Significance
Kaposi's sarcoma-associated herpesvirus and Epstein–Barr virus latent antigens, latency-associated nuclear antigen 1 (LANA1) and Epstein–Barr nuclear antigen 1 (EBNA1), are multifunctional proteins involved in the maintenance of episome, latency, regulation of transcription, cell cycle, and immune surveillance. These latent antigens generate +1/−2 frameshifted alternative reading frame (ARF) isoforms by programmed ribosomal frameshifting. EBNA1 recoding generates a LANA1-like glutamine- and glutamic acid-rich EBNA1ARF, implicating a crucial role for these sequences in both viruses, whereas high recoding ability of LANA1 generates a serine/arginine-rich repeat sequence protein similar to those found in neurodegenerative disorders. Here we show that repeat recoding in oncogenic human herpesviruses increases the genetic coding capacity of their latent viral proteins. Repetitive elements may be an unexpected source for human and virus protein expression diversity.
Keywords: PRF, EBV, POLY-Q, HHV8, HHV4
Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV) and Epstein–Barr virus (EBV) are human DNA tumor viruses that express nuclear antigens [latency-associated nuclear antigen 1 (LANA1) and Epstein–Barr nuclear antigen 1 (EBNA1)] necessary to maintain and replicate the viral genome. We report here that both LANA1 and EBNA1 undergo highly efficient +1/−2 programmed ribosomal frameshifting to generate previously undescribed alternative reading frame (ARF) proteins in their repeat regions. EBNA1ARF encodes a KSHV LANA-like glutamine- and glutamic acid-rich protein, whereas KSHV LANA1ARF encodes a serine/arginine-like protein. Repeat sequence recoding has not been described previously for human DNA viruses. Programmed frameshifting (recoding) to generate multiple proteins from one RNA sequence can increase the coding capacity of a virus, without incurring a selective penalty against increased capsid size. The presence of similar repeat sequences in cellular genes, such as huntingtin, suggests that a comparison of repeat recoding in virus and human systems may provide functional and mechanistic insights for both systems.
Recoding refers to a dynamic reprogramming of translation that includes programmed frameshifting, programmed translational bypass, and codon redefinition (1). Most frequently, recoding involves a −1 frameshift that is triggered by specific slippery sequences such as poly-adenine–thymine stretches, or mRNA structures including pseudoknots or stem loops (1, 2). Another feature that contributes to frameshifting is ribosome stalling during translation, allowing ribosomal slippage from the A to P position (3). The mechanisms for −2 frameshifting are less well defined than for −1 frameshifting, in part because there are fewer examples (4, 5).
Kaposi’s sarcoma-associated herpesvirus (KSHV) is a human herpesvirus causing cancers particularly among immunosuppressed and elderly populations (6). The KSHV ORF73 encodes a major latency-associated nuclear antigen 1 (LANA1) that was first discovered as a latent viral antigen recognized by KS patient sera in infected cells (7). The LANA1 protein has three recognizable domains: a basic N-terminal region (N), an acidic central repeat (CR) region (further divisible into CR1, CR2, and CR3), and another basic C-terminal region (C) (8, 9). This multifunctional protein is involved in the maintenance of KSHV episomes, regulation of viral latency, transcriptional regulation of viral and cellular genes, and impairment of cell-cycle checkpoints (10–12).
LANA1 is comprised of multiple high- and low-molecular weight isoforms, seen as a “LANA ladder” banding pattern by immunoblotting. Initially, LANA1 was described as a doublet (13, 14) migrating at 222 and 234 kDa. The shorter form of the doublet is due to an alternative C-terminal polyadenylation site (15). More recently, even faster migrating isoforms have been characterized to result from in-frame, internal translation initiation at sites in the N-terminal and CR1 regions (16). All of these known isoforms have the same amino acid sequence as canonical LANA1 and differ only in being N- or C-terminally truncated.
LANA1 has evolved protein processing-based mechanisms to evade immune surveillance through its central repeat region (17–19) similar to those reported for another related herpesvirus protein, the Epstein–Barr virus (EBV) latent nuclear antigen, Epstein–Barr nuclear antigen 1 (EBNA1) (20, 21), which has a central repeat region composed of glycine–alanine residues (GArs). Although KSHV and EBV have limited overall homology to each other (9), the repeat sequences of EBNA1 and LANA1 are nearly identical on the nucleotide level but are frameshifted relative to each other so that they generate different peptide sequences. Frameshift recoding within the EBNA1 mRNA generates a peptide in its repeat region, having peptide sequences similar to canonical LANA1 repeats (19, 22).
Simple repeat sequence elements are also found in human trinucleotide repeat expansion disorders (e.g., Huntington disease, spinocerebellar ataxia). We find that programmed ribosomal frameshifting (PRF) occurs in the LANA1 repeat sequence, generating steganographic changes similar to translational frameshifting within the expanded polyQ stretch in some neurodegenerative disorders. These findings suggest that recoding can be commonly associated with highly repetitive sequences and that viral oncoproteins may provide valuable models to examine repeat-related frameshifting.
Results
LANA1 Generates −2 Alternative Reading Frame (LANA1ARF) Protein(s).
