Significance
Nature is full of species that cooperate in mutually beneficial interactions to survive. Some are completely dependent on such relationships. How and why does this specialization evolve? We show that as the bacterium Desulfovibrio vulgaris evolved for 1,000 generations in conditions forcing cooperation with the archaeon Methanococcus maripaludis, it lost a key metabolic trait that would be required for it to grow alone in most environments. Large subpopulations lacking the capacity to respire sulfate evolved in 13 of 21 replicates. Such striking parallel evolution suggests a trade-off between performance in the mutualistic environment and maintaining the flexibility to survive alone. This result may explain why sulfate reducers share a common ancestor with many species specialized for cooperation with methanogens.
Keywords: trade-offs, sulfate-reducing prokaryote, syntrophy, coevolution, experimental evolution
Abstract
Many species have evolved to function as specialized mutualists, often to the detriment of their ability to survive independently. However, there are few, if any, well-controlled observations of the evolutionary processes underlying the genesis of new mutualisms. Here, we show that within the first 1,000 generations of initiating independent syntrophic interactions between a sulfate reducer (Desulfovibrio vulgaris) and a hydrogenotrophic methanogen (Methanococcus maripaludis), D. vulgaris frequently lost the capacity to grow by sulfate respiration, thus losing the primary physiological attribute of the genus. The loss of sulfate respiration was a consequence of mutations in one or more of three key genes in the pathway for sulfate respiration, required for sulfate activation (sat) and sulfate reduction to sulfite (apsA or apsB). Because loss-of-function mutations arose rapidly and independently in replicated experiments, and because these mutations were correlated with enhanced growth rate and productivity, gene loss could be attributed to natural selection, even though these mutations should significantly restrict the independence of the evolved D. vulgaris. Together, these data present an empirical demonstration that specialization for a mutualistic interaction can evolve by natural selection shortly after its origin. They also demonstrate that a sulfate-reducing bacterium can readily evolve to become a specialized syntroph, a situation that may have often occurred in nature.
From flowering plants and their pollinators to the microbial endosymbionts of insects, there are many examples in nature of obligate mutualists (1, 2), or species dependent upon a mutually beneficial interaction for their survival or reproduction. How these interactions evolve is a mystery because much theory predicts that cooperative interactions should be unstable (3) and because of the difficulty of inferring evolutionary events that occurred in the distant past (4). Although there are few, if any, empirical observations of evolution toward dependence on mutualism, there are now several examples of mutualisms evolving de novo in the laboratory (5–8). This advancement has provided researchers an experimental framework to study populations and ecological conditions in the early stages of evolution (5–8).
Here, we describe our observations of rapid and repeated evolution of increased dependency on a mutualism through natural selection. This interaction is similar to a widespread relationship between prokaryotes that plays a pivotal role in the decomposition of carbon in many oxygen-free environments. In these syntrophic mutualisms, bacteria ferment organic acids, producing hydrogen or formate as by-products, which are then used by hydrogen-consuming species, often methanogenic archaea (9). Removal of hydrogen and formate benefits the bacteria because the free energy (ΔG) available decreases with increasing concentrations of these products (9). A variety of bacterial species have been described that seem to be specialized for fermenting organic acids in syntrophic association with hydrogen-consuming species (10–13). Notably, most clades of characterized syntrophs share a recent common ancestry with sulfate reducers (10, 14). Some retain vestiges of the sulfate-reducing pathway, and several lines of evidence hint at the possibility that specialized syntrophs were once sulfate reducers (11, 14).
Sulfate-reducing bacteria gain energy from organic acids, such as lactate, in the absence of oxygen by coupling their oxidation to the reduction of sulfate to sulfide. These bacteria play a critical role in sulfur and carbon cycling, contribute to corrosion in the petroleum industry and wastewater treatment plants, and have been used for bioremediation of toxic heavy metals (15). The ability of sulfate reducers to grow in syntrophic association with methanogens was first demonstrated in laboratory studies (16) and is now generally recognized to be of environmental relevance (17–19). Many sulfate reducers would therefore be better described as facultative syntrophs, well adapted to environments of fluctuating electron acceptor availability (17, 20). Past evolutionary transitions of sulfate-reducing bacteria between obligate and facultative syntrophs is also indicated by comparative analyses indicating horizontal transfer of genes in the pathway of sulfate respiration (21, 22). Thus, the evolutionary adaptive flexibility of sulfate-reducing bacteria suggests that they offer an attractive experimental system to study the evolution of mutualism.
