Significance
Phosphate (Pi) is a primary nutrient for plant growth. Because of the low availability of soil Pi, the Pi starvation signaling in plants is gaining great interest. Arabidopsis AtPHR1 and its rice homologue OsPHR2 are known to be central transcription factors in Pi homeostasis; however, the mechanism of how plants sense external Pi fluctuation to regulate the activity of AtPHR1/OsPHR2 has been elusive. Here, we identify rice SPX1 and SPX2 as Pi-dependent inhibitors of PHR2, implicating SPX1 and SPX2 in the Pi-sensing mechanism. We also show that the SPX domain of SPX1 and SPX2 is critical for repressing PHR2 binding to cis elements by protein interaction. The discovery of cellular nutrient concentration-dependent fine-tuning sheds light on a novel mechanism of plant adaption to environmental cues.
Keywords: SPX-domain protein, PHR2, Pi signaling, Pi-dependent inhibition
Abstract
In plants, sensing the levels of external and internal nutrients is essential for reprogramming the transcriptome and adapting to the fluctuating environment. Phosphate (Pi) is a key plant nutrient, and a large proportion of Pi starvation-responsive genes are under the control of PHOSPHATE STARVATION RESPONSE REGULATOR 1 (PHR1) in Arabidopsis (AtPHR1) and its homologs, such as Oryza sativa (Os)PHR2 in rice. AtPHR1 and OsPHR2 expression is not very responsive to Pi starvation, raising the question as to how plants sense changes in cellular Pi levels to activate the central regulator. SPX [named after SYG1 (suppressor of yeast gpa1), Pho81 (CDK inhibitor in yeast PHO pathway), and XPR1 (xenotropic and polytropic retrovirus receptor)] proteins that harbor only the SPX domain are reported to be involved in the negative regulation of Pi starvation responses. Here, we show that the nuclear localized SPX proteins SPX1 and SPX2 are Pi-dependent inhibitors of the activity of OsPHR2 in rice. Indeed, SPX1 and SPX2 proteins interact with PHR2 through their SPX domain, inhibiting its binding to P1BS (the PHR1-binding sequence: GNATATNC). In vivo data, as well as results from in vitro experiments using purified SPX1, SPX2, and OsPHR2 proteins, showed that SPX1 and SPX2 inhibition of OsPHR2 activity is Pi-dependent. These data provide evidence to support the involvement of SPX1 and SPX2 in the Pi-sensing mechanism in plants.
Phosphorus (P) is an essential macroelement for plant growth and development. Because of high chemical fixation, slow diffusion, and substantial fractions of organic compounds by microorganisms, phosphate (Pi) limitation is usually a constraint for crop production in cultivated soils (1). However, intensive application of P fertilizer to increase agricultural production results in higher cost and environmental pollution and aggravates the shortage of nonrenewable resources worldwide for P fertilizer production (2). Therefore, improving effective Pi use by crops to reduce agricultural dependence on heavy Pi fertilizer application is an important challenge for sustainable agricultural production.
The role of Arabidopsis PHOSPHATE STARVATION RESPONSE REGULATOR 1 (AtPHR1) and its orthologs as important regulators in Pi signaling and homeostasis through binding to the PHR1-binding sequence (P1BS) is well established in plants. AtPHR1 binds as a dimer to an imperfect palindromic sequence (GNATATNC), and this DNA-binding ability is dependent on the MYB and coiled-coil (CC) domains present in AtPHR1 and related proteins (3, 4). Orthologs of AtPHR1 have also been described in rice [Oryza sativa (Os)PHR2], common bean [Phaseolus vulgaris (Pv)PHR1], rape [Brassica napus (Bn)PHR1], and common wheat [Triticum aestivum (Ta)PHR1] (5–8), indicating a conserved function of the central regulator in Pi signaling and homeostasis in plants.
The SPX domain (Pfam PF03105) is named after the suppressor of yeast gpa1 (SYG1), the yeast cyclin-dependent kinase inhibitor (PHO81), and the human xenotropic and polytropic retrovirus receptor 1 (XPR1). In yeast (Saccharomyces cerevisiae), the SPX domain forms part of the competitive dual-transporter system that prolongs preparation for starvation and facilitates subsequent recovery of cellular Pi. This competitive system optimizes sensing of nutrient depletion by integrating internal and external information about nutrient availability (9, 10). SPX proteins are referenced as exclusively harboring the SPX domain (11). Four SPX proteins in Arabidopsis (named AtSPX1–AtSPX4) and six SPX proteins in rice (named OsSPX1–OsSPX6) have been identified, and all of them, except SPX4, are responsive to Pi starvation (12–14). In rice, genetic analysis has demonstrated that OsSPX1 (hereafter SPX1) counteracts the function of OsPHR2 (hereafter PHR2) in inducing the expression of PT2, which plays a major role in Pi translocation and accumulation (15, 16). However, the mechanisms of the negative regulation of SPX proteins on the activity of PHR2 master regulator remain to be elucidated.
Phylogenetic analysis showed that SPX1 and SPX2 have the highest homology among six SPX proteins (11). In addition, SPX1 and SPX2 are exclusively localized in the nucleus as PHR2, whereas the other SPX proteins are not (14). Genetic analysis indicates that SPX1 has a negative effect on PHR2 (16). Therefore, it is important to investigate whether SPX1 and SPX2 act on PHR2 in regulating Pi signaling. Here, we show that SPX1 and SPX2 are indeed Pi-dependent inhibitors of PHR2 activity. SPX1 and SPX2 interact with the C terminus of PHR2 containing the MYB-CC domain, which is responsible for binding to P1BS. Through genetic analysis, we found that SPX1 and SPX2 have a redundant function in repressing Pi starvation responses through PHR2 under Pi-sufficient conditions. In vivo data, as well as in vitro experiments using purified SPX1, SPX2, and PHR2 proteins, showed that the inhibition of SPX1 and SPX2 on PHR2 binding to P1BS is Pi-dependent, revealing an SPX1- and SPX2-mediated Pi-sensing mechanism in plants.
Results
SPX1 and SPX2 Interact with PHR2.
