Significance
The role of conformational fluctuations in protein reactions has been frequently mentioned to discuss the reaction mechanism. Supporting evidence for the importance of the fluctuation has been reported by showing the relationship between the flexibility of the reactant structure and reaction efficiency. However, there has been no direct evidence showing that the fluctuation is indeed enhanced during the reaction, although recent molecular dynamic simulations pointed out the importance. Here, we focused our attention on the experimental proof of enhancement by the time-resolved transient grating method, which is a unique and powerful method. Our results showed that fluctuation is a key to understanding why light-stimulated proteins can transfer the signal without changing the averaged conformation.
Abstract
Knowledge of the dynamical behavior of proteins, and in particular their conformational fluctuations, is essential to understanding the mechanisms underlying their reactions. Here, transient enhancement of the isothermal partial molar compressibility, which is directly related to the conformational fluctuation, during a chemical reaction of a blue light sensor protein from the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1 (TePixD, Tll0078) was investigated in a time-resolved manner. The UV-Vis absorption spectrum of TePixD did not change with the application of high pressure. Conversely, the transient grating signal intensities representing the volume change depended significantly on the pressure. This result implies that the compressibility changes during the reaction. From the pressure dependence of the amplitude, the compressibility change of two short-lived intermediate (I1 and I2) states were determined to be +(5.6 ± 0.6) × 10−2 cm3⋅mol−1⋅MPa−1 for I1 and +(6.6 ± 0.7)×10−2 cm3⋅mol−1⋅MPa−1 for I2. This result showed that the structural fluctuation of intermediates was enhanced during the reaction. To clarify the relationship between the fluctuation and the reaction, the compressibility of multiply excited TePixD was investigated. The isothermal compressibility of I1 and I2 intermediates of TePixD showed a monotonic decrease with increasing excitation laser power, and this tendency correlated with the reactivity of the protein. This result indicates that the TePixD decamer cannot react when its structural fluctuation is small. We concluded that the enhanced compressibility is an important factor for triggering the reaction of TePixD. To our knowledge, this is the first report showing enhanced fluctuations of intermediate species during a protein reaction, supporting the importance of fluctuations.
Proteins often transfer information through changes in domain–domain (or intermolecular) interactions. Photosensor proteins are an important example. They have light-sensing domains and function by using the light-driven changes in domain–domain interactions (1). The sensor of blue light using FAD (BLUF) domain is a light-sensing module found widely among the bacterial kingdom (2). The BLUF domain initiates its photoreaction by the light excitation of the flavin moiety inside the protein, which changes the domain–domain interaction, causing a quaternary structural change and finally transmitting biological signals (3, 4). It has been an important research topic to elucidate how the initial photochemistry occurring in the vicinity of the chromophore leads to the subsequent large conformation change in other domains, which are generally apart from the chromophore.
It may be reasonable to consider that the conformation change in the BLUF domain is the driving force in its subsequent reaction; that is, the change in domain–domain interaction. However, sometimes, clear conformational changes have not been observed for the BLUF domain; its conformation is very similar before and after photo-excitation (5–13). The circular dichroism (CD) spectra of BLUF proteins AppA and PixD from thermophilic cyanobacterium Thermosynechococcus elongatus BP-1 (TePixD) did not change on illumination (5, 13). Similarly, solution NMR studies of AppA and BlrB showed only small chemical shifts on excitation (9, 10). The solution NMR structure of BlrP1 showed a clear change, but this was limited in its C-terminal extension region and not core BLUF (11). Furthermore, the diffusion coefficient (D) of the BLUF domain of YcgF was not changed by photo-excitation (12), although D is sensitive to global conformational changes. These results imply that a minor structural change occurs in the BLUF domain. In such cases, how does the BLUF domain control its interdomain interaction? Recently, a molecular dynamics (MD) simulation on another light-sensing domain, the light-oxygen-voltage (LOV) sensing domain, suggested that fluctuation of the LOV core structure could be a key to understanding the mechanism of information transfer (14–16).
Because proteins work at room temperature, they are exposed to thermal fluctuations. The importance of such structural fluctuations for biomolecular reactions has been also pointed out: for example, enzymatic activity (17–20). Experimental detections of such conformation fluctuations using single molecular detection (21) or NMR techniques such as the hydrogen-deuterium (H-D) exchange, relaxation dispersion method, and high-pressure NMR (22–24) have succeeded. However, these techniques could not detect the fluctuation of short-lived transient species. Indeed, single molecule spectroscopy can trace the fluctuation in real time, but it is still rather difficult to detect rapid fluctuations for a short-lived intermediate during a reaction. Therefore, information about the fluctuation of intermediates is thus far limited.
A thermodynamic measurement is another way to characterize the fluctuation of proteins. In particular, the partial molar isothermal compressibility is essential, because this property is directly linked to the mean-square fluctuations of the protein partial molar volume by (25). (Here, <X> means the averaged value of a quantity of X.) Therefore, isothermal compressibility is thought to reflect the structural fluctuation of molecules (26). However, experimental measurement of this parameter of proteins in a dilute solution is quite difficult. Indeed, this quantity has been determined indirectly from the theoretical equation using the adiabatic compressibility of a protein solution, which was determined by the sound velocity in the solution (26–31). Although the relation between volume fluctuations and isothermal compressibility is rigorously correct only with respect to the intrinsic part of the volume compressibility, and not the partial molar volume compressibility (32), we considered that this partial molar volume compressibility is still useful for characterizing the fluctuation of the protein structure including its interacting water molecules. In fact, the relationship between and the volume fluctuation has been often used to discuss the fluctuation of proteins (17, 26–28), and the strong correlation of of reactants with the functioning for some enzymes (17, 33, 34) has been reported. These studies show the functional importance of the structural fluctuation represented by . However, thermodynamic techniques lack time resolution, and it has been impossible to measure the fluctuations of short-lived intermediate species.