To understand how mutualisms, and syntrophic interactions in particular, might evolve from their origin, we paired the sulfate-reducing bacterium, Desulfovibrio vulgaris Hildenborough, with the archaeon, Methanococcus maripaludis S2, and propagated 22 initially isogenic planktonic cocultures for 1,000 generations in medium with lactate but no sulfate or added hydrogen. In this environment, neither species can gain energy from the oxidation of lactate without syntrophic cooperation.
Within the first 300 generations of evolution, the cocultures evolved increased stability, higher yields, and higher growth rates, with both species contributing to these changes (6), a trend that continued through 1,000 generations. We describe a common evolutionary outcome of these experiments. Many of the independently evolved D. vulgaris accumulated loss-of-function mutations in genes required for the reduction of sulfate, suggesting strong selection for mutations resulting in loss of the ability to respire sulfate during evolution in syntrophy.
Results
Changes in Growth Rate and Yield of 1,000-Generation–Evolved Cocultures.
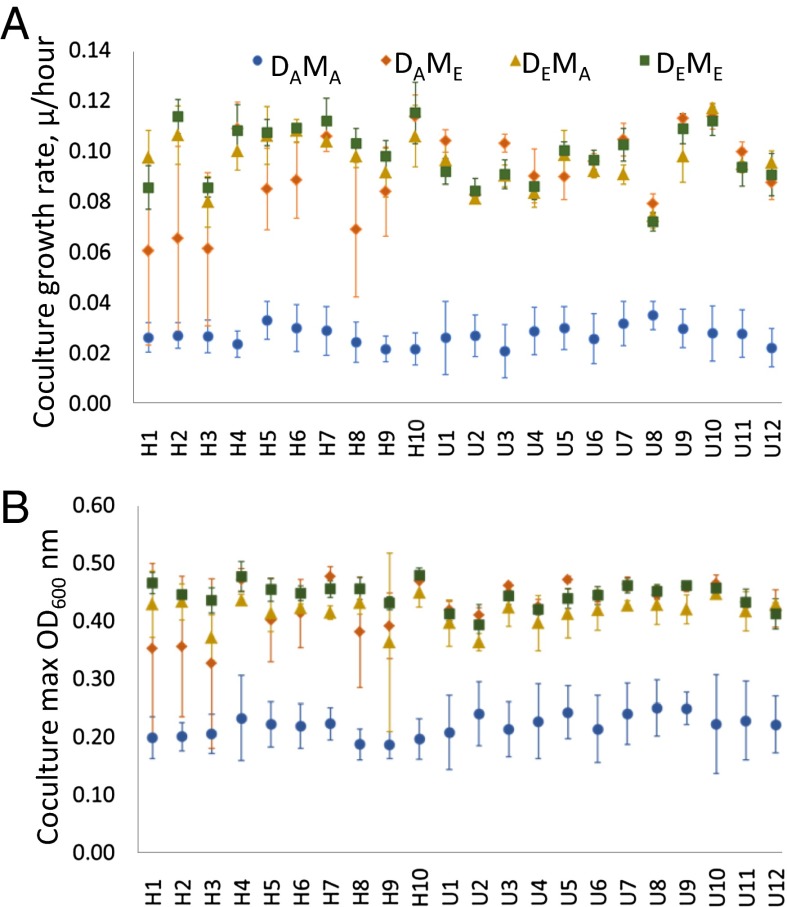
Within the first 300 generations of evolution, both D. vulgaris and M. maripaludis populations acquired mutations allowing them to improve the growth rate and yield of cocultures (6). We tested whether there were further improvements in these populations after 700 additional generations of evolution by pairing D. vulgaris and M. maripaludis populations from 1,000-generation cocultures with their unevolved ancestors. Sulfite was used to isolate the D. vulgaris populations (23) instead of sulfate, which had been used in previous studies (6). The 1,000-generation–evolved cocultures had much faster growth rates and higher yields than the ancestor, whether D. vulgaris only (DEMA), M. maripaludis only (DAME), or both populations (DEME) were evolved (Fig. 1; P < 0.0001 for each of the six comparisons of DAMA with DAME, DEMA, or DEME in each evolution environment). The doubling times of 1,000-generation–evolved cocultures were much faster than those observed previously at 300 generations. The average DEME doubling times at 1,000 generations for all 22 evolved cocultures was 7.0 h (highest, 9.6; lowest, 6.0), whereas at 300 generations, evolved cocultures doubled every 13 h, on average (6). The composition of cocultures (DAMA, DAME, DEMA, or DEME) had a significant effect on growth rate [Table S1; F(3,60) = 348.3, P < 0.0001] and yield [Table S1; F(3,321) = 474.0, P < 0.0001], and the relative