rosebush PI, Mazurek MF. Haloperidol induces persistent down-regulation of tyrosine hydroxylase immunoreactivity in substantia nigra but not ventral tegmental area in the rat. Neuroscience. 1998;84:201–211. doi: 10.1016/S0306-4522(97)00447-8. [DOI] [PubMed] [Google Scholar]
- Li H, Li SH, Yu ZX, Shelbourne P, Li XJ. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington's disease mice. J Neurosci. 2001;21:8473–8481. doi: 10.1523/JNEUROSCI.21-21-08473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CL, Sinton CM, German DC. Midbrain dopaminergic neurons in the mouse: co-localization with Calbindin-D28K and calretinin. Neuroscience. 1996;75:523–533. doi: 10.1016/0306-4522(96)00228-X. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning-Bog AB, McCormack AL, Li J, Uversky VN, Fink AL, Di Monte DA. The herbicide paraquat causes up-regulation and aggregation of alpha-synuclein in mice: paraquat and alpha-synuclein. J Biol Chem. 2002;277:1641–1644. doi: 10.1074/jbc.C100560200. [DOI] [PubMed] [Google Scholar]
- Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis EB, Lock H, Chefer VI, Shippenberg TS, Hjelmstad GO, Fields HL. Kappa opioids selectively control dopaminergic neurons projecting to the prefrontal cortex. Proc Natl Acad Sci U S A. 2006;103:2938–2942. doi: 10.1073/pnas.0511159103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda W, Furuta T, Nakamura KC, Hioki H, Fujiyama F, Arai R, Kaneko T. Single nigrostriatal dopaminergic neurons form widely spread and highly dense axonal arborizations in the neostriatum. J Neurosci. 2009;29:444–453. doi: 10.1523/JNEUROSCI.4029-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GW, Staley JK, Heilman CJ, Perez JT, Mash DC, Rye DB, Levey AI. Immunochemical analysis of dopamine transporter protein in Parkinson's disease. Ann Neurol. 1997;41:530–539. doi: 10.1002/ana.410410417. [DOI] [PubMed] [Google Scholar]
- Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K. alpha-synuclein and mitochondria: partners in crime? Neurotherapeutics. 2013;10:391–399. doi: 10.1007/s13311-013-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Nemani VM, Azarbal F, Skibinski G, Levy JM, Egami K, Munishkina L, Zhang J, Gardner B, Wakabayashi J, Sesaki H, Cheng Y, Finkbeiner S, Nussbaum RL, Masliah E, Edwards RH. Direct membrane association drives mitochondrial fission by the Parkinson disease-associated protein alpha-synuclein. J Biol Chem. 2011;286:20710–20726. doi: 10.1074/jbc.M110.213538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff H, Neu A, Liss B, Roeper J. I(h) channels contribute to the different functional properties of identified dopaminergic subpopulations in the midbrain. J Neurosci. 2002;22:1290–1302. doi: 10.1523/JNEUROSCI.22-04-01290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirenberg MJ, Chan J, Vaughan RA, Uhl GR, Kuhar MJ, Pickel VM. Immunogold localization of the dopamine transporter: an ultrastructural study of the rat ventral tegmental area. J Neurosci. 1997;17:5255–5262. doi: 10.1523/JNEUROSCI.17-14-05255.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Wiederkehr A, Kirkpatrick C, Mattenberger Y, Martinou JC, Marchetti P, Demaurex N, Wollheim CB. Selective actions of mitochondrial fission/fusion genes on metabolism-secretion coupling in insulin-releasing cells. J Biol Chem. 2008;283:33347–33356. doi: 10.1074/jbc.M806251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak D, Berthet A, Nakamura K. Energy failure-does it contribute to neurodegeneration? Ann Neurol. 2013;74:506–516. doi: 10.1002/ana.24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham AH, Meng S, Chu QN, Chan DC. Loss of Mfn2 results in progressive, retrograde degeneration of dopaminergic neurons in the nigrostriatal circuit. Hum Mol Genet. 2012;21:4817–4826. doi: 10.1093/hmg/dds311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickel VM, Johnson E, Carson M, Chan J. Ultrastructure of spared dopamine terminals in caudate-putamen nuclei of adult rats neonatally treated with intranigral 6-hydroxydopamine. Brain Res Dev Brain Res. 1992;70:75–86. doi: 10.1016/0165-3806(92)90105-6. [DOI] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheff SW, Price DA, Schmitt FA, DeKosky ST, Mufson EJ. Synaptic alterations in CA1 in mild Alzheimer disease and mild cognitive impairment. Neurology. 2007;68:1501–1508. doi: 10.1212/01.wnl.0000260698.46517.8f. [DOI] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Stout AK, Lund S, Baptista M, Panov AV, Cookson MR, Greenamyre JT. An in vitro model of Parkinson's disease: linking mitochondrial impairment to altered alpha-synuclein metabolism and oxidative damage. J Neurosci. 2002;22:7006–7015. doi: 10.1523/JNEUROSCI.22-16-07006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer TB, Kim JH, Betarbet R, Greenamyre JT. Subcutaneous rotenone exposure causes highly selective dopaminergic degeneration and alpha-synuclein aggregation. Exp Neurol. 2003;179:9–16. doi: 10.1006/exnr.2002.8072. [DOI] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL, Dawson TM. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with Lewy bodies. Proc Natl Acad Sci U S A. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Grüenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Vila M, Vukosavic S, Jackson-Lewis V, Neystat M, Jakowec M, Przedborski S. Alpha-synuclein up-regulation in substantia nigra dopaminergic neurons following administration of the parkinsonian toxin MPTP. J Neurochem. 2000;74:721–729. doi: 10.1046/j.1471-4159.2000.740721.x. [DOI] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY. Dopaminergic neurons in the rat ventral tegmental area. Brain Res Rev. 1981;3:123–140. doi: 10.1016/0165-0173(81)90002-3. [DOI] [Google Scholar]
- Wang X, Petrie TG, Liu Y, Liu J, Fujioka H, Zhu X. Parkinson's disease-associated DJ-1 mutations impair mitochondrial dynamics and cause mitochondrial dysfunction. J Neurochem. 2012a;121:830–839. doi: 10.1111/j.1471-4159.2012.07734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yan MH, Fujioka H, Liu J, Wilson-Delfosse A, Chen SG, Perry G, Casadesus G, Zhu X. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum Mol Genet. 2012b;21:1931–1944. doi: 10.1093/hmg/dds003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zala D, Hinckelmann MV, Yu H, Lyra da Cunha MM, Liot G, Cordelières FP, Marco S, Saudou F. Vesicular glycolysis provides on-board energy for fast axonal transport. Cell. 2013;152:479–491. doi: 10.1016/j.cell.2012.12.029. [DOI] [PubMed] [Google Scholar]
- Zhang J, Pho V, Bonasera SJ, Holtzman J, Tang AT, Hellmuth J, Tang S, Janak PH, Tecott LH, Huang EJ. Essential function of HIPK2 in TGFbeta-dependent survival of midbrain dopamine neurons. Nat Neurosci. 2007;10:77–86. doi: 10.1038/nn1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TA, Placzek AN, Dani JA. In vitro identification and electrophysiological characterization of dopamine neurons in the ventral tegmental area. Neuropharmacology. 2010;59:431–436. doi: 10.1016/j.neuropharm.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Palmiter RD. Dopamine-deficient mice are severely hypoactive, adipsic, and aphagic. Cell. 1995;83:1197–1209. doi: 10.1016/0092-8674(95)90145-0. [DOI] [PubMed] [Google Scholar]
- Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]