Recently, we developed a time-resolving method for assessing thermodynamic properties using the pulsed laser induced transient grating (TG) method. Using this method, we thus far succeeded in measuring the enthalpy change (ΔH) (35–38), partial molar volume change () (12, 35, 37), thermal expansion change () (12, 37), and heat capacity change (ΔCp) (36–38) for short-lived species. Therefore, in principle, the partial molar isothermal compressibility change () of a short-lived intermediate become observable if we conduct the TG experiment under the high-pressure condition and detect with varying external pressure.
There are several difficulties in applying the traditional high-pressure cell to the TG method to measure thermodynamic parameters quantitatively. The most serious problem is ensuring the quantitative performance of the intensity of TG signals measured under the high-pressure condition. On this point, our group has developed a new high-pressure cell specially designed for TG spectroscopy (39) and overcome this problem. In this paper, by applying this high-pressure TG system to the BLUF protein TePixD, we report the first measurement, to our knowledge, of of short-lived intermediates to investigate the mechanism underlying signal transmission by BLUF proteins, from the view point of the transient fluctuation.
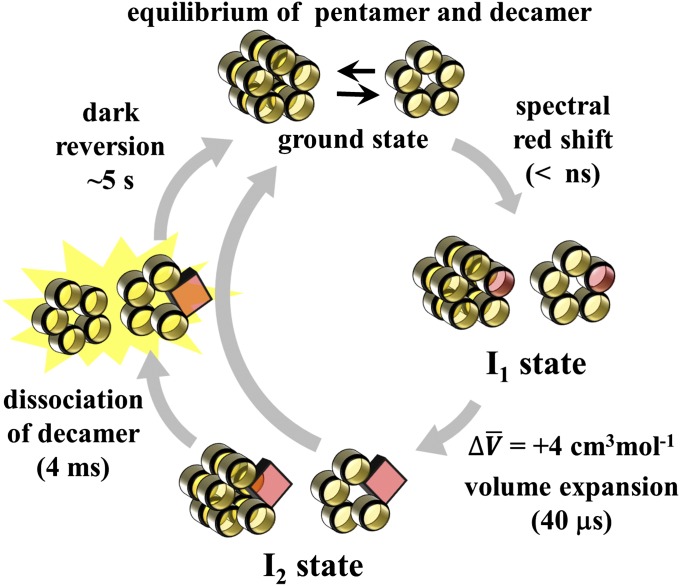
TePixD is a homolog of the BLUF protein PixD, which regulates the phototaxis of cyanobacterium (40) and exists in a thermophilic cyanobacterium Thermocynechococcus elongates BP-1 (Tll0078). TePixD is a relatively small (17 kDa) protein that consists only of the BLUF domain with two extended helices in the C-terminal region. In crystals and solutions, it forms a decamer that consists of two pentameric rings (41). The photochemistry of TePixD is typical among BLUF proteins (42–45); on blue light illumination, the absorption spectrum shifts toward red by about 10 nm within a nanosecond. The absorption spectrum does not change further, and the dark state is recovered with a time constant of ∼5 s at room temperature (40, 43). The spectral red shift was explained by the rearrangement of the hydrogen bond network around the chromophore (6, 46–48). The TG method has revealed the dynamic photoreaction mechanism, which cannot be detected by conventional spectroscopic methods. The TG signal of TePixD (Fig. S1) showed that there are two spectrally silent reaction phases: a partial molar volume expansion with the time constant of ∼40 μs and the diffusion coefficient (D) change with a time constant of ∼4 ms. Furthermore, it was reported that the pentamer and decamer states of TePixD are in equilibrium and that the final photoproduct of the decamer is pentamers generated by its dissociation (13, 49). On the basis of these studies, the reaction scheme has been identified as shown in Fig. 1. Here, I1 is the intermediate of the spectrally red-shifted species (generated within a nanosecond) and I2 is the one created on the subsequent volume expansion process of +4 cm3⋅mol−1 (∼40 μs). Furthermore, an experiment of the excitation laser power dependence of its TG signal revealed that the TePixD decamer undergoes the original dissociation reaction when only one monomer in the decamer is excited (50). In this study, we investigated the transient compressibility of the intermediates I1 and I2 of the photoreaction of TePixD and found a direct link between their fluctuation and reactivity.
Fig. 1.
Schematic illustration of the photoreaction of TePixD. Yellow circles represent the TePixD monomer in the ground state, which constructs the decamer and pentamer states. In the dark state, these two forms are in equilibrium. The excited, spectral red-shifted state of the TePixD monomer is indicated by a red circle. The square represents the I2 state of the monomer, which is created by the volume expansion process.
Results
Reaction Detected by Absorption at High Pressures.
Before measuring changes in compressibility, we first investigated the effects of pressure on the UV-Vis absorption spectra of TePixD in the dark state (Fig. S2). Here, the spectrum was corrected to allow for the increase in density (i.e., concentration) of the solution owing to the increase in pressure (51). The absorption spectrum of TePixD was almost independent of the pressure. In addition, we checked the permanent pressure denaturation of TePixD by comparing the CD spectrum before and after applying the high pressure. The spectrum in a range of 200–250 nm recovered completely after the pressurization of 200 MPa. These results indicated that permanent pressure denaturation of TePixD did not occur in this pressure range.
The effect of pressure on the photochemistry of TePixD was investigated by the transient absorption (TA) method. The pressure dependence of the TA spectrum measured at 10 μs after excitation is shown in Fig. S3. Here, the intensities were corrected using the absorbance change from the UV-Vis spectra at the excitation wavelength (462 nm). It is clear that the spectrum was not altered by pressure except for a slight decrease in amplitude. In addition, the time profiles of the TA signal of TePixD were probed at 483 nm under various pressures (Fig. S4). The amplitude of the signal decreased slightly at high pressures, and the decay rate was increased. Because the TA spectrum was not altered by pressure, this slight decrease in amplitude was attributed to the quantum yield change. The quantum yield change (as the relative parameter ϕ/ϕ0; ϕ0: the quantum yield at 0.1 MPa) and the lifetime (τ) of the dark recovery at various pressures were plotted (Fig. S4 B and C). The acceleration of the dark recovery and slight decrease in the quantum yield observed indicate the pressure effect to the transition state of the reaction. The pressure dependence of the rate is related with the activation volume along the reaction coordinate, and this value is negative in this case. More importantly for this study, we should point out that the pressure does not affect the reaction scheme of TePixD. Hence, we can discuss the fluctuation by measuring the volume change at various pressures.