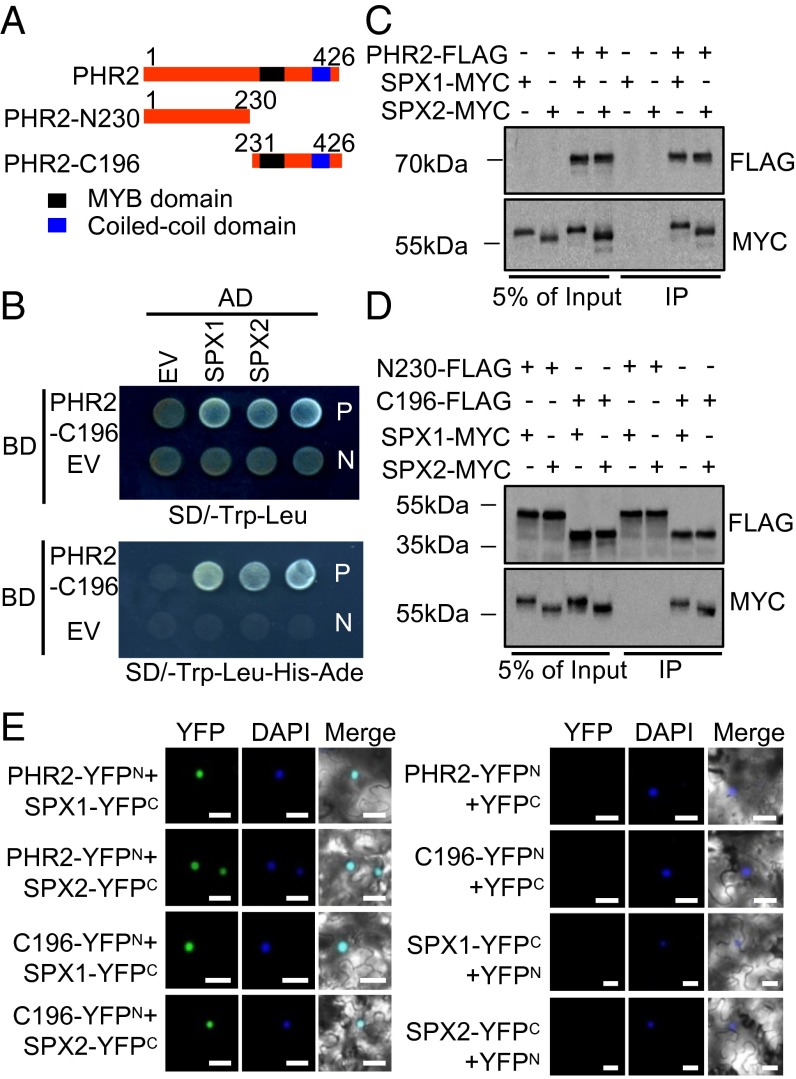
To examine the possible interaction of SPX1 and SPX2 with PHR2, we performed yeast two-hybrid (Y2H) assays using Matchmaker GAL4 two-hybrid systems (Clontech). The PHR2 N terminus has a transactivation domain that is active in yeast (4, 5); therefore, in the Y2H assay, we used the PHR2 C terminus containing the MYB-CC domains (PHR2-C196aa) responsible for specific binding to the P1BS sequence motif (3) (Fig. 1A). We observed an interaction between SPX1 and SPX2 and the PHR2 C terminus in yeast cells (Fig. 1B). To verify these results, we performed in vivo coimmunoprecipitation (co-IP) assays using tobacco leaves cotransformed with PHR2-FLAG and SPX1-MYC or SPX2-MYC, and found that SPX1 and SPX2 interact with PHR2 in plant cells (Fig. 1C). The co-IP assay also confirmed that the PHR2 C terminus is the region that interacts with SPX1 and SPX2 (Fig. 1D). Meanwhile, the interaction of SPX1 and SPX2 with PHR2 in the nucleus was shown using bimolecular fluorescence complementation (BiFC) assays in tobacco leaves. The empty vector controls showed no detectable fluorescence, but coexpression of PHR2-YFPN or PHR2-C196-YFPN with SPX1-YFPC or SPX2-YFPC led to a fluorescence signal in the nucleus (Fig. 1E). These results demonstrated that SPX1 and SPX2 interact with the PHR2-C196aa fragment containing the MYB-CC domain in the nucleus.
Fig. 1.
SPX1 and SPX2 interact with the PHR2 C terminus. (A) Scheme of full-length PHR2 and deletion derivatives. Numbers above each truncation indicate the PHR2 amino acid coordinates. (B) SPX1 and SPX2 interact with PHR2, as indicated by yeast two-hybrid assays. EV, empty vector; N, negative control; P, positive control. (C) SPX1 and SPX2 interact with PHR2, as indicated by co-IP assays. Protein extracts (Input) were immunoprecipitated with anti-FLAG antibody (IP). Immunoblots were developed with anti-FLAG antibody to detect PHR2 and with anti-MYC to detect SPX1 and SPX2. Molecular mass markers are shown (kDa). (D) SPX1 and SPX2 interact with the PHR2 C terminus, as indicated by co-IP assays. Immunoblots were developed with anti-FLAG and anti-MYC antibodies. Molecular mass markers are shown (kDa). (E) SPX1 and SPX2 interact with PHR2 in the nucleus, as indicated by BiFC analysis. The nucleus was stained with DAPI. (Scale bars: 50 μm.)