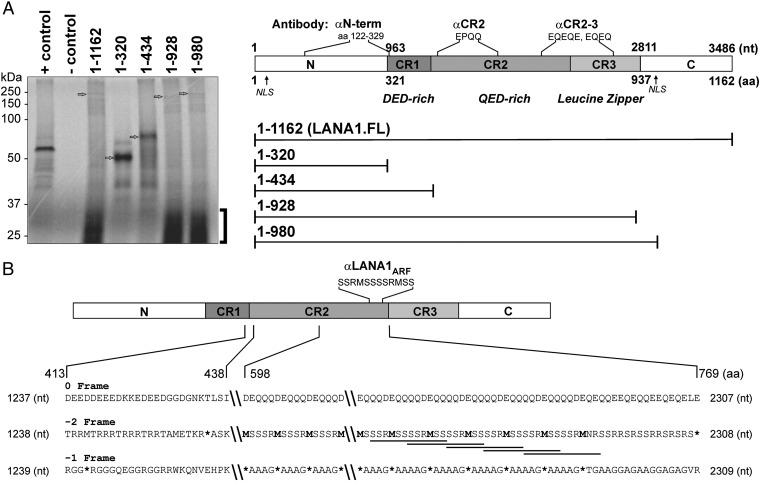
During our studies of LANA1 translation (17, 18), we noted that in vitro transcription and translation reactions of LANA1 RNAs containing the CR2 domain incorporate [35S]-methionine into low molecular-weight products below 37 kDa (Fig. 1A and Fig. S1). No methionines are predicted to be present in the CR2 peptide sequence based on the canonical ORF73; however, the −2 (or +1) reading frame of the LANA1 mRNA sequence in CR2 would encode numerous methionines (Fig. 1B). In this study, we refer to this frameshift as −2 frameshifting as opposed to +1 frameshifting, although both processes produce identical peptide sequences beyond the frameshift codon. If translation of LANA1 shifts into a −2 frame in the CR2 region, this would have to occur after a −2 frame stop codon at nt 1307–1309. Translation of the frameshifted peptide would then be terminated by a −2 frame stop codon at nt 2306–2308 (Fig. 1C).
Fig. 1.
Anomalous [S35]-methionine incorporation in the LANA1 repeat region. (A, Left) In uncoupled in vitro translation assay, equimolar amounts of RNA transcribed from full-length LANA1 (LANA1.FL, 1–1,162) and successive C-terminally deleted constructs (1–320, 1–434, 1–928, and 1–980; Right) were used. Luciferase RNA was used as a positive control, and negative control reaction was performed without any RNA. Arrows indicate expected canonical translational products for each lane. Unexpected [S35]-Met incorporation into small molecular-weight translation products is shown by a bracket. (A, Right) Map of LANA1 domains and antibody epitopes used in this study are designated by amino acid sequences or repeat sequences based on the BC-1 KSHV strain template (U75698). Full-length LANA1 comprises N-terminal (N), C-terminal (C), and central repeat (CR1, CR2, and CR3) domains and two nuclear localization signals (NLSs). The αN-term [ORF73/HHV8 (4C11), Novus] antibody and αCR2 (CM-A810) recognize the N terminus of LANA1 (aa 122–329) and the repetitive epitope EPQQ in CR2, respectively. αCR2–3 (Novus, αLNA1) recognizes repetitive EQEQ and EQEQE epitope found in CR2 and CR3 domains. See also Fig. S1. (B) Three-frame translation of LANA1 in the CR2 repeat region shows peptides containing MSR repeat sequences in the −2 frame. Overlapping antibody epitopes of LANA1ARF (SSRMSSSSRMSS, underlined) localize to the C terminus of CR2. Stop codons are denoted by asterisks. Recoding boundaries for the CR2 −2 frame are constrained by two stop codons (nt 1307–1309 and 2306–2308). See also Fig. S3.
Translational Recoding Occurs in the CR2 Repeat Domain.
To demonstrate that the in vitro translation results can be replicated in vivo and to determine whether LANA1 CR2 is the source of LANA1ARF proteins, we generated a set of translational reporters. Enhanced green fluorescence protein (eGFP) was fused to the C terminus of the LANA1 NCR1CR2 coding sequence in all three reading frames at nt 2304, before the predicted C-terminal −2 frame stop codon. Protein expression was detected by red fluorescence using a LANA1 N-terminal antibody (αN-term; epitope shown in Fig. 1A, column 1), and recoding was assessed by C-terminal eGFP green fluorescence (Fig. 2A, column 2). As expected, the eGFP 0 frame fusion reporter showed strong green fluorescence entirely colocalizing with αN-term staining. No eGFP fluorescence was seen for the −1 reporter, but the −2 reporter produced intense, coarsely speckled nuclear fluorescence from the −2 frame translation. To identify the LANA1 domain required for −2 frameshifting, different LANA1 regions were tested. In agreement with in vitro translation results, mapping coupled with fluorimetry demonstrated that −2 LANA1ARF requires the CR2 region (Fig. 2B and Fig. S2).
Fig. 2.
Recoding in the LANA1 CR2 domain generates LANA1ARF proteins. (A) LANA1 NCR1CR2 domain-based 0, −1, and −2 eGFP reporter constructs were expressed in U2OS cells. Protein expression from all reporter constructs was detected with the αN-term antibody (red, first column) in the nucleus. The −2 reporter produced speckled nuclear fluorescence (green, second column). (B) The −2 eGFP reporter constructs LANA1 N, NCR1, NCR1CR2, and CR2 were expressed in 293 cells. Fluorescence from GFP-positive cells standardized to nontransfected control cells (fold, 1) was examined by fluorimetry 48 h after transfection. Data are represented as mean ± SEM (n = 3). (C) CR2 domain-based two-color in-frame control and −2 reporters were expressed in 293 cells. CR2 expression (0 frame) was detected in the cytoplasm (0 reporter, green), whereas CR2ARF (−2 reporter) was visualized in the nucleus (−2 reporter, red). (D) CR2 repeat sequences of KSHV show high recoding efficiency compared with known viral HIV (gag-pol) and MMTV (gag-pro) and cellular (AZ1) frameshifting sequences in 293 cells. Error bars represent ± SEM (n = 3). See also Fig. S2.