Transient Fluctuation During the Reaction.
We measured the TG signal for TePixD under the high-pressure condition to investigate the fluctuation of its intermediates at a weak light intensity, 1.02 ± 0.02 mJ⋅cm−2, which is weak enough to excite only one monomer unit in the decamer (50). The time evolution of the TG signal of TePixD after photo-excitation has been described previously (49). Here, we briefly summarize its essential feature. A typical TG signal at q2 = 3.5 × 1012 m−2 in a wide time range is depicted in Fig. S1. The signal consists of the thermal grating component (∼1 μs), a volume expansion process (weak decay after thermal diffusion; ∼40 μs), and a peak of the molecular diffusion signal (2–20 ms), which represents the diffusion coefficient change. Analyzing the TG signal, we determined the reaction scheme for TePixD (Fig. 1). In the present study, we applied high pressure and measured the TG signal representing the volume expansion process from an intermediate I1 to I2 (the amplified signal shown in the Inset of Fig. S1) to detect their fluctuations.
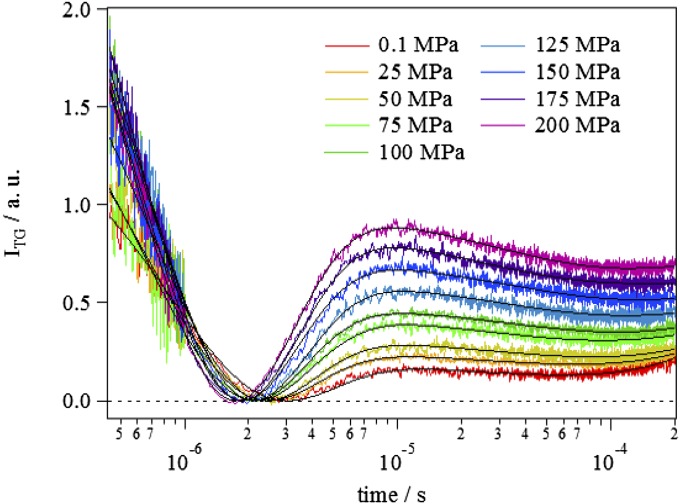
Fig. 2 shows the pressure dependence of the TG signal of the volume expansion process at q2 = 3.5 × 1012 m−2. It is clear that the TG signal of TePixD depended significantly on the pressure, in contrast to the results of UV-Vis and transient absorptions. As shown in SI Text, the TG signal in a longer time range of Fig. 2 represents the protein diffusion signal, which has been analyzed by a sum of three exponential functions (49). However, in this study, we need only the amplitude of the volume grating signal and not the time profiles of the diffusion. Hence, to reduce the ambiguity of the fitting, we analyzed the diffusion signal by expanding the exponential function in the early time range and neglecting higher-order terms of t (SI Text). The resultant fitting function is
| [1] |
Here, α is a proportional constant, the first term of Eq. 1 represents the thermal diffusion process (δnth; thermal grating, Dth; diffusion coefficient of the heat), the second term represents the volume expansion process (δnV; amplitude of the volume grating, kV; reaction rate of the volume change), and the last term (A+Bt) represents the contribution of the molecular diffusion signal. The TG signals at different pressures were fitted by Eq. 1, and fitting curves are shown by the solid lines in Fig. 2. The fitting curves almost perfectly reproduced the signal, and we could uniquely determine the parameters.
Fig. 2.
Typical TG signals of TePixD in the submillisecond time region, which represents the volume expansion process from the intermediate I1 to I2, recorded at every 25 MPa from 0.1 to 200 MPa (from Lower to Upper) with q2 = 3.5 × 1012 m−2. Fitting curves based on the fitting function Eq. 1 are shown by black solid lines. Pressures are indicated by the legend in the figure.
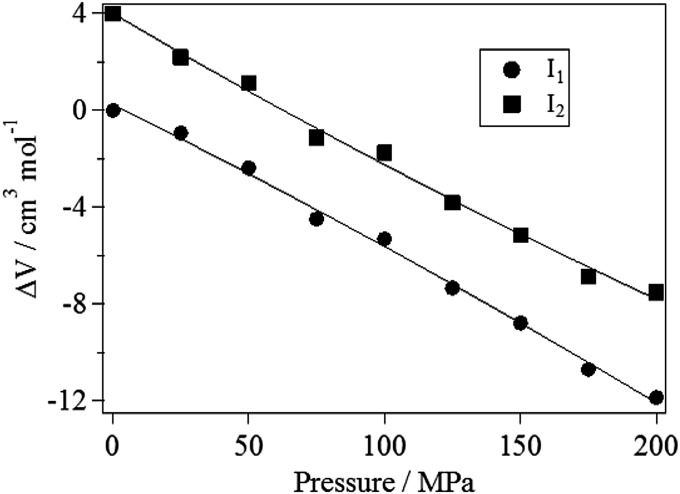
From the pressure dependence of the amplitude of species grating of I1 and I2 states, the pressure dependences of the volume changes () for the I1 and I2 states were determined by a method described in SI Text and shown in Fig. 3, where for I1 at 0.1 MPa was used as the reference value. We fitted the data by the following quadratic function:
| [2] |
where is the pressure, is the volume difference at 0.1 MPa, is the partial molar compressibility change compared with the ground state, and the last term is for the correction of the slight compressibility change by the pressure. From this fitting, we determined each parameter for I1 and I2 as follows: = +(5.6 ± 0.6) × 10−2 cm3⋅mol−1⋅MPa−1 (for I1) and = +(6.6 ± 0.7) × 10−2 cm3⋅mol−1⋅MPa−1 (for I2). Therefore, using the relationship between the compressibility and the volume fluctuation [i.e., ], the volume fluctuation change from the ground state to the excited state [] was obtained to be 140 ± 20 (cm3⋅mol−1)2 for I1 and 160 ± 20 (cm3⋅mol−1)2 for I2. (Here mol means the number of excited monomers.) This result showed that the partial molar volume fluctuation of the short-lived intermediate states is larger than that of the ground state.