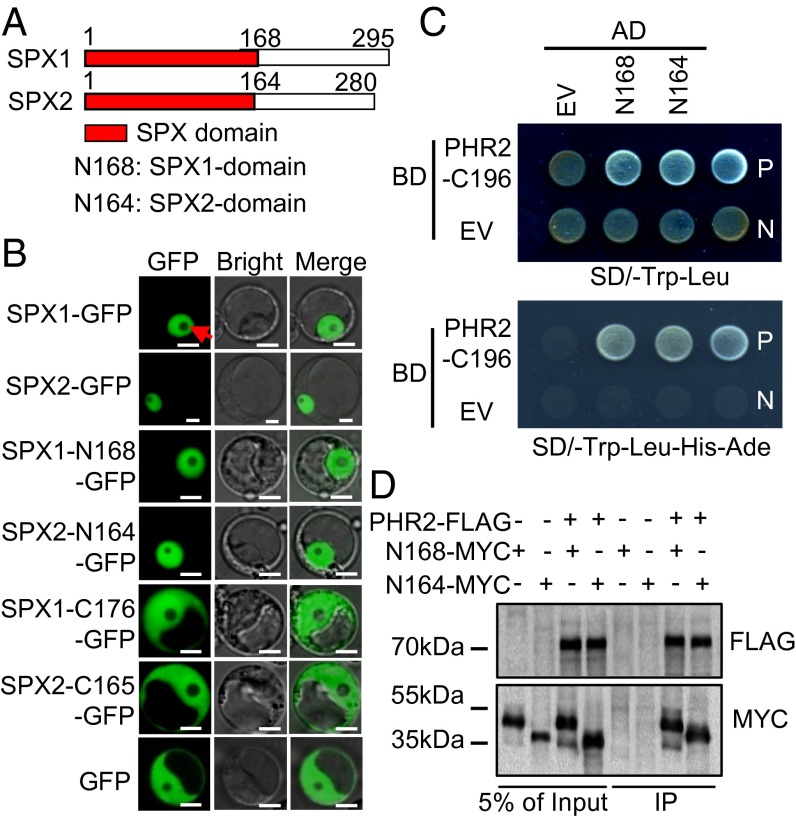
Previous reports showed that the SPX domain is involved in protein–protein interaction in plants (17–19). To determine whether the SPX domain is crucial for SPX1 and SPX2 interactions with PHR2, we first examined the subcellular localization of SPX domains of SPX1 and SPX2 and SPX1 and SPX2 C termini lacking the partial SPX domain. Exclusive nuclear localization of SPX1 and SPX2 N termini and the full-length SPX1 and SPX2 proteins were observed, whereas the SPX1 and SPX2 C termini were localized to the cytosol, as for GFP alone (Fig. 2 A and B). In vitro and in vivo assays verified that the SPX domains interact with PHR2 (Fig. 2 C and D). The genetic evidence for the function of SPX1 and SPX2 domains as repressors of PHR2 in rice was provided by the plants harboring simultaneously overexpressed SPX domains and PHR2 (Fig. S1). The results showed that the two SPX domains counteract PHR2 function, similar to full-length SPX1 (16).
Fig. 2.
Interaction of SPX domains of SPX1 and SPX2 with PHR2. (A) Schematics of SPX1 and SPX2 protein structures. (B) Subcellular localization of full-length and deletion derivatives of SPX1 and SPX2 in rice protoplasts. All truncations were fused with GFP at their C termini under control of the 35S promoter. (Scale bars: 5 μm.) The arrowhead points to the nucleolus. (C) SPX domains of SPX1 and SPX2 interact with PHR2, as indicated by yeast two-hybrid assays. AD, activation domain; BD, binding domain. (D) SPX domains of SPX1 and SPX2 interact with PHR2, as indicated by co-IP assays. Protein extracts (Input) were immunoprecipitated with anti-FLAG antibody (IP) and resolved by SDS/PAGE. The immunoblots shown were developed with anti-FLAG antibody to detect PHR2 and with anti-MYC antibody to detect SPX1-N168 and SPX2-N164. Molecular mass markers are shown (kDa).
SPX1 and SPX2 Have Redundant Functions in Repressing the Activity of PHR2.
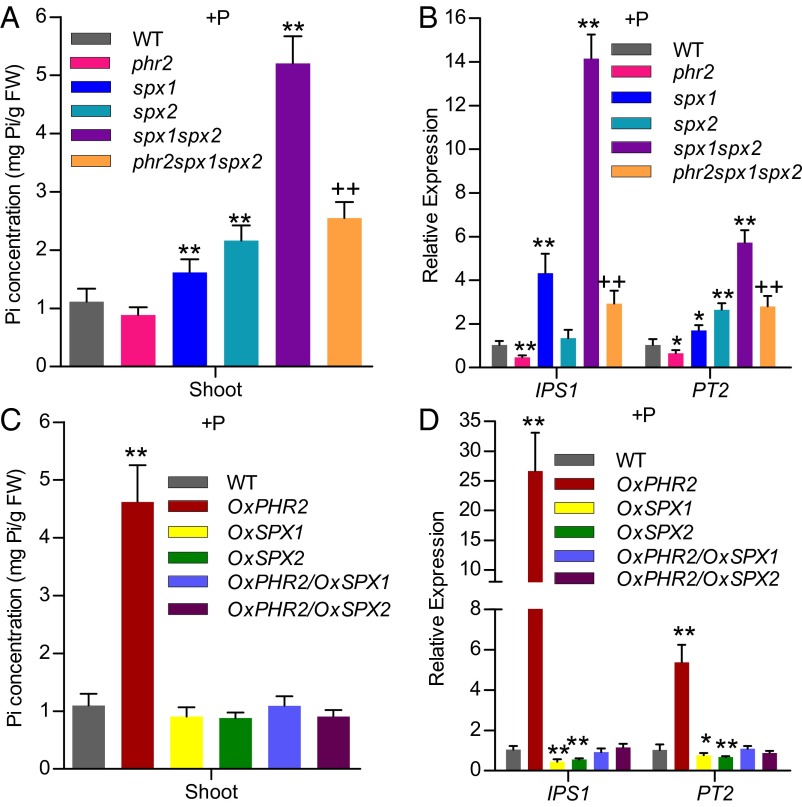
To investigate the function of SPX1 and SPX2 in vivo, we isolated spx1 and spx2 single mutants from public rice transfer DNA (T-DNA) insertional mutant libraries and developed a spx1spx2 double mutant on a similar background (Nipponbare, japonica cultivar) by successive backcrossing using Nipponbare as the recurrent parent (Fig. S2 A–D). Compared with WT plants, spx1 and spx2 single mutants had significantly higher Pi concentrations, and the spx1spx2 double mutant showed a synergistic effect on shoot Pi accumulation and lower biomass in high-Pi growth conditions (200 μM Pi) (Fig. 3A and Fig. S2 G and H). Consistently, the necrosis symptoms on the leaf tips of the spx2 mutant and spx1spx2 double mutant, predominantly in the older leaves, were also observed (Fig. S2I). In addition, the root hairs are longer in the spx1spx2 mutant than in WT (Fig. S2 E and F). Furthermore, the spx1spx2 mutant displayed a shorter primary root and an increased root-to-shoot growth ratio (Fig. S2 H and J). However, no significant difference in anthocyanin content was found between WT and the spx1spx2 mutant (Fig. S2K). The significantly higher Pi uptake ability and shoot-to-root ratio in spx1 and spx2 mutants and the synergistic effect in spx1spx2 were also indicated by 33P-labeled Pi accumulation (Fig. S2 L and M). The synergistic effects in the spx1spx2 double mutant indicate that SPX1 and SPX2 are proteins with redundant functions.