Using CR2 fused to C-terminal, dual-color fluorescence reporters containing both eGFP and dsRed (Fig. 2C), CR20 expressed in the canonical frame shows colocalization of both in-frame fluorescent reporters in the cytoplasm (Fig. 2C, Upper panels). In contrast, −2 frameshifted CR2 products (−2 reporter) represented by red fluorescence are localized to the nucleus (Fig. 2C, Lower panels), suggesting that the −2 CR2 sequence generates a new nuclear targeting or retention sequence. Analysis of the −2 CR2 sequence tested in this construct (aa 598–768) by NLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) (23) does not show conserved nuclear localization sequences.
To quantitate the recoding efficiency of LANA1 CR2, a cell-based Renilla/Firefly dual luciferase reporter system was used (24). The KSHV CR2 (nt 1288–2304) sequence shows ∼39% recoding efficiency (Fig. 2D), an extraordinary degree of recoding compared with known recoding sites from antizyme 1 (AZ1), MMTV gag-pro, and HIV gag-pol. The contribution of alternative initiation at abundant −2 frame methionines may, in part, account for this finding, as the quantitative recoding assay used here cannot distinguished between alternative initiation and programmed frameshifting.
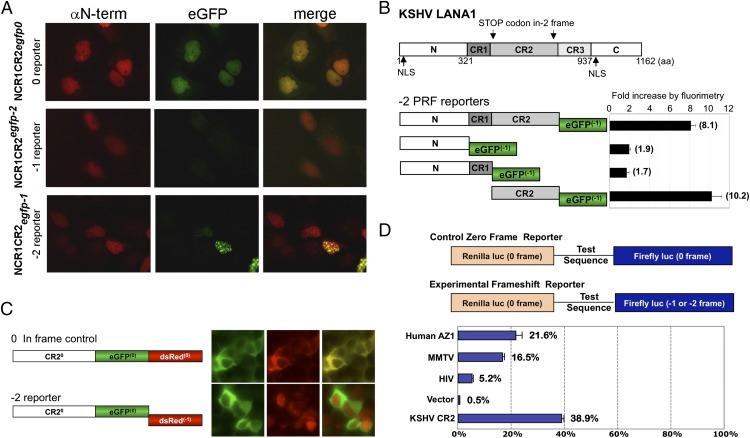
LANA1ARF Is Generated by Both PRF and Alternative Translation Initiation.
To confirm LANA1ARF expression in the context of full-length LANA1, we generated mouse (MαLANA1ARF) and rabbit (RαLANA1ARF) LANA1ARF antibodies against the −2 frame repeat sequence epitope in CR2 (Fig. 3A). We overexpressed various LANA1 constructs (LANA1.FL, LANA1ΔCR2, CR2) in HEK293 cells and used these two antibodies sequentially in immunoprecipitation–immunoblot (IP–IB) analysis. This circumvented antibody cross-reactivity to methionine/serine/arginine (MSR)-rich cellular proteins seen in direct immunoblotting using each antibody alone (Fig. 3B, Left). We detected alternative translation initiation products (∼23 kDa) from both full-length LANA1 and CR2 alone (Fig. 3B). In addition, higher molecular-weight forms migrating at ∼150 kDa were also seen. These higher molecular-weight LANA1ARF proteins were detectable by immunoblotting using MαLANA1ARF only in CR2-containing constructs when first immunoprecipitated by an antibody against the N terminus canonical frame of LANA1 (αN-term) (Fig. 3D). This suggests that the 150-kDa product contains the N-terminal region of LANA1 and undergoes downstream PRF to the ARF in CR2.
Fig. 3.
LANA1ARF is generated by PRF in vivo. (A) A forced −2 frame of CR2 [pCMV-tag2A.CR2(−2)] was expressed in U2OS cells and stained with MαLANA1ARF (green) and RαLANA1ARF (red). Both antibodies detected the speckled nuclear pattern of LANA1ARF. (B) LANA1.FL, LANA1ΔCR2, and CR2 constructs were transfected into 293 cells. Lysates were subjected to RαLANA1ARF or rabbit IgG isotype control for IP and IB with MαLANA1ARF. Two main LANA1ARF products (150 kDa, black bracket; 23 kDa, black arrow) were detected by IP–IB analysis. None of the LANA1ARF forms were detected from CR2-deleted LANA1 (LANA1ΔCR2). An asterisk denotes nonspecific bands. (C) Lysates from 293 cells expressing LANA1.FL, LANA1ΔCR2, and NCR1CR2 constructs were immunoprecipitated with αN-term and immunoblotted with MαLANA1ARF. The higher molecular-weight form LANA1ARF (100–150 kDa) is due to PRF and has an intact N terminus (brackets). An asterisk denotes nonspecific bands including immunoglobulins. (D) Lysates from PEL cells were subjected to RαLANA1ARF for IP and immunoblotted with MαLANA1ARF or αCR2–3. The LANA1ARF protein is detected in KSHV-positive BC-1 and BCBL-1 but not JSC-1, depending on the presence of antibody epitope (Fig. S3). An asterisk denotes nonspecific bands including immunoglobulins. (E) IP of LANA1 from KSHV-positive BC-1 was performed using various 0 frames of the LANA1 protein (LANA1 Abs: αN-term, αCR2, and αCR2–3) and RαLANA1ARF as a control. LANA1ARF (bracket) was detected with MαLANA1ARF. Predicted LANA1ARF generated by PRF comprises the following: N, CR1, C-term of CR2, and serine (S), arginine (R), and methionine (M)-rich repeat sequences. The putative frameshift site within the CR2 domain is between aa 598 and 768.