Fig. 3.
Pressure dependence of the volume change from the ground state (g) to the excited state (e) (i.e., ) for I1 and I2 states. Because the absolute value of of I1 and I2 are not known, their pressure dependences in this figure are plotted by relative values from of I1 at 0.1 MPa. Solid lines represent the best fitting results by a quadratic function of Eq. 2.
Compressibility of Multiexcited Species.
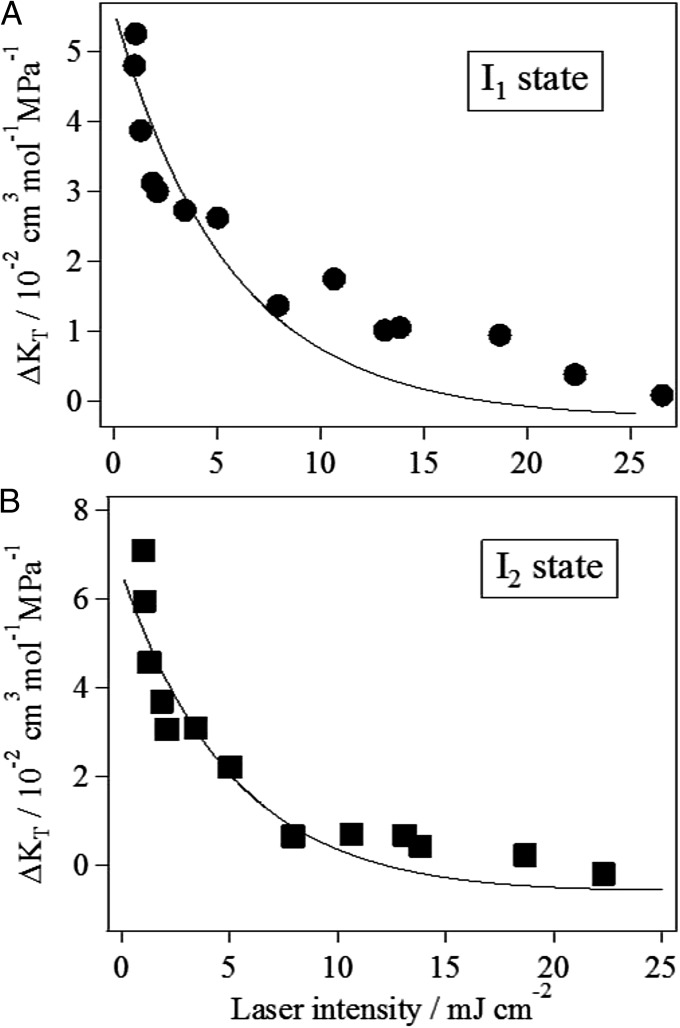
To further examine the importance of the compressibility in the intermediate state of the reaction, we studied the laser power dependence of the compressibility changes. Previously, it was shown that photo-excitation of a monomer of TePixD yields I1 and I2 intermediates at any laser power but does not produce the final product when multiple monomers in the decamer unit were excited. Therefore, if the structural fluctuation correlates with the reactivity of TePixD, examining the excitation power dependence of the compressibility will be a good test for it.
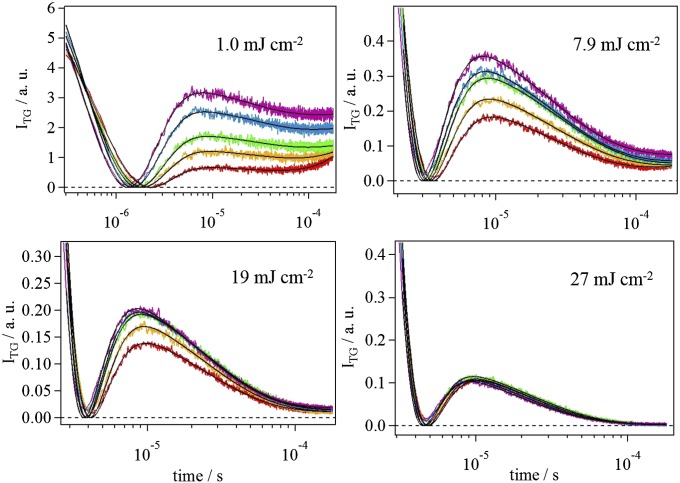
Fig. 4 shows the pressure dependence of the TG signal at q2 = 4.4 × 1012 m−2 under four different excitation laser powers: 1.0, 7.9, 19, and 27 mJ⋅cm−2. [For a negative control experiment, we measured the TG signal of a photo-inactive mutant (Q50A) under the same conditions (Fig. S5). Any volume change reaction was not observed for this mutant, confirming that the above experimental conditions did not cause any artifact.] From the results shown in Fig. 4, it is clear that the TG signals became less sensitive to pressure with increasing excitation laser power. We fitted these TG signals at different powers by Eq. 1 and determined the compressibility of the intermediates in the similar way. The laser power dependence of the apparent compressibility change () for I1 and I2 states obtained from the fitting is shown in Fig. 5 A and B, respectively. For both states, the compressibility decreased monotonically with increasing the excitation laser power. Therefore, it is qualitatively apparent that a TePixD decamer (or pentamer) containing multiple excited monomers possesses smaller compressibilities than does a decamer containing only one excited monomer.
Fig. 4.