impact of evolved D. vulgaris or M. maripaludis on coculture growth differed depending on whether they evolved in the uniform or heterogeneous environment [Table S1; F(3,60) = 7.59, P = 0.0002; and yield, F(3,321) = 7.17, P = 0.0001]. In the uniform environment, there was no difference in growth rate between cocultures when one or both populations were evolved (DAME vs. DEME: P = 0.4272; DEMA vs. DEME: P = 0.6788; DEMA vs. DAME P = 0.2289), but in the heterogeneous environment DAME cocultures grew slower, on average, than DEMA or DEME cocultures (P = 0.0001 for both comparisons) and DEMA cocultures grew similarly to fully evolved cocultures (P = 0.6788). Fully evolved cocultures had the highest yield, on average in the heterogeneous environment (DAME vs. DEME: P = 0.4423; DEMA vs. DEME: P = 0.0236; DEMA vs. DAME: P = 0.0025), but uniform-evolved M. maripaludis could cause higher yield with the ancestor than with a coevolved partner (DAME vs. DEME: P < 0.0001; DEMA vs. DEME: P = 0.0002; DEMA vs. DAME: P = 0.2134).
Fig. 1.
Average growth rate (A) and yield (B) of ancestral cocultures (DAMA; blue dots), cocultures where only D. vulgaris (DEMA; mustard triangles) or M. maripaludis (DAME; orange triangles) was 1,000-generation evolved, and cocultures containing both evolved populations (DEME; green squares). Error bars indicate the SD of four measurements.
Summary of Observed Mutations in Evolved Cocultures.
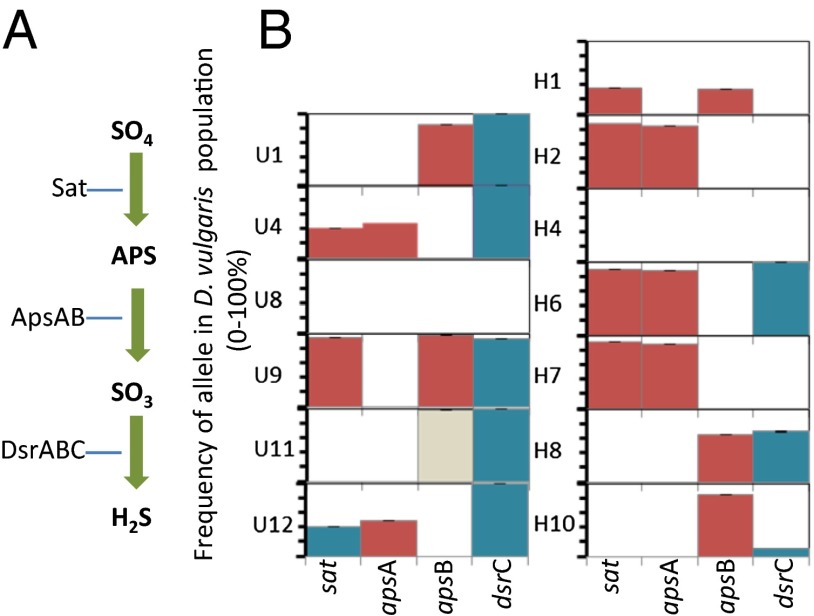
To identify the mutations responsible for these increases in absolute fitness after 1,000 generations of evolution, metagenomic sequences of 13 of the 22 evolved cocultures were determined. One of the most striking results from this sequence data was repeated evolution across populations, of mutations in genes that are required for sulfate reduction, and are hence important for an independent lifestyle in D. vulgaris (Fig. 2). Ten out of the 13 evolved cocultures characterized by resequencing contained D. vulgaris populations with nonsense or frameshift mutations that would effectively inactivate one or more of three genes required for the first two steps of sulfate reduction: (i) activation of sulfate with ATP by the sulfate adenylyl transferase (Sat), and (ii) reduction of the activated sulfate to sulfite by the adenylyl-sulfate reductase (ApsAB; Fig. 2). Fig. 2 shows that most of these loss-of-function mutations were at high frequency in their respective populations (>40%) and some of the evolved populations had loss-of-function mutations in more than one of these three genes.