Fig. 3.
SPX1 and SPX2 have redundancy on repressing the Pi concentration and Pi signaling. Shoot cellular Pi concentrations (A) and relative expression of IPS1 and PT2 (B) of 15-d-old WT, phr2, spx1, spx2, spx1spx2, and phr2spx1spx2 plants in Pi-sufficient conditions (+P, 200 μM Pi). Shoot cellular Pi concentrations (C) and relative expression of IPS1 and PT2 (D) of 15-d-old WT, OxPHR2, OxSPX1, OxSPX2, OxPHR2/OxSPX1, and OxPHR2/OxSPX2 plants in Pi-sufficient conditions (+P, 200 μM Pi). Data show mean + SD (n = 5 for A and C, n = 3 for B and D). FW, fresh weight. Data that differ significantly from the corresponding controls are indicated (plants vs. WT: *P < 0.05, **P < 0.01; triple mutant vs. the spx1spx2 double mutant: ++P < 0.01; Student t test).
Furthermore, we developed the triple mutant phr2spx1spx2 and found that the phr2 mutation partially counteracted the effect of spx1spx2 on Pi accumulation and up-regulation of Pi starvation-induced (PSI) genes (Fig. 3 A and B). Additionally, we found that overexpression of SPX2 could prevent the increased Pi concentration and up-regulation of PSI genes caused by PHR2 overexpression (Fig. 3 C and D), as was previously reported for the counteraction between overexpression of SPX1 and PHR2 (16). On the other hand, EMSA using nuclear protein extracts indicates that the P1BS motif-binding activity was higher in spx1spx2 than in WT and the spx1 or spx2 single mutant, whereas it was lower in the phr2 mutant (Fig. S3). These data are in agreement with the notion that SPX1 and SPX2 repress PHR2 activity.
PHR2 Directly Up-Regulates SPX1 and SPX2 Expression.
Because expression of SPX1 and SPX2 is responsive to Pi starvation (14) and these genes contain P1BS sites in their 5′ UTR or promoter region, we predicted that SPX1 and SPX2 are direct targets of PHR2. To analyze this prediction, we generated a transgenic plant expressing the functional PHR2-FLAG fusion from its own promoter (Fig. S4). EMSA and ChIP-PCR analysis supported our hypothesis, indicating that PHR2, SPX1, and SPX2 constitute a regulatory feedback loop in Pi signaling (Fig. S5 A–C).
SPX1 and SPX2 Inhibition of PHR2 Binding to P1BS Motif Is Pi-Dependent.
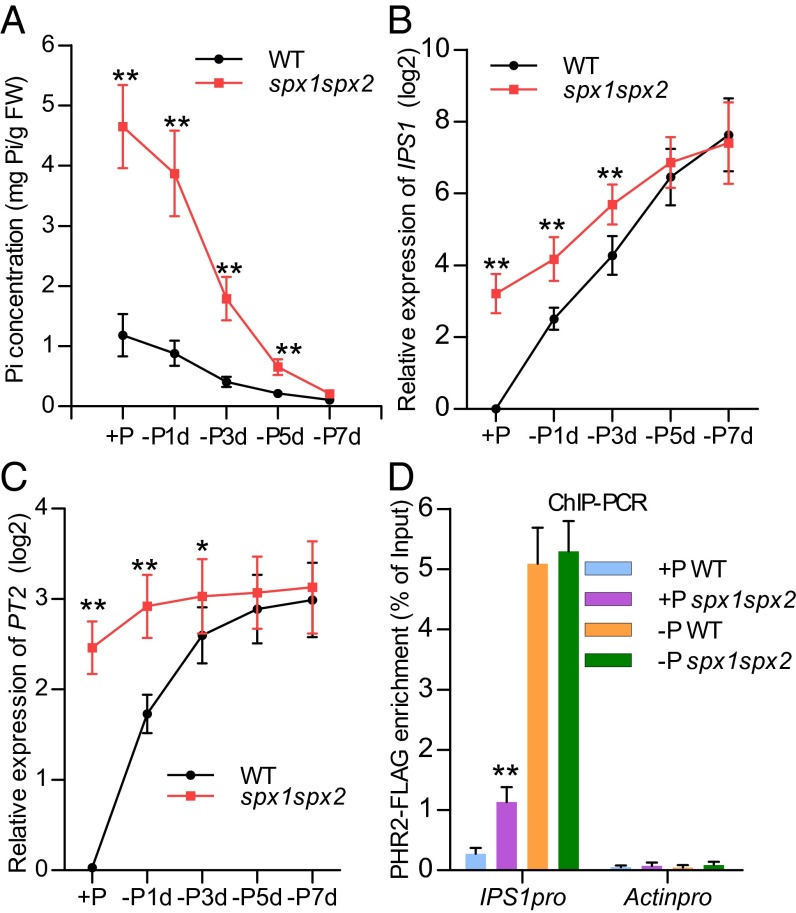
The increased expression and accumulation of SPX1 and SPX2 proteins in Pi starvation conditions (Fig. S5D) was somewhat paradoxical in light of the fact that PHR2 activity is higher in −P conditions. We performed a Pi starvation time-course experiment to evaluate the Pi effect on SPX1 and SPX2 activity in vivo, as indicated by expression of PSI long noncoding RNA IPS1 and the Pi transporter PT2, both controlled by PHR2 (3, 4, 16). Notably, the increased expression of these genes in the spx1spx2 double mutant vs. the WT was greatly reduced when cellular Pi levels decreased (Fig. 4 A–C). Genome-wide expression profiling also showed that the up-regulated PSI genes in the spx1spx2 mutant vs. WT were greatly reduced under −P conditions compared with those genes under +P conditions (Fig. S6). The inhibitory effect of SPX1 and SPX2 on PHR2 thus appeared to depend on cellular Pi concentration. Furthermore, we performed ChIP-PCR analyses of PHR2 activity in plants grown in +Pi and −Pi conditions. In plants grown in +P conditions, in vivo PHR2 binding to its targets was higher in the spx1spx2 double-mutant plant than in the WT plant, whereas in −Pi conditions, PHR2 binding to its targets was similar in mutant and WT plants (Fig. 4D). Consistently, the increase in the Pi concentration in spx1, spx2, and spx1spx2 mutant plants compared with WT plants diminished under Pi-deficient conditions (Fig. S7). These data thus indicate that SPX1 and SPX2 inhibition of PHR2 is Pi-dependent.