Similar TG signals for TePixD to those in Fig. 3 under four different laser power conditions of 1.0, 7.9, 19, and 27 mJ /cm2. Applied pressures were 0.1 (red), 50 (orange), 100 (green), 150 (blue), and 200 MPa (magenta). The grating wave number was q2 = 4.4 × 1012 m−2. Signal intensities with different excitation laser powers were normalized by the obtained fitting parameter δnv of Eq. 1 at 0.1 MPa, which is proportional to the number of excited species. Fitting curves based on the fitting function Eq. 1 are shown by solid lines.
Fig. 5.
Laser power dependence of the volume fluctuation change from the ground state to the excited state I1 (A) and I2 (B). Best-fit curves by Eq. 4 are shown by the solid lines.
The observed compressibility change is the sum of contributions from a decamer having different numbers of excited monomers. To extract the compressibility change of multiexcited species, we fitted the results of Fig. 5 by the function of laser power as follows. The observed volume fluctuation is the sum of contributions from oligomers having different numbers of excited monomers. The apparent compressibility () may be expressed as
| [3] |
Here, fn denotes the fraction of oligomers having n excited monomer units, and is the compressibility change of a monomer in that decamer. was determined in the former section, but other parameters () are unknown. Hence, if we use this function to fit the laser power dependence in all power ranges, there are too many adjustable parameters to be uniquely determined. To avoid ambiguity for the fitting, we analyzed the data in a relatively weak laser power region as follows. In a laser power range of <8 mJ⋅cm−2, the fraction of the triple excited species (f3) is estimated to be smaller than 15% of the total excited decamers (Figs. S6 and S7). The fraction of the species having n > 3 should be much smaller. Therefore, it may be reasonable to consider only n = 1 and 2 for the fitting in a weak laser power region. In this case, Eq. 3 becomes
| [4] |
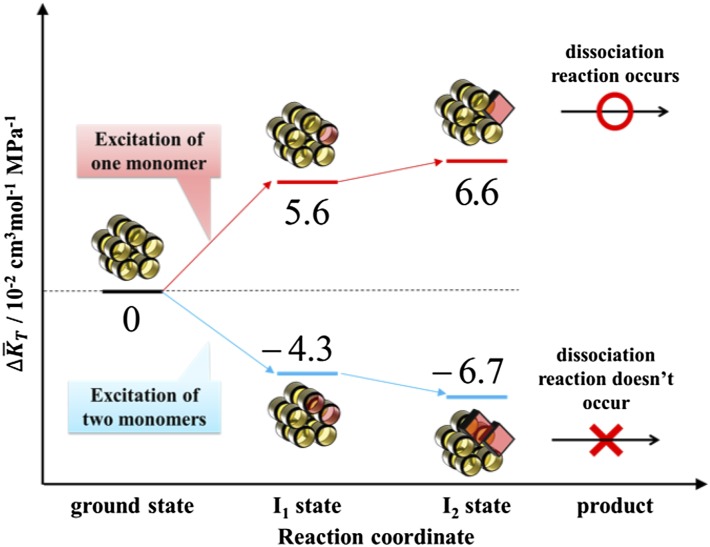
Here, the fractions fn are given by Eq. S12, and the parameters c and Is were fixed to the predetermined values described in SI Text. For , we also fixed it to the values determined in the former section: = 5.6 × 10−2 cm3⋅mol−1⋅MPa−1 for I1 and 6.6 × 10−2 cm3⋅mol−1⋅MPa−1 for I2. Hence, Eq. 4 now contains only one adjustable parameter, . By using Eq. 4, we fitted the data in the laser power region below 8 mJ⋅cm−2, and the results are shown in Fig. 5. Although the adjustable parameter is only , the fitting curve well reproduced the laser power dependence in this region. From this fitting, the compressibility change of double-excited species () was uniquely determined as –(4.3 ± 1.5) × 10−2 cm3⋅mol−1⋅MPa−1 for I1 and –(6.7 ± 2.4) × 10−2 cm3⋅mol−1⋅MPa−1 for I2. The compressibility of both I1 and I2 of two-excited decamer was found to be much smaller than that of the one excited species and even smaller than that of its ground state. Therefore, we concluded that the enhanced compressibility is important to lead to the dissociation reaction of TePixD decamer.
These results are schematically illustrated in Fig. 6.
Fig. 6.
Schematic illustration of the volume fluctuation change from the ground state, depicted along the reaction coordinate of TePixD for both cases in which one monomer is excited (red lines) or multiple monomers are excited (blue lines). In the figure, volume fluctuation change is expressed per mole of TePixD monomers.
Discussion
Traditionally, compressibility has been measured from the pressure dependence of the equilibrium constant at a pressure P, K(P), which may be expressed by
Therefore, for a reaction under equilibrium between two states, the compressibility may be measured by the second-order expansion of P of the pressure-dependent K. However, this traditional method cannot be applied to the short-lived intermediate species during chemical reactions in principle. Furthermore, higher-pressure data are more important for determining the quadratic behavior of K. Therefore, this method may easily suffer from the effects of high pressure on protein structure (not the volumetric effect); that is, artifact. Conversely, the present TG technique is more advanced; the volumetric data are directly determined from the signal intensity; and the compressibility can be determined from the pressure effect in a low-pressure range.
The detected enhancement of the compressibility was 5.6 × 10−2 cm3⋅mol−1⋅MPa−1 for I1 and 6.6 × 10−2 cm3⋅mol−1⋅MPa−1 for I2. Although the compressibility in the ground stable state of TePixD has not yet been reported, we can roughly estimate how large the enhancement is compared with the ground state as follows. According to the studies of Gekko et al., the square root of the volume fluctuation of globular proteins is about 0.3% of their partial molar volume (26), and the partial specific volumes of many globular proteins are very similar, ranging from 0.7 to 0.75 cm3⋅g−1. Using these data, the partial molar volume of the TePixD monomer is estimated to be ∼13,000 cm3⋅mol−1, assuming a partial specific volume of 0.75 cm3⋅g−1. Therefore, its square root of the volume fluctuation in the ground state is calculated to be ∼39 cm3 mol−1. This value corresponds to the compressibility of 60 × 10−2 cm3⋅mol−1⋅MPa−1 in the ground state. Therefore, the observed enhancement of the compressibility (5.6 × 10−2 and 6.6 × 10−2 cm3⋅mol−1⋅MPa−1 for I1 and I2, respectively) in the intermediate states is about 10% of the ground state compressibility for both I1 and I2.