Although our data demonstrate that LANA1 undergoes recoding after heterologous expression, we also sought evidence for LANA1ARF proteins in natural infection to confirm that recoding is not an artifact of cloning nor of the various expression systems used. LANA1 has a central repeat region that varies in the number of repeats among different KSHV strains. This can be detected by immunoblotting by differences in LANA1 migration in naturally KSHV-infected primary effusion lymphoma (PEL) B-cell lines (25). The BC-1 PEL cell line has four overlapping LANA1ARF antibody epitopes, whereas BCBL-1 has three; no complete epitope is present in JSC-1 (Fig. S3). IP was performed with RαLANA1ARF followed by immunoblotting with MαLANA1ARF on these various PEL cell lines. Consistent with the variability of LANA1 repeat sequences in different KSHV strains, LANA1ARF differs in size between each of the different PEL cell lines—migrating at ∼150 kDa in BC-1 and ∼100 kDa in BCBL-1—and is not detectable in JSC-1 (Fig. 3D). ARF products lower than 40 kDa seen in in vitro translation reactions and with overexpression constructs are not detectable in PEL cells (Fig. 3D). To confirm that N-terminal sequences are present in LANA1ARF, LANA1 antibodies recognizing different epitopes throughout the canonical LANA1 protein were used for IP testing (see Fig. 1A for epitope locations of antibodies). LANA1ARF could be identified in pull-down reactions with αN-term, αCR2, and RαLANA1ARF antibodies but not with a αCR2–3 antibody that recognizes an epitope in a region spanning the CR2 and CR3 junction (Fig. 3E). Thus, LANA1ARF translation starts at the canonical initiation site and differs from LANA1 after translational recoding in CR2.
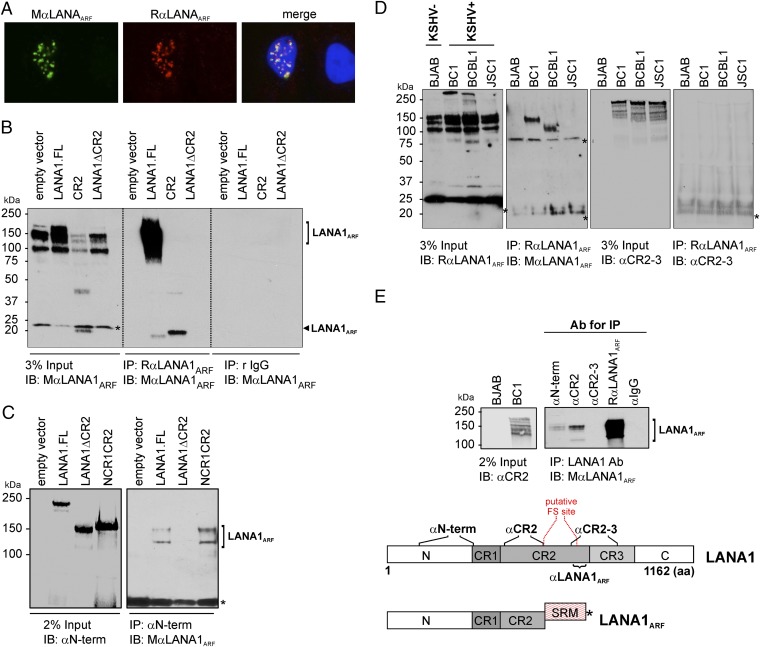
EBV EBNA1 Generates EBNA1ARF Proteins by Both Ribosomal Frameshifting and Internal Translation Initiation.
The −2 frame of the EBV EBNA1 GAr domain with high glutamine and glutamic acid (QE) repeats has ∼35% amino acid identity to the 0 reading frame of the LANA1 CR2 protein (Fig. 4A). It has been proposed that an embedded reading frame of EBNA1 might encode for an ARF starting at methionine (nt 116–118) (22). This EBNA1ARF would be predicted to terminate at −2 frame stop codon nt 1225–1228 (Fig. 4A). To determine whether EBNA1 actually generates frameshifted proteins, EBNA1 N terminus, N terminus plus GAr (NGAr), or GAr regions were cloned into 0, −1, and −2 eGFP reporters. As a group, the EBNA1 N terminus reporters showed a ∼3.6- and 16-fold higher level of translation compared with the array of reporters associated with either NGAr or with GAr alone, respectively, likely due to GAr inhibition of translation in cis (22) (Fig. 4B). The −2 eGFP reporter of EBNA1 N terminus shows 5% frameshifting efficiency compared with 0 reporter (Fig. 4C, Bottom). The −2 reporter construct of NGAr showed ∼10% recoding when standardized against canonical translation in the 0 reporter, whereas the −1 reporter only showed 2% recoding. Very low protein expression was detectable from the 0 reporter of GAr alone (22%) compared with the 0 reporter of NGAr; nevertheless, the −2 reporter for GAr showed 6% recoding relative to the canonical frame translation of the NGAr protein (100%) and ∼28% recoding compared with its corresponding GAr 0 reporter.
Fig. 4.
Recoding of EBNA1 generates QE-rich EBNA1ARF that localizes to the cytoplasm. (A) Map of KSHV LANA1 and EBV EBNA1 and amino acid sequence alignment comparing LANA1.CR2 and EBNA1.GAr in the −2 frame (EBNA1ARF). EBNA1 comprises the N terminus, GA-rich central domain, and C-terminal DNA binding domain. Stop codons of the −2 frame are indicated with arrows. Although the EBNA1 GAr (0 frame) has no amino acid similarity to the LANA1 CR, EBNA1ARF has ∼35% similarity to the 0 frame of the LANA1 CR2 domain, and both sequences contain highly acidic QE-rich repeats. (B) NGAr and GAr domain-based 0, −1, and −2 eGFP reporter constructs were expressed in 293 cells. Percentage of GFP fluorescence was calculated relative to the 0 frame control of NGAr (100%). Error bars represent SEM (n = 10). Representative confocal images are shown in the Lower panel. (C) Three isoforms were produced by −2 ARF in the EBNA1 N-terminal sequence. The 293 cells transfected with the GST–EBNA1.N-MH(−1) construct expresses three isoforms (∼75, ∼45, and ∼41 kDa). In the Left panel, all three isoforms are eluted from a nickel column. In the Right panel, GST is intact only in the ∼75-kDa isoform after purification (Top panel). The N domain of EBNA1 eGFP reporter constructs was examined. Approximately 5% of the eGFP signal was detected in the −2 reporter by fluorimetry (Bottom panel). See also Figs. S4 and S5. (D) Full-length EBNA1–eGFP was transfected into 293 cells, and EBNA1ARF was detected by confocal microscopy. Full-length canonical EBNA1 localizes to the nucleus (green), whereas EBNA1ARF is visualized in the cytoplasm (red). (Scale bar, 5 μm.) See also Fig. S6.