Fig. 4.
SPX1 and SPX2 inhibition of PHR2 activity is Pi-dependent in vivo. (A) Shoot cellular Pi concentrations of 15-d-old WT and spx1spx2 double-mutant seedlings in Pi-sufficient (+P, 200 μM Pi) and Pi starvation (−P, no Pi) conditions for 7 d. Relative expression of IPS1 (B) and PT2 (C) in 15-d-old WT and spx1spx2 seedlings grown in Pi-sufficient conditions (+P, 200 μM Pi) and treated under Pi starvation conditions (−P, no Pi) for 7 d more were calculated and plotted on a semilog graph. (D) Enrichment in PHR2-FLAG at the P1BS region (percentage of Input) of the IPS1 promoter (IPS1pro), as indicated by ChIP-PCR assay. Chromatin prepared from WT and spx1spx2 double-mutant seedlings with PHR2pro-PHR2-FLAG fusion grown in +P and −P conditions for 7 d was immunoprecipitated with anti-FLAG antibody. Actin promoter (Actinpro) was used as a negative control. Data show (D) mean + SD and (A–C) mean ± SD (n = 5 for A, n = 3 for B–D). Data that differ significantly from those data for WT plants are indicated (*P < 0.05, **P < 0.01; Student t test).
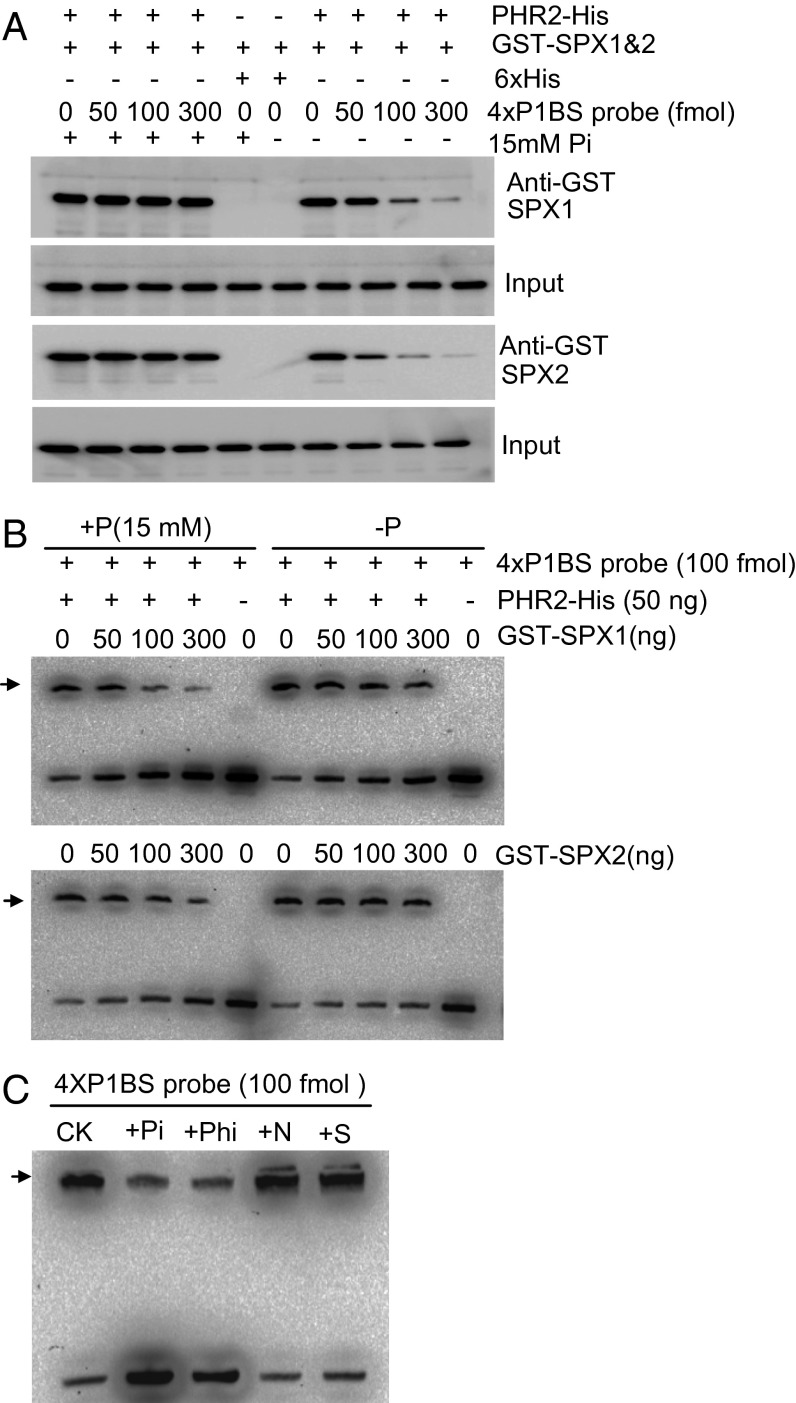
To verify the Pi dependence of SPX1 and SPX2 inhibition of PHR2 binding to DNA, and to examine whether the Pi effect was direct, we used pull-down experiments to determine whether the P1BS motif competed with SPX1 and SPX2 for PHR2 interaction and DNA-binding assays to examine the effect of SPX1 and SPX2 on the PHR2–P1BS interaction in vitro. These competition experiments were performed with affinity-purified PHR2, SPX1, and SPX2 proteins expressed in Escherichia coli tagged with hexahistidine (6× His) and GST, respectively. Results from pull-down experiments showed that P1BS motif-displaced SPX1 and SPX2 interacted with PHR2 only in the absence of Pi (Fig. 5A). Reciprocally, EMSA experiments showed that SPX1 and SPX2 inhibited PHR2 binding to P1BS in a dose-dependent manner, but only in the presence of Pi (Fig. 5B). EMSA and pull-down assays under various Pi concentrations also showed the positive correlation between the Pi concentration and the inhibition of SPX1 on the P1BS-binding ability of PHR2 (Fig. S8 A and B). The plant cytoplasmic Pi concentration was found to be 0.3–0.5 mM under soil culture or hydroponics by in vivo 31P-NMR analysis (20). In Fig. S8 A and B, significant inhibition of SPX1 on the PHR2-binding P1BS was detected when the Pi concentration was reduced to 0.25 mM, which is in the range of the cellular Pi concentration reported by Rouached et al. (20). To determine the specificity of the Pi effect on the SPX–PHR2 interaction, we tested the effect of other anions, such as nitrate (N), sulfate (S), and the nonmetabolizable Pi analog phosphite (Phi) (21, 22), on PHR2 binding to P1BS. EMSA results showed that only Phi, like Pi, allowed SPX to compete with the P1BS motif for PHR2 binding (Fig. 5C). This finding strengthens the idea that Pi acts as a direct signal. We also analyzed the effect of the SPX domains of SPX1 and SPX2 on PHR2 binding to P1BS in the presence of Pi; these domains were sufficient to inhibit PHR2 binding to P1BS (Fig. S8 C–E). We conclude that SPX1 and SPX2 inhibition of PHR2 binding to P1BS is Pi-dependent and is mediated by the SPX domain.
Fig. 5.