We consider that the estimated increase of 10% in compressibility is large, because the fluctuation change may not be spread over the whole protein but rather is localized in a small area, in particular, around the interface of TePixD pentamer rings, which must be important for the dissociation reaction. The light-induced structural change of the BLUF domain has been expected to occur in the C-terminal extension region of the BLUF domain; that is, from the β4-β5 loop to the α4 helix (11, 52–54). However, in the case of TePixD, these regions are far from the interface of pentamer rings. Therefore, a structural change in these regions is insufficient to explain the dissociation of the decamer. Instead, it may use the enhanced fluctuation of interface region to help achieve the dissociation reaction. In our previous study (50), we reported the discovery of the strange light intensity dependence. In this paper, we found that the two-photon excitation suppresses the fluctuation and concluded that this smaller fluctuation is a cause of the suppression of the reaction.
In conclusion, we succeeded in detecting isothermal partial molar compressibility of two short-lived intermediates during the photoreaction of TePixD. The enhancement of the volume fluctuation was observed for both I1 and I2 intermediate states, and this enhancement should be the trigger for the dissociation reaction of the TePixD decamer. To our knowledge, this is the first direct experimental report to connect protein reactivity and fluctuations of reaction intermediates.
Methods
Sample Preparation.
TePixD was expressed using a pET28a vector transformed into Escherichia coli BL21 (DE3) and purified by nickel affinity column chromatography, as reported previously (40). In all measurements, the sample was prepared by dissolving in Hepes buffer (20 mM Hepes-NaOH, pH 7.5, and 500 mM NaCl). The concentration of TePixD was determined by UV-Vis absorption measurement, using the extinction coefficient of FAD; ε = 11,300 M−1⋅cm−1 at 450 nm. In most cases, the sample concentration used was ∼530 μM.
High-Pressure Equipment.
Details of the high-pressure apparatus used in this study have been described elsewhere (39). The pressure resistance of this cell is up to 500 MPa. In all measurements, the internal temperature was set to be 295.5 K, and the applied pressure range was from 0.1 to 200 MPa. It has been validated that this high-pressure cell can achieve the complete reproducibility of a TG signal by applying high pressure and the sample replacement operation (39).
TA Measurement.
The TA signals were monitored after photo-excitation by a XeCl excimer laser-pumped dye laser beam (Lambda Physik CompexPro102; λ = 308 nm, Lumonics Hyper Dye 300; λ = 462 nm). An Xe lamp was used to measure the TA spectra. The probe light passing through the sample was focused on an optical fiber, leading to a monochrometer (ACTON Research Corporation SpectraPro 2300i). The temporal profile of the TA signal was monitored by a probe light from a light-emitting diode (LED Luminar; Nissin Electronic) at a wavelength of 483 nm, with a full width at half maximum of 16 nm, which was selected by long-pass glass filters. This light was detected by a photomultiplier tube (R1477; Hamamatsu). The signal was fed into a digital oscilloscope (TDS-7104; Tektronix) and averaged 20 times. The repetition rate for excitation was set to 0.025 Hz.
TG Measurement Under High Pressure.
Detailed descriptions on the TG method are described in SI Text. Briefly, in the TG method, two laser pulses are introduced into the sample solution to trigger the photoreaction. The intensity (ITG) is proportional to the square of the generated refractive index change (δn) arising from the volume change, temperature change, and absorption change. The experimental setup for TG measurement was similar to that reported before (12, 35–39, 49, 50). The excitation laser pulse and detection systems (a photomultiplier tube and digital oscilloscope) were all the same as those used for measuring the time profile of the TA signal. A CW diode laser (835 nm; Crysta Laser) was used as a probe beam. The grating wave number q in the experimental condition was determined from the thermal grating signal of a calorimetric reference sample (bromocresol purple in water) measured under the same conditions. The repetition rate for excitation was set to 0.04 Hz, which is slower than the dark recovery time of TePixD (∼5 s). Whenever we applied high pressure, we always reset the pressure to 0.1 MPa after every compression to check the recovery of the signal. It was confirmed that the TG signals were completely reversible. The excitation laser power was monitored using a pyroelectric Joulemeter (J3-09; Coherent).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413222111/-/DCSupplemental.