To determine whether ARF products also resulted from alternative translation initiation in the N terminus of EBNA1 (22), we generated an EBNA1 N construct tagged at the N terminus with GST and at the C terminus with maltose binding protein (MBP)/6×Histidine (MH). This construct has a 0 frame N-terminal GST and a C-terminal MH fused such that the MH peptide will only be expressed when translation into the −2 frame occurs. Overexpressed GST–EBNA.N-MH(−1) shows three prominent bands (∼75 kDa, ∼45 kDa, and ∼41 kDa) when immunoblotted with αMBP (Fig. 4C). After His-tag purification, all three bands are eluted, suggesting that all three proteins are C-terminally tagged to −2 frameshifted MH. When the same lysate is applied to a GST column, only the slowest migrating band (∼75 kDa) is eluted (Fig. 4C) and detected with anti-MBP Ab. This band is consistent in size with a programmed frameshift product (75 kDa: ∼26 kDa GST, ∼9 kDa EBNA1.N, and ∼40 kDa MH). Mutation to alanine of the two −2 frame methionines in EBNA1.N (nt 116–119 and nt 203–205), individually and in combination, results in corresponding disappearance of one or both of the remaining two faster migrating bands (Figs. S4 and S5). These data suggest that −2 EBNA1ARF products can be generated through both frameshift recoding and alternative translational initiation from the N terminus of EBNA1.
In the context of full-length EBNA1 (EBNA1–eGFP), the EBNA1ARF was readily detected by immunofluorescence microscopy using an antibody (αEBNA1ARF) generated against a −2 frame peptide repeat epitope in GAr (Fig. 4D). Similar results are seen with a GAr construct, GAr–eGFP (Fig. S6). The EBNA1ARF emitted a diffuse cytoplasmic signal, which is likely due to loss of the canonical nuclear localization signal (aa 379–386) in the EBNA1 C terminus (26).
Discussion
A large body of functional studies has been based on the assumption that single EBNA1 and LANA1 proteins are expressed in cells. In fact, the names of both proteins specify, and restrictively imply, a nuclear localization. Recently, we have shown that cytoplasmic forms of truncated LANA1 exist (16). In this report, our data demonstrate that the major EBV and KSHV oncogenes generate additional, highly divergent frameshifted protein products. At least one of these EBV EBNA1ARF products is found in the cytoplasmic compartment as well and has high similarity to the canonical KSHV LANA1 QE-rich domain.
The general mechanisms of −2 and −1 programmed frameshifting have largely been elucidated from studies of RNA viruses, retrotransposons, insertional elements, and bacteriophage genes (27, 28). PRF from wild-type human DNA virus genomes has not been previously described. We find that both KSHV and EBV nuclear antigen repeat sequences undergo efficient frameshifting. Both viral frameshifted proteins are detectable when heterologously expressed, and LANA1ARF is present in naturally KSHV-infected PEL cell lines. The only previous examples of frameshift products reported in DNA viruses have resulted from DNA mutations in some herpesvirus simplex type 2 and EBV strain isolates in the thymidine kinase (TK) (29) and LF3 early genes (30), respectively.
The −2 frame LANA1ARF proteins can be generated by both PRF and internal translation initiation in the CR2 domain using in vitro translation and by transfection/overexpression experiments (Figs. 1 and 2); however, IP studies in PEL cells suggest that the predominant LANA1ARF isoform produced by KSHV infection of its natural host cell results from PRF (Fig. 3D). IP studies also show that the putative −2 frameshift site occurs between aa 598 and 768 within the CR2 domain (Fig. 3E). Difficulties in subcloning the LANA1 CR2 repeat structure limits our ability to precisely localize the specific frameshift site, and the repeat nature of these sequences precludes direct mass spectroscopy analysis. It is possible that there is no single frameshift site, but instead, frameshifting occurs at multiple repeat sites in this delineated span of CR2 viral repeats. This may explain the observation of LANA1ARF in PEL cells as multiple molecular forms (100–150 kDa). Alternatively, noncanonical LANA1 forms, starting in the N term (16), might also generate some of these LANA1ARF forms. FSFinder (http://wilab.inha.ac.kr/fsfinder2) (31) sequence analysis failed to identify known motifs such as slippery sequences or pseudoknots that can generate programmed frameshifting in LANA1 CR2.
LANA1ARF encodes a highly repetitive SR-rich peptide with a distinctive subnuclear localization pattern. BLAST search analysis (National Center for Biotechnology Information protein BLAST) of LANA1ARF (protein query sequence, SSRMSSSSRMSS) shows some low-significance similarity with several cellular SR proteins (32) involved in mRNA splicing such as TR150, SRRM2, SFRS16, Acinus, and Pinin. We sought a possible role for LANA1ARF in mRNA splicing using colocalization studies, however initial results show poor overlap not only with nuclear splicesome proteins, such as SC-35, but also with other candidate subnuclear structures, such as nucleolin and promyelocytic leukemia bodies (Fig. S7). Our assays based on the original observation of anomalous 35S methionine incorporation in in vitro translation assays are directed at the demonstration of a −2 frame LANA1ARF isoform and do not preclude even more complicated and possible sequential frameshifting events to produce −1 frame GA-rich peptides from the LANA1 repeats.