SPX1 and SPX2 inhibition of PHR2 binding to P1BS is Pi-dependent in vitro. (A) Pull-down assays indicate that the interaction of SPX1 and SPX2 with PHR2 is displaced by the P1BS probe in vitro in −P conditions but not in +P conditions (with addition of 15 mM NaH2PO4). Purified bacterially expressed PHR2-His (50 ng), 6× His (50 ng), and GST-SPX1 and GST-SPX2 (250 ng) proteins were used in each lane, with varying amounts of 4× P1BS probe. (B) EMSA showing that SPX1 and SPX2 inhibit PHR2 binding to the 4× P1BS probe in a dose-dependent manner in +P conditions but not in −P conditions. We used PHR2-His (50 ng) with varying amounts of GST-SPX1 and GST-SPX2 (0, 50, 100, and 300 ng) proteins and biotin-labeled 4× P1BS probe (100 fmol). (C) EMSA showing specificity of the Pi effect on SPX1 inhibition of PHR2 binding to P1BS. Each reaction contains PHR2-His (50 ng), GST-SPX1 (250 ng), and biotin-labeled 4× P1BS probe (100 fmol). For controls (CK), 50 mM NaCl was added to EMSA buffer; for +Pi treatment, 15 mM NaH2PO4 and 5 mM NaCl were added; for +Phi treatment, 15 mM NaH2PO3 and 5 mM NaCl were added; for +N treatment, 45 mM NaNO3 and 5 mM NaCl were added; and for +S treatment, 22.5 mM Na2SO4 and 5 mM NaCl were added. In B and C, the PHR2–4× P1BS complex is indicated (black arrow).
Discussion
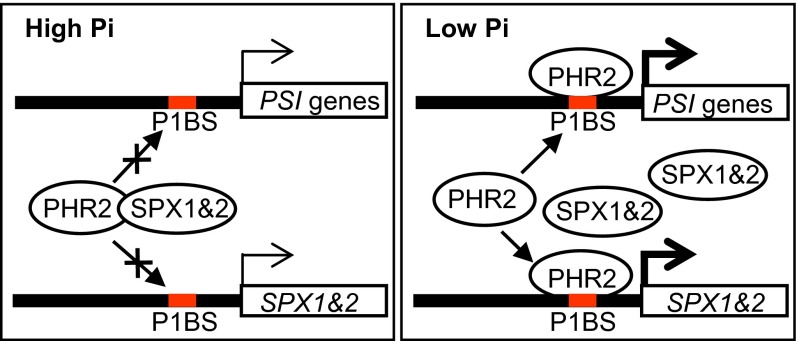
In this report, we demonstrate that SPX1 and SPX2 function as key components in the Pi-sensing mechanism to control the activity of PHR2, a central regulator of Pi starvation responses, as depicted in our working model (Fig. 6). Under high cellular Pi conditions, SPX1 and SPX2 interact with PHR2 with high binding affinity in the nucleus and prevent PHR2 from binding to the P1BS motif in the promoters of PSI genes. Thus, the transcription levels of PSI genes, including SPX1 and SPX2, are basal. However, under low cellular Pi conditions, interaction of SPX1 and SPX2 with PHR2 was weakened and the P1BS motif of PSI genes competes with SPX1 and SPX2 to interact with PHR2, allowing PHR2 to up-regulate PSI genes, including SPX1 and SPX2. In turn, the accumulation of SPX1 and SPX2 under Pi-deficient conditions allows plants to shut down the PHR2-dependent Pi starvation response rapidly after Pi repletion. In other organisms, such as yeast, SPX domain proteins also participate in Pi sensing, although the precise role of the SPX domain in this process has not been elucidated. The SPX-bearing yeast protein PHO81 acts as a cellular Pi-dependent inhibitor of the Pho80/Pho85 cyclin-dependent kinase complex, which inhibits activity of PHO4, the master transcription factor in Pi starvation responses (23–26), but PHO81 sensing activity is at least partially independent of its SPX domain (27). Our study elucidates a novel feedback regulatory loop on Pi starvation signaling formed by SPX1, SPX2, and PHR2, whose output is dependent on the cellular Pi concentration, providing plants with a delicate mechanism to maintain Pi homeostasis when the environmental Pi concentration fluctuates. Results similar to those results reported here have been obtained by Puga et al. (28) in the Arabidopsis system, indicating the ubiquitous function of SPX1 and SPX2 in plants.