References
- 1.Zoltowski BD, Gardner KH. Tripping the light fantastic: Blue-light photoreceptors as examples of environmentally modulated protein-protein interactions. Biochemistry. 2011;50(1):4–16. doi: 10.1021/bi101665s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Losi A, Gartner W. Bacterial bilin- and flavin-binding photoreceptors. Photochem Photobiol Sci. 2008;7(10):1168–1178. doi: 10.1039/b802472c. [DOI] [PubMed] [Google Scholar]
- 3.Masuda S. Light detection and signal transduction in the BLUF photoreceptors. Plant Cell Physiol. 2013;54(2):171–179. doi: 10.1093/pcp/pcs173. [DOI] [PubMed] [Google Scholar]
- 4.Losi A, Gärtner W. The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors. Annu Rev Plant Biol. 2012;63:49–72. doi: 10.1146/annurev-arplant-042811-105538. [DOI] [PubMed] [Google Scholar]
- 5.Kraft BJ, et al. Spectroscopic and mutational analysis of the blue-light photoreceptor AppA: A novel photocycle involving flavin stacking with an aromatic amino acid. Biochemistry. 2003;42(22):6726–6734. doi: 10.1021/bi030055o. [DOI] [PubMed] [Google Scholar]
- 6.Masuda S, Hasegawa K, Ishii A, Ono TA. Light-induced structural changes in a putative blue-light receptor with a novel FAD binding fold sensor of blue-light using FAD (BLUF); Slr1694 of synechocystis sp. PCC6803. Biochemistry. 2004;43(18):5304–5313. doi: 10.1021/bi049836v. [DOI] [PubMed] [Google Scholar]
- 7.Anderson S, et al. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 2005;44(22):7998–8005. doi: 10.1021/bi0502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majerus T, Kottke T, Laan W, Hellingwerf K, Heberle J. Time-resolved FT-IR spectroscopy traces signal relay within the blue-light receptor AppA. ChemPhysChem. 2007;8(12):1787–1789. doi: 10.1002/cphc.200700248. [DOI] [PubMed] [Google Scholar]
- 9.Grinstead JS, et al. The solution structure of the AppA BLUF domain: Insight into the mechanism of light-induced signaling. ChemBioChem. 2006;7(1):187–193. doi: 10.1002/cbic.200500270. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, Ko WH, Gardner KH. Structural requirements for key residues and auxiliary portions of a BLUF domain. Biochemistry. 2008;47(39):10271–10280. doi: 10.1021/bi8011687. [DOI] [PubMed] [Google Scholar]
- 11.Wu Q, Gardner KH. Structure and insight into blue light-induced changes in the BlrP1 BLUF domain. Biochemistry. 2009;48(12):2620–2629. doi: 10.1021/bi802237r. [DOI] [PubMed] [Google Scholar]
- 12.Nakasone Y, Ono TA, Ishii A, Masuda S, Terazima M. Temperature-sensitive reaction of a photosensor protein YcgF: Possibility of a role of temperature sensor. Biochemistry. 2010;49(10):2288–2296. doi: 10.1021/bi902121z. [DOI] [PubMed] [Google Scholar]
- 13.Kuroi K, et al. Anomalous diffusion of TePixD and identification of the photoreaction product. Photochem Photobiol Sci. 2013;12(7):1180–1186. doi: 10.1039/c3pp25434h. [DOI] [PubMed] [Google Scholar]
- 14.Freddolino PL, Dittrich M, Schulten K. Dynamic switching mechanisms in LOV1 and LOV2 domains of plant phototropins. Biophys J. 2006;91(10):3630–3639. doi: 10.1529/biophysj.106.088609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freddolino PL, Gardner KH, Schulten K. Signaling mechanisms of LOV domains: New insights from molecular dynamics studies. Photochem Photobiol Sci. 2013;12(7):1158–1170. doi: 10.1039/c3pp25400c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peter E, Dick B, Baeurle SA. Signals of LOV1: A computer simulation study on the wildtype LOV1-domain of Chlamydomonas reinhardtii and its mutants. J Mol Model. 2012;18(4):1375–1388. doi: 10.1007/s00894-011-1165-6. [DOI] [PubMed] [Google Scholar]
- 17.Gekko K, Obu N, Li J, Lee JC. A linear correlation between the energetics of allosteric communication and protein flexibility in the Escherichia coli cyclic AMP receptor protein revealed by mutation-induced changes in compressibility and amide hydrogen-deuterium exchange. Biochemistry. 2004;43(13):3844–3852. doi: 10.1021/bi036271e. [DOI] [PubMed] [Google Scholar]
- 18.Eisenmesser EZ, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438(7064):117–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 19.Bhabha G, et al. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science. 2011;332(6026):234–238. doi: 10.1126/science.1198542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitahara R, et al. A delicate interplay of structure, dynamics, and thermodynamics for function: A high pressure NMR study of outer surface protein A. Biophys J. 2012;102(4):916–926. doi: 10.1016/j.bpj.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deniz AA, Mukhopadhyay S, Lemke EA. Single-molecule biophysics: At the interface of biology, physics and chemistry. J R Soc Interface. 2008;5(18):15–45. doi: 10.1098/rsif.2007.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang T-L, van Zijl PC, Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J Biomol NMR. 1998;11(2):221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 23.Mittermaier A, Kay LE. New tools provide new insights in NMR studies of protein dynamics. Science. 2006;312(5771):224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 24.Akasaka K, Kitahara R, Kamatari YO. Exploring the folding energy landscape with pressure. Arch Biochem Biophys. 2013;531(1-2):110–115. doi: 10.1016/j.abb.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Cooper A. Thermodynamic fluctuations in protein molecules. Proc Natl Acad Sci USA. 1976;73(8):2740–2741. doi: 10.1073/pnas.73.8.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gekko K. Compressibility gives new insight into protein dynamics and enzyme function. Biochim Biophys Acta. 2002;1595(1-2):382–386. doi: 10.1016/s0167-4838(01)00358-2. [DOI] [PubMed] [Google Scholar]
- 27.Gekko K, Noguchi H. Compressibility of globular proteins in water at 25.degree.C. J Phys Chem. 1979;83(21):2706–2714. [Google Scholar]
- 28.Gekko K, Hasegawa Y. Compressibility-structure relationship of globular proteins. Biochemistry. 1986;25(21):6563–6571. doi: 10.1021/bi00369a034. [DOI] [PubMed] [Google Scholar]