EBNA1 undergoes frameshifting to generate a distinct −2 EBNA1ARF protein that is diffusely localized to the cytoplasm with ∼35% identity to repeat regions of the canonical, QE-rich 0 frame LANA1 protein. Ossevoort et al. proposed that an embedded reading frame of EBNA1 might encode for a protein with amino acid similarity to LANA1 repeats starting at methionine (nt 116–118) (22). They further engineered such a protein by introducing two nucleotides at the beginning of the GAr domain to shift this region into the −2 frame. This construct, when expressed, was capable of inhibiting antigen presentation in cis. Our data demonstrate that EBNA1ARF recoding occurs by both PRF as well as at alternative translational initiation (at −2 methionine nt 116–119 and −2 methionine 203–205) in the EBNA1 N-terminal unique sequences. Furthermore, the GAr sequences alone also show significant recoding activity, ∼28% of 0 frame GAr translation, in quantitative assays (Fig. 4B). Consistent with this, we have been able to detect EBNA1ARF peptides by liquid chromotography (LC)-MS/MS analysis of EBV-positive BC-1 cells (33).
Although EBV and KSHV diverged over 80 million y ago, they belong to closely related genera of gammaherpesviruses that display similar genome organizations (34). Conservation of the glutamine-, glutamic acid-, and aspartic acid (QED)-rich sequence in both LANA1 and EBNA1ARF implies a fundamental biological importance of this motif to these viruses. One function attributable to both EBNA1 and LANA1 repeat sequences is inhibition of major histocompatibility complex (MHC) peptide presentation (17–21). Both EBNA1 GAr and LANA1 QED-rich CR2CR3 have been shown to retard protein synthesis in cis and to enhance protein stability by reducing defective ribosomal product processing. Specific peptide sequences in repeat regions appear to be responsible for retardation of LANA1 synthesis and antigen presentation (17, 18), and RNA structure and/or peptides from different EBNA1 frames have been proposed to be involved in EBNA1-mediated immune evasion (35, 36). The contribution of QE-rich sequences in comparison with GA-rich sequences, as related to these functions, remains to be elucidated in light of our finding for efficient frameshifting activity in these two latency-associated proteins. Other gammaherpesviruses also encode for latency-associated proteins harboring repeat sequences. However, due to the repetitive nature of these repeats, few have been amenable to analysis or even deposited. Out of seven representative strains for which sequences are available, we find repeats to differ widely in length, yet in silico frameshift analysis shows common glycine (G)-, serine (S)-, arginine (R)-, or glutamic acid (E)-rich repeat motifs (Table S1).
The mechanism of repeat-related recoding in human genes is associated most notably with the codon reiteration disorders or trinucleotide repeat disorders. These hereditary disorders, defined by the presence of abnormal and unstable expansion of DNA triplet repeats include Huntington disease (CAG repeats, poly-Q), fragile X syndrome (CGG repeats, poly-R), and oculopharyngeal muscular dystrophy (GCN repeats, poly-A) (37, 38). The function of repeat regions in these proteins remains obscure, but they have been suggested to mediate the assembly of protein complexes required in chromatin packaging or transcription (38, 39). In Huntington disease, ribosomal frameshifting in its poly-Q sequence produces proteins containing poly-A or poly-SR tracts (40–43). These frameshift products, although clinically and functionally aberrant, have been shown to modulate aggregation (40–44). The physiological implications of expression of these proteins in vivo remain to be elucidated and functionally confirmed. An examination of how repeat recoding cross-compares in virus and human systems may provide valuable functional and mechanistic insights for both. The viral models are tractable systems to define and manipulate factors controlling repeat-associated frameshifting.
ARF proteins have been invisible to investigators studying KSHV and EBV yet may be critical to virus-induced tumor cell phenotypes. Our study underscores the existence of both nuclear and cytoplasmic isoforms for these important viral oncoproteins and reveals new aspects of protein coding diversity involving frameshifting. Further, identification and immunodetection of these LANA1ARF and EBNA1ARF proteins may provide new clinical diagnostic markers for associated tumors. Highly repetitive sequences are frequently found in eukaryote coding regions, and our study suggests that repeat recoding may be a more common event in protein translation than previously appreciated.
Materials and Methods
Expanded methods are presented in SI Materials and Methods.
Plasmids.
LANA1 constructs were generated by PCR from the BC-1 KSHV strain DNA template (U75698) using primers described previously (17). eGFP or dsRed reporter sequences were amplified from pEGFP-C1 or pDs-Red-Monomer-Hyg-N1 (Clontech). To measure recoding efficiency, test sequences were cloned into p2-Luc and p2-Luci vectors. For the −2 frame CR2 (1288–2304 aa) expression plasmid [pCMV-tag2A.CR2(−2)] (Fig. 3A), the CR2 insert from pCMV-tag2B.LANA1 CR2 (17) was ligated into the pCMV-tag2A vector to facilitate a frame change. A dual N-terminal GST and C-terminal MBP/His-tagged EBNA.N was cloned into a modified pSPORT vector between the GST (0 frame) and MBP/His genes (−1 frame) to investigate −2 frameshifting of EBNA1. Constructs containing repeat sequences are propagated in recA1– Escherichia coli strains NEB Stable (NEB) and One Shot Top10 (Invitrogen).
Cell Lines.
HEK293 and U2OS cells were maintained in DMEM (Cellgro, #10–013), and BJAB, BC-1, BCBL-1, and JSC-1 cells (ATCC) were grown in RPMI 1640 (Cellgro, #10–040) supplemented with 10% (vol/vol) FBS (Sigma-Aldrich).
In Vitro Transcription/Translation.