Fig. 6.
Model of interaction of SPX1 and SPX2 with PHR2 in response to the cellular Pi concentration for PSI transcription. In high cellular Pi, SPX1 and SPX2 interact with PHR2 at a high affinity in the nucleus, inhibiting PHR2 from binding to the P1BS motif of PSI genes. Thus, the transcription of PSI genes, including SPX1 and SPX2, is basal. However, in low cellular Pi, SPX1 and SPX2 showed a low affinity to PHR2 and P1BS motifs compete with SPX1 and SPX2 on interacting with PHR2, resulting in the up-regulation of PSI genes, including SPX1 and SPX2. Accumulated SPX1 and SPX2 proteins allow quick repression of the PHR2-dependent Pi signaling with Pi resupply. The thick lines with arrows represent enhancement.
It is well known that there are two types of response to Pi deprivation in plants. One is systemically controlled by whole-plant Pi status, and the other is governed by local Pi status. It has been revealed that the PSI expression of IPS1 and several Pi transporters is systemically controlled (29, 30), although the root growth response to Pi deficiency, such as an increase in the length of the root hair, is locally regulated (31). SPX1 and SPX2 repress the expression of IPS1 and PT2 through inhibition of PHR2 activity under Pi-sufficient conditions (Fig. 3 B and D), indicating the role of SPX–PHR2 interaction on the regulation of systemically controlled Pi starvation responses. In addition, the longer and denser root hairs in the spx1spx2 double mutant, in accordance with those root hairs observed in the roots of OsPHR2-overexpressing lines (5), indicate that SPX–PHR2 interaction is also involved in local Pi signaling.
This study also demonstrates that the SPX domain interacts with PHR2 and acts as a negative regulator on PHR2 activity. It is possible that other SPX proteins might also play negative roles in PHR2 activity and that there is functional redundancy among SPX proteins. Recently, we found that SPX4 is also a negative regulator of PHR2 activity (32). At high cellular Pi, SPX4 reduces PHR2 traffic into the nucleus through trapping it within the cytoplasm and preventing its binding to the P1BS motif. While at a low cellular Pi concentration, SPX4 is degraded through the proteasome pathway, resulting in increased targeting of PHR2 to the nucleus and enhancing its PIBS-binding ability, which eventually leads to the up-regulation of PSI genes. Additionally, SPX3 and SPX5 were indicated to be repressors of PHR2, although the precise mechanism remains to be determined (33). We note that the induced transcript levels of PSI genes and the enrichment of PHR2 binding on the IPS1 promoter in spx1spx2 under +P conditions have not reached their induced levels under −P conditions in WT (Fig. 4). Furthermore, the microarray data showed that only a portion of PSI genes were up-regulated in the spx1spx2 double mutant compared with those genes in WT under Pi-sufficient conditions (Fig. S6). These results indicate that the inhibitory mechanism of SPX1 and SPX2 at the nuclear level is complemented by other inhibitory mechanism acting in the cytoplasm based on SPX4 or possibly other SPX proteins.
In addition, we found that the mutation of both SPX1 and SPX2 still had an effect on Pi accumulation and expression of PSI genes in the phr2 background (Fig. 3 A and B), yet to a lesser extent than in the WT plants. This finding can be explained by functional redundancy between PHR2- and PHR2-related genes in rice, as was reported for AtPHR1 and its homolog PHR1-LIKE1 in Arabidopsis (4). In such case, we would expect that in addition to PHR2, SPX1 and SPX2 would repress the activity of PHR2-related transcriptional factors. On the other hand, it has been reported that SPX1 of rice is involved in cold stress (34). From the microarray data, we also found that SPX1 and SPX2 affect the expression of many genes other than PSI genes (Fig. S6), implying the broad functions of SPX1 and SPX2 in addition to Pi signaling.
Experimental Procedures
A detailed description of the different methods used in this study can be found in SI Experimental Procedures.
Plant Materials and Growth Conditions.
The spx1 mutant (M0101661) was identified from the Taiwan Rice Insertional Mutants Database (http://trim.sinica.edu.tw/), and the spx2 mutant (PFG_3A-02559) was bought from the Rice T-DNA Insertion Sequence Database (http://cbi.khu.ac.kr/RISD_DB.html). The primers used for the identification of mutants are listed in Table S1. Additional details of genetic materials and plant growth conditions can be found in SI Experimental Procedures.
Yeast Two-Hybrid Assays, Co-IP Assays, and BiFC Assays.
The Matchmaker GAL4 two-hybrid system (Clontech) was used for yeast two-hybrid assays. The co-IP assays were performed as described (35). For BiFC assay, YFP fluorescence of tobacco leaves was assayed 3 d postinfiltration under a Zeiss LSM710 confocal microscope. The details are described in SI Experimental Procedures.
ChIP-PCR Analysis.
To generate PHR2pro-PHR2-FLAG transgenic plants, the 3×FLAG coding sequence was amplified from 35S-FLAG to generate FLAG-pBI101.3. The PHR2pro-PHR2-FLAG construct was introduced into the phr2 mutant (36). The spx1spx2 plants harboring PHR2pro-PHR2-FLAG for ChIP-PCR were obtained by crossing spx1spx2 plants with PHR2pro-PHR2-FLAG plants. ChIP-PCR assays were performed as described (37). Primers used for the constructs and ChIP-PCR assays are listed in Table S1.
Microarray Analysis.
Fourteen-day-old plants were treated with +P or −P for another 7 d, and shoot of plants from three biological repeats were sampled for Affymetrix microarray analysis. Microarray and data analyses were performed as described (4). The raw microarray data can be accessed in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo) via accession no. GSE60823.
Measurements of Pi Concentration and Pi Uptake Ability in Plants.
Measurements of Pi concentration and Pi uptake ability and distribution in plants were performed as described previously (5, 38).
In Vitro Pull-Down Assay and EMSA.
Fusion protein purification and pull-down assays followed the standard protocol. Pull-down assays were performed as described (35). Nuclear protein was extracted using a Plant Nuclei Isolation/Extraction Kit (Sigma) with 3% (vol/vol) Triton X-100 added to the extraction buffer. EMSA was performed with a Light Shift Chemiluminescent EMSA Kit (Pierce). The details are described in SI Experimental Procedures.