Fig. 2.
(A) Enzymes in the sulfate reduction pathway. APS is adenosine 5′-phosphosulfate. (B) Occurrence, frequency, and predicted effects of mutations in these genes after 1,000 generations of evolution in syntrophy. Height of bars indicates the frequency of the allele within the population. The predicted effect of the mutation on the amino acid sequence or functionality of the protein is indicated by color. Predicted loss-of-function mutations (red) include both premature stop codons and frameshift mutations. Missense mutations are blue and synonymous mutations are tan.
We then determined whether mutations in these genes were also common in the nine evolved cocultures that were not characterized by genome resequencing. The genes for ApsA, ApsB, and Sat were sequenced by the Sanger method from PCR products. This method is unlikely to detect mutations at low frequency in the population, but high-frequency mutations could be detected. At least four additional evolved cocultures had acquired loss-of-function mutations (Table S2). One of these (U10) had three sulfate respiration (SR)-negative mutations that were nearly identical to those observed in U9, suggesting the possibility of cross-contamination, and is therefore not considered a completely independent evolutionary event. Thus, a total of 13 of 21 independently evolved cocultures accumulated at significant frequency loss-of-function mutations in the pathway of sulfate respiration.
The final step in the pathway for sulfate reduction is the reduction of sulfite to sulfide by the dissimilatory sulfite reductase, composed of three proteins. DsrA and DsrB are the active subunits of this enzyme, and DsrC is thought to play a key role in electron transfer from a transmembrane complex (DsrMKJOP) to the DsrAB (24). Interestingly, no population had any kind of mutation in either the dsrA or dsrB gene. However, 8 of the 13 populations initially sequenced had missense mutations in dsrC, and Sanger sequencing detected mutations in 2 of the 9 remaining lines. No nonsense or frameshift mutations were detected in dsrC (Fig. 2 and Table S2). We also looked for mutations in several other genes thought to be involved in sulfate reduction, including three permeases (DVU0053, DVU0279, DVU1999), PpaC (DVU1636), QmoABCD (DVU0848-0851), DsrMKJOP (DVU1290-1286), and a regulator thought to be involved in sulfate reduction (DVU0916). A missense mutation in the regulator DVU0916 was at 88% frequency in population U8 and 45% of population U4 harbored a missense mutation in the permease DVU1999 (Table S2). No mutations were found in any of the other 13 genes.
To put these data about parallel evolution of the sulfate respiration pathway in perspective, each D. vulgaris population had, on average, about 34 genes (highest, 48; lowest, 15) that harbored nonsynonymous mutations and 5 genes with synonymous mutations (11 highest; 2 lowest) at a frequency of 50% or higher. Some mutations were exactly the same in all or most populations, suggesting they were present in the ancestral population. Apart from this standing genetic variation and the alleles in Fig. 2, parallel evolution of new alleles across six or more populations (frequency of >20% per population) was observed for nine genes having a variety of functions. None of these genes were part of the same metabolic pathway. Thus, the sulfate reduction pathway was the metabolic pathway most commonly targeted by evolution in syntrophy in these experiments, comprising 25% of the 12 genes showing highly parallel evolution (across 6 of the 13 populations sequenced).