RNAs from LANA1 constructs were generated and used in uncoupled in vitro translation with the Rabbit Reticulocyte Lysate System (Promega). pcDNA.LANA1 C-terminal truncations were generated by digesting and linearizing a pcDNA full-length LANA1 construct at the following restriction sites: HincII (320 aa), AclI (434 aa), BsmbI (928 aa), NruI (980 aa), and XhoI (1,162 aa).
Luciferase Assay for Recoding Efficiency and GFP Fluorescence Analysis.
Renilla and Firefly luminescence activity were analyzed using the dual luciferase reporter assay (Promega) according to the manufacturer’s protocol. eGFP fluorescence was analyzed by a Symergy 2 fluorescence reader (Biotek).
Generation of Antibodies.
Rabbit polyclonal antibodies for LANA1ARF (RαLANA1ARF, CM826) and EBNA1ARF (RαEBNA1ARF) were generated commercially (Covance) by inoculating rabbits with peptides corresponding to the predicted −2 frames of LANA1 CR2 (CKK-ACP-SSRMSSSSRMSS) and GAr (CKK-ACP-QEQEEGQEGQEQEG). The mouse monoclonal IgM antibody (MαLANA1ARF) was generated by standard protocol in the Epitope Recognition Immunoreagent Core facility (University of Alabama). Antibodies were further purified with either SulfoLink immobilization kit (Pierce) or Nab protein A/G Spin kit (Pierce) according to the manufacturers’ protocol.
Immunofluorescence and Confocal Microscopy.
For conventional immunofluorescence microscopy, cells were rinsed and fixed with 4% (wt/vol) paraformaldehyde for 20 min, permeabilized with PBS with 0.1% Triton X-100, and blocked with 10% (vol/vol) normal goat serum. For confocal images, cells were fixed and permeabilized with ice-cold methanol for 10 min at −20 °C. For staining of nuclear organelles, cells were treated with 2 M HCl at room temperature for 20 min followed by a PBS wash and neutralization with 0.1 M boric acid, pH 8.5, for 10 min. Following an additional PBS wash, cells were blocked with 10% (vol/vol) FBS in PBS. Cells were counterstained with DAPI and examined under a fluorescent microscope (AX70, Olympus). For confocal microscopy, the nuclei were stained with DRAQ5 (Molecular Probes) for 15 min at room temperature, rinsed in PBS, and mounted with Gelvatol mounting medium for fluorescence (Center for Biologic Imaging, University of Pittsburgh). Images were acquired with a fluorescent microscope (AX70, Olympus) or a Leica TCS SP confocal microscope and processed using Adobe Photoshop and Leica imaging software.
Detection of LANA1ARF by IP–IB.
LANA1-expressing cells (BC-1, BCBL-1, JSC-1) and negative control cell lines (HEK293, BJAB) were lysed with IP lysis buffer (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 2 mM NaF, 1 mM NaVO3 with protease inhibitors), and proteins were precleared and immunoprecipitated with various antibodies. IB analysis was performed with MαLANA1ARF using secondary mouse anti-IgM (Santa Cruz).
EBNA1 N-Terminus Protein Expression and Purification.
The EBNA1 N-terminus expression plasmid (pSport-GST-EBNA1.N-MH) was expressed in 293F cells (Invitrogen) using Freestyle Max reagent (Invitrogen) as described by the manufacturer. Cells were lysed in PBS+0.5% Triton X-100 with protease inhibitors by sonication 6 d after transfection. The fusion protein was purified by affinity chromatography using Glutathione Sepharose 4B (GE Healthcare) and Ni-NTA Agarose (Novagen).
Supplementary Material
Acknowledgments
We thank Ronit Sarid for helpful discussion and Xi Liu for preliminary technical assistance. We thank Neil Blake for providing EBNA1 expression plasmids and Lisa Moore and Donnacha Dennehy for help with the manuscript. J.F.A. was personally supported by Science Foundation Ireland. This study was supported by National Institutes of Health (NIH) Grants CA136363, CA170354, and CA120726 (to P.S.M. and Y.C.). P.S.M. and Y.C. are also funded as American Cancer Society Research Professors. This project used University of Pittsburgh Cancer Institute resources supported in part by NIH Grant P30CA047904.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416122111/-/DCSupplemental.