Supplementary Material
Acknowledgments
Regretfully, Prof. Ping Wu died during the review process of this paper. We dedicate this article to his memory. We thank Prof. Javier Paz-Ares for exchanging results and ideas during the late stages of this study and during the drafting of the manuscript, Dr. Fangliang Huang and Dr. Xin Chen for microarray and data analysis, Dr. Xueping Zhou for providing the vectors for BiFC assays, Dr. Shuqun Zhang and Dr. Fuquan Liu for critically reading the manuscript, and Yunrong Wu and Minxiu Chen for developing genetic materials and the transgenic plants. This work was supported by the National Basic Research and Development Program of China (Grant 2011CB100303), Ministry of Science and Technology of China (Grants 2012AA10A302 and 2010DFA31080), and the Ministry of Education and Bureau of Foreign Experts of China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
3Deceased June 12, 2014.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE60823).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404680111/-/DCSupplemental.
References
- 1.Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–693. doi: 10.1146/annurev.arplant.50.1.665. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert GA, Knight JD, Vance CP, Allan DL. Acid phosphatase activity in phosphorus-deficient white lupin roots. Plant Cell Environ. 1999;22(7):801–810. [Google Scholar]
- 3.Rubio V, et al. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15(16):2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustos R, et al. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6(9):e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J, et al. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 2008;146(4):1673–1686. doi: 10.1104/pp.107.111443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valdés-López O, et al. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 2008;31(12):1834–1843. doi: 10.1111/j.1365-3040.2008.01883.x. [DOI] [PubMed] [Google Scholar]
- 7.Ren F, et al. Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS ONE. 2012;7(8):e44005. doi: 10.1371/journal.pone.0044005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, et al. A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann Bot (Lond) 2013;111(6):1139–1153. doi: 10.1093/aob/mct080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hürlimann HC, Pinson B, Stadler-Waibel M, Zeeman SC, Freimoser FM. The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep. 2009;10(9):1003–1008. doi: 10.1038/embor.2009.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy S, Kafri M, Carmi M, Barkai N. The competitive advantage of a dual-transporter system. Science. 2011;334(6061):1408–1412. doi: 10.1126/science.1207154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Secco D, et al. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012;193(4):842–851. doi: 10.1111/j.1469-8137.2011.04002.x. [DOI] [PubMed] [Google Scholar]
- 12.Duan K, et al. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008;54(6):965–975. doi: 10.1111/j.1365-313X.2008.03460.x. [DOI] [PubMed] [Google Scholar]
- 13.Wang C, et al. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 2009;57(5):895–904. doi: 10.1111/j.1365-313X.2008.03734.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, et al. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J Integr Plant Biol. 2009;51(7):663–674. doi: 10.1111/j.1744-7909.2009.00834.x. [DOI] [PubMed] [Google Scholar]
- 15.Ai P, et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009;57(5):798–809. doi: 10.1111/j.1365-313X.2008.03726.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu F, et al. OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 2010;62(3):508–517. doi: 10.1111/j.1365-313X.2010.04170.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Ni M. SHORT HYPOCOTYL UNDER BLUE1 truncations and mutations alter its association with a signaling protein complex in Arabidopsis. Plant Cell. 2010;22(3):703–715. doi: 10.1105/tpc.109.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu TY, et al. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell. 2012;24(5):2168–2183. doi: 10.1105/tpc.112.096636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang X, Li W, Zhou Y, Ni M. A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLoS Genet. 2013;9(3):e1003347. doi: 10.1371/journal.pgen.1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouached H, et al. Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J. 2011;65(4):557–570. doi: 10.1111/j.1365-313X.2010.04442.x. [DOI] [PubMed] [Google Scholar]
- 21.Carswell C, et al. The fungicide phosphonate disrupts the phosphate-starvation response in Brassica nigra seedlings. Plant Physiol. 1996;110(1):105–110. doi: 10.1104/pp.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ticconi CA, Delatorre CA, Abel S. Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol. 2001;127(3):963–972. [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider KR, Smith RL, O’Shea EK. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science. 1994;266(5182):122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- 24.O’Neill EM, Kaffman A, Jolly ER, O’Shea EK. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science. 1996;271(5246):209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y-S, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316(5821):109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y-S, Huang K, Quiocho FA, O’Shea EK. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol. 2008;4(1):25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S, Jeffery DA, Anthony MD, O’Shea EK. Functional analysis of the cyclin-dependent kinase inhibitor Pho81 identifies a novel inhibitory domain. Mol Cell Biol. 2001;21(19):6695–6705. doi: 10.1128/MCB.21.19.6695-6705.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puga, et al. SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:14947–14952. doi: 10.1073/pnas.1404654111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou XL, et al. Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ. 2005;28(3):353–364. [Google Scholar]
- 30.Thibaud M-C, et al. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 2010;64(5):775–789. doi: 10.1111/j.1365-313X.2010.04375.x. [DOI] [PubMed] [Google Scholar]
- 31.Bates TR, Lynch JP. Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ. 1996;19(5):529–538. [Google Scholar]
- 32.Lv Q, et al. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell. 2014;26(4):1586–1597. doi: 10.1105/tpc.114.123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi J, et al. The paralogous SPX3 and SPX5 genes redundantly modulate Pi homeostasis in rice. J Exp Bot. 2014;65(3):859–870. doi: 10.1093/jxb/ert424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L, et al. Increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana. Plant Biotechnol J. 2009;7(6):550–561. doi: 10.1111/j.1467-7652.2009.00423.x. [DOI] [PubMed] [Google Scholar]
- 35.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451(7177):475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, et al. OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 2011;157(1):269–278. doi: 10.1104/pp.111.181669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saleh A, Alvarez-Venegas R, Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc. 2008;3(6):1018–1025. doi: 10.1038/nprot.2008.66. [DOI] [PubMed] [Google Scholar]
- 38.Wu Z, Ren H, McGrath SP, Wu P, Zhao FJ. Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol. 2011;157(1):498–508. doi: 10.1104/pp.111.178921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.