These data strongly suggested that most of the D. vulgaris population in several lineages was no longer able to reduce sulfate, but the sequence data also showed that some genotypes with functional sulfate reduction machinery were still present in most of the evolved cocultures. We could readily isolate sulfate-reducing D. vulgaris from every evolved coculture even after 1,300 generations of evolution. Some of these SR-positive subpopulations grew faster and had higher yield than the ancestor on lactate and sulfate at 1,000 generations (Fig. S1). To verify that nonsense mutations in sulfate reduction genes actually result in a SR-negative phenotype, dilution-to-extinction experiments were used to obtain cocultures likely to have only one genotype of each species. Eight evolved cocultures (H2, H4, H6, H8, H10, U9, U8, and U12) that varied in the combination of mutated sulfate reduction genes (including one predicted to have no mutations in these genes) were diluted to extinction using a 10-fold dilution series of stationary-phase cultures (containing ∼5 × 108 cells per mL). The highest dilution showing growth (generally 1 × 107- to 1 × 1010-fold diluted) was diluted again to extinction and the coculture originating from the highest second dilution series was studied. These simplified evolved cocultures were used to verify losses in sulfate reduction by transferring inocula to medium with sulfate and antibiotic to suppress methanogen growth and then monitoring growth for at least 4 wk. We obtained dilution lines that were unable to reduce sulfate from four of the eight cocultures diluted. An additional coculture (H8) had a dilution line that gave mixed results from multiple tests of growth on sulfate.
The presence and genetic linkages of the mutations in the simplified cultures was evaluated by Sanger sequencing of PCR-amplified products (primer sequences in Table S3). The SR-negative phenotypes observed in end-point dilutions (EPDs) from H2, H6, U9, and U12 were correlated with the presence of loss-of-function mutations in genes for sulfate reduction in these same dilution lines. Every EPD-coculture from evolved line H10 had a missense mutation in dsrC, but no mutation in apsB, and these cocultures grew on lactate and sulfate. In populations harboring more than one mutation in genes required for sulfate reduction (U9, U12, H2, H6, H8, H10), all of these mutations were linked on the same genetic background, except in cocultures U12 and H10. This result is consistent with the frequency data (Fig. 2), which shows that, in U9, H2, H6, and H8, both sulfate respiration mutations are over 50% frequency and therefore must be on the same genetic background, but the frequencies in U12 and H10 (DsrC mutation is 10%) are much lower, adding up to 100% or lower.
Comparison of Growth of Evolved and Constructed SR-Positive and SR-Negative Cocultures.
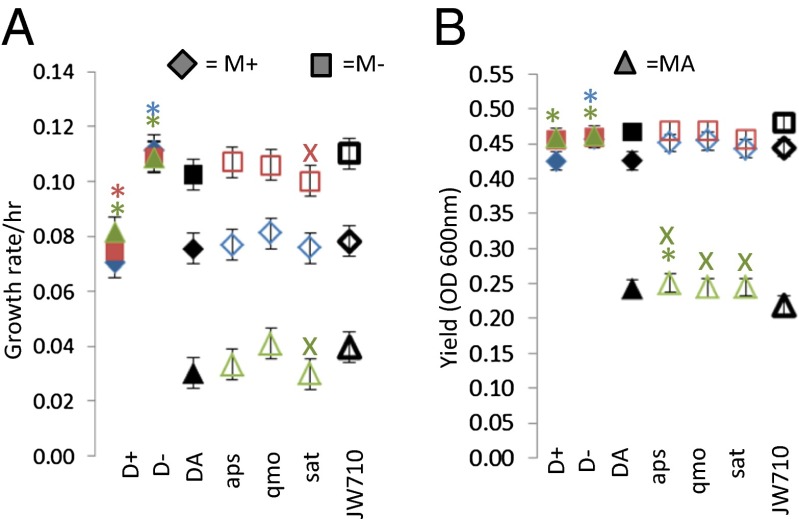
Independent losses of sulfate respiration capability during evolution in syntrophy suggested that loss is linked to a beneficial trait. To test this possibility, D. vulgaris mutants containing markerless deletions in apsA, apsB, sat, and qmoABCD [an operon that is essential for sulfate reduction (25)] were constructed and paired with either ancestral M. maripaludis