References
- 1.Atkins JF, Gesteland RF. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Springer Verlag; New York: 2010. [Google Scholar]
- 2.Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441(7090):244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flanagan JF, 4th, Namy O, Brierley I, Gilbert RJ. Direct observation of distinct A/P hybrid-state tRNAs in translocating ribosomes. Structure. 2010;18(2):257–264. doi: 10.1016/j.str.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanov IP, Gesteland RF, Atkins JF. Antizyme expression: A subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 2000;28(17):3185–3196. doi: 10.1093/nar/28.17.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Y, et al. Efficient -2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc Natl Acad Sci USA. 2012;109(43):E2920–E2928. doi: 10.1073/pnas.1211145109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Moore PS, et al. Primary characterization of a herpesvirus agent associated with Kaposi’s sarcomae. J Virol. 1996;70(1):549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo JJ, et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93(25):14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman PM, Hu J, Renne R. Gammaherpesvirus maintenance and replication during latency. In: Arvin A, Mocarski A, Moore PS, Roizman B, Whitley RJ, editors. Human Herpesviruses: Biology, Therapy and Immunoprophylaxis. Cambridge Univ Press; Cambridge, UK: 2007. pp. 379–402. [PubMed] [Google Scholar]
- 10.Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284(5414):641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 11.Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402(6764):889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 12.Radkov SA, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat Med. 2000;6(10):1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 13.Gao SJ, et al. Seroconversion to antibodies against Kaposi’s sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi’s sarcoma. N Engl J Med. 1996;335(4):233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 14.Rainbow L, et al. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71(8):5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canham M, Talbot SJ. A naturally occurring C-terminal truncated isoform of the latent nuclear antigen of Kaposi’s sarcoma-associated herpesvirus does not associate with viral episomal DNA. J Gen Virol. 2004;85(Pt 6):1363–1369. doi: 10.1099/vir.0.79802-0. [DOI] [PubMed] [Google Scholar]
- 16.Toptan T, Fonseca L, Kwun HJ, Chang Y, Moore PS. Complex alternative cytoplasmic protein isoforms of the Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 generated through noncanonical translation initiation. J Virol. 2013;87(5):2744–2755. doi: 10.1128/JVI.03061-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwun HJ, et al. Kaposi’s sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mimics Epstein-Barr virus EBNA1 immune evasion through central repeat domain effects on protein processing. J Virol. 2007;81(15):8225–8235. doi: 10.1128/JVI.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwun HJ, et al. The central repeat domain 1 of Kaposi’s sarcoma-associated herpesvirus (KSHV) latency associated-nuclear antigen 1 (LANA1) prevents cis MHC class I peptide presentation. Virology. 2011;412(2):357–365. doi: 10.1016/j.virol.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaldumbide A, Ossevoort M, Wiertz EJ, Hoeben RC. In cis inhibition of antigen processing by the latency-associated nuclear antigen I of Kaposi sarcoma herpes virus. Mol Immunol. 2007;44(6):1352–1360. doi: 10.1016/j.molimm.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Levitskaya J, et al. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375(6533):685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 21.Yin Y, Manoury B, Fåhraeus R. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science. 2003;301(5638):1371–1374. doi: 10.1126/science.1088902. [DOI] [PubMed] [Google Scholar]
- 22.Ossevoort M, et al. The nested open reading frame in the Epstein-Barr virus nuclear antigen-1 mRNA encodes a protein capable of inhibiting antigen presentation in cis. Mol Immunol. 2007;44(14):3588–3596. doi: 10.1016/j.molimm.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106(25):10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4(4):479–486. [PMC free article] [PubMed] [Google Scholar]
- 25.Gao SJ, et al. Molecular polymorphism of Kaposi’s sarcoma-associated herpesvirus (Human herpesvirus 8) latent nuclear antigen: Evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J Infect Dis. 1999;180(5):1466–1476. doi: 10.1086/315098. [DOI] [PubMed] [Google Scholar]
- 26.Ambinder RF, Mullen MA, Chang YN, Hayward GS, Hayward SD. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65(3):1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baranov PV, Fayet O, Hendrix RW, Atkins JF. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22(3):174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J Gen Virol. 2012;93(Pt 7):1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang CB, et al. A net +1 frameshift permits synthesis of thymidine kinase from a drug-resistant herpes simplex virus mutant. Proc Natl Acad Sci USA. 1994;91(12):5461–5465. doi: 10.1073/pnas.91.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue SA, Jones MD, Lu QL, Middeldorp JM, Griffin BE. Genetic diversity: Frameshift mechanisms alter coding of a gene (Epstein-Barr virus LF3 gene) that contains multiple 102-base-pair direct sequence repeats. Mol Cell Biol. 2003;23(6):2192–2201. doi: 10.1128/MCB.23.6.2192-2201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon S, Byun Y, Kim HJ, Jeong S, Han K. Predicting genes expressed via -1 and +1 frameshifts. Nucleic Acids Res. 2004;32(16):4884–4892. doi: 10.1093/nar/gkh829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ankö ML. Regulation of gene expression programmes by serine-arginine rich splicing factors. Semin Cell Dev Biol. 2014;32C:11–21. doi: 10.1016/j.semcdb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Dresang LR, et al. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: Insights on the detection and discovery of viral genes. BMC Genomics. 2011;12:625. doi: 10.1186/1471-2164-12-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGeoch DJ. Molecular evolution of the gamma-Herpesvirinae. Philos Trans R Soc Lond B Biol Sci. 2001;356(1408):421–435. doi: 10.1098/rstb.2000.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tellam JT, Lekieffre L, Zhong J, Lynn DJ, Khanna R. Messenger RNA sequence rather than protein sequence determines the level of self-synthesis and antigen presentation of the EBV-encoded antigen, EBNA1. PLoS Pathog. 2012;8(12):e1003112. doi: 10.1371/journal.ppat.1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tellam J, et al. Regulation of protein translation through mRNA structure influences MHC class I loading and T cell recognition. Proc Natl Acad Sci USA. 2008;105(27):9319–9324. doi: 10.1073/pnas.0801968105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown LY, Brown SA. Alanine tracts: The expanding story of human illness and trinucleotide repeats. Trends Genet. 2004;20(1):51–58. doi: 10.1016/j.tig.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Faux NG, et al. Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res. 2005;15(4):537–551. doi: 10.1101/gr.3096505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerber HP, et al. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263(5148):808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 40.Berger Z, et al. Deleterious and protective properties of an aggregate-prone protein with a polyalanine expansion. Hum Mol Genet. 2006;15(3):453–465. doi: 10.1093/hmg/ddi460. [DOI] [PubMed] [Google Scholar]
- 41.Davies JE, Rubinsztein DC. Polyalanine and polyserine frameshift products in Huntington’s disease. J Med Genet. 2006;43(11):893–896. doi: 10.1136/jmg.2006.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Girstmair H, et al. Depletion of cognate charged transfer RNA causes translational frameshifting within the expanded CAG stretch in huntingtin. Cell Reports. 2013;3(1):148–159. doi: 10.1016/j.celrep.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 43.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501(7465):45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stochmanski SJ, et al. Expanded ATXN3 frameshifting events are toxic in Drosophila and mammalian neuron models. Hum Mol Genet. 2012;21(10):2211–2218. doi: 10.1093/hmg/dds036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.