- 29.Sarvazyan AP. Ultrasonic velocimetry of biological compounds. Annu Rev Biophys Biophys Chem. 1991;20(1):321–342. doi: 10.1146/annurev.bb.20.060191.001541. [DOI] [PubMed] [Google Scholar]
- 30.Kharakoz DP. Partial volumes and compressibilities of extended polypeptide chains in aqueous solution: Additivity scheme and implication of protein unfolding at normal and high pressure. Biochemistry. 1997;36(33):10276–10285. doi: 10.1021/bi961781c. [DOI] [PubMed] [Google Scholar]
- 31.Taulier N, Chalikian TV. Compressibility of protein transitions. Biochim Biophys. 2002;1595(1-2):48–70. doi: 10.1016/s0167-4838(01)00334-x. [DOI] [PubMed] [Google Scholar]
- 32.Son I, Selvaratnam R, Dubins DN, Melacini G, Chalikian TV. Ultrasonic and densimetric characterization of the association of cyclic AMP with the cAMP-binding domain of the exchange protein EPAC1. J Phys Chem B. 2013;117(37):10779–10784. doi: 10.1021/jp406451p. [DOI] [PubMed] [Google Scholar]
- 33.Kamiyama T, Gekko K. Effect of ligand binding on the flexibility of dihydrofolate reductase as revealed by compressibility. Biochim Biophys Acta. 2000;1478(2):257–266. doi: 10.1016/s0167-4838(00)00019-4. [DOI] [PubMed] [Google Scholar]
- 34.Gekko K, Yamagami K. Compressibility and volume changes of lysozyme due to inhibitor binding. Chem Lett. 1998;27(8):839–840. [Google Scholar]
- 35.Inoue K, Sasaki J, Morisaki M, Tokunaga F, Terazima M. Time-resolved detection of sensory rhodopsin II-transducer interaction. Biophys J. 2004;87(4):2587–2597. doi: 10.1529/biophysj.104.043521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan JS, Imamoto Y, Kataoka M, Tokunaga F, Terazima M. Time-resolved thermodynamics: Heat capacity change of transient species during photoreaction of PYP. J Am Chem Soc. 2006;128(3):1002–1008. doi: 10.1021/ja055584p. [DOI] [PubMed] [Google Scholar]
- 37.Eitoku T, et al. Photochemical intermediates of Arabidopsis phototropin 2 LOV domains associated with conformational changes. J Mol Biol. 2007;371(5):1290–1303. doi: 10.1016/j.jmb.2007.06.035. [DOI] [PubMed] [Google Scholar]
- 38.Hazra P, Inoue K, Laan W, Hellingwerf KJ, Terazima M. Energetics and role of the hydrophobic interaction during photoreaction of the BLUF domain of AppA. J Phys Chem B. 2008;112(5):1494–1501. doi: 10.1021/jp0767314. [DOI] [PubMed] [Google Scholar]
- 39.Hoshihara Y, Kimura Y, Matsumoto M, Nagasawa M, Terazima M. An optical high-pressure cell for transient grating measurements of biological substance with a high reproducibility. Rev Sci Instrum. 2008;79(3):034101. doi: 10.1063/1.2894331. [DOI] [PubMed] [Google Scholar]
- 40.Okajima K, et al. Biochemical and functional characterization of BLUF-type flavin-binding proteins of two species of cyanobacteria. J Biochem. 2005;137(6):741–750. doi: 10.1093/jb/mvi089. [DOI] [PubMed] [Google Scholar]
- 41.Kita A, Okajima K, Morimoto Y, Ikeuchi M, Miki K. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J Mol Biol. 2005;349(1):1–9. doi: 10.1016/j.jmb.2005.03.067. [DOI] [PubMed] [Google Scholar]
- 42.Laan W, van der Horst MA, van Stokkum IH, Hellingwerf KJ. Initial characterization of the primary photochemistry of AppA, a blue-light-using flavin adenine dinucleotide-domain containing transcriptional antirepressor protein from Rhodobacter sphaeroides: A key role for reversible intramolecular proton transfer from the flavin adenine dinucleotide chromophore to a conserved tyrosine? Photochem Photobiol. 2003;78(3):290–297. doi: 10.1562/0031-8655(2003)078<0290:icotpp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 43.Fukushima Y, Okajima K, Shibata Y, Ikeuchi M, Itoh S. Primary intermediate in the photocycle of a blue-light sensory BLUF FAD-protein, Tll0078, of Thermosynechococcus elongatus BP-1. Biochemistry. 2005;44(13):5149–5158. doi: 10.1021/bi048044y. [DOI] [PubMed] [Google Scholar]
- 44.Gauden M, et al. Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry. 2005;44(10):3653–3662. doi: 10.1021/bi047359a. [DOI] [PubMed] [Google Scholar]
- 45.Okajima K, et al. Fate determination of the flavin photoreceptions in the cyanobacterial blue light receptor TePixD (Tll0078) J Mol Biol. 2006;363(1):10–18. doi: 10.1016/j.jmb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 46.Bonetti C, et al. The role of key amino acids in the photoactivation pathway of the Synechocystis Slr1694 BLUF domain. Biochemistry. 2009;48(48):11458–11469. doi: 10.1021/bi901196x. [DOI] [PubMed] [Google Scholar]
- 47.Domratcheva T, Grigorenko BL, Schlichting I, Nemukhin AV. Molecular models predict light-induced glutamine tautomerization in BLUF photoreceptors. Biophys J. 2008;94(10):3872–3879. doi: 10.1529/biophysj.107.124172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gauden M, et al. Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc Natl Acad Sci USA. 2006;103(29):10895–10900. doi: 10.1073/pnas.0600720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka K, et al. Oligomeric-state-dependent conformational change of the BLUF protein TePixD (Tll0078) J Mol Biol. 2009;386(5):1290–1300. doi: 10.1016/j.jmb.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka K, et al. A way to sense light intensity: Multiple-excitation of the BLUF photoreceptor TePixD suppresses conformational change. FEBS Lett. 2011;585(5):786–790. doi: 10.1016/j.febslet.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Saul A, Wagner W. A fundamental equation for water covering the range from the melting line to 1273 K at pressures up to 25 000 MPa. J Phys Chem Ref Data. 1989;18(4):1537–1564. [Google Scholar]
- 52.Hasegawa K, Masuda S, Ono TA. Spectroscopic analysis of the dark relaxation process of a photocycle in a sensor of blue light using FAD (BLUF) protein Slr1694 of the cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 2005;46(1):136–146. doi: 10.1093/pcp/pci003. [DOI] [PubMed] [Google Scholar]
- 53.Khrenova M, Domratcheva T, Grigorenko B, Nemukhin A. Coupling between the BLUF and EAL domains in the blue light-regulated phosphodiesterase BlrP1. J Mol Model. 2011;17(7):1579–1586. doi: 10.1007/s00894-010-0842-1. [DOI] [PubMed] [Google Scholar]
- 54.Barends TR, et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459(7249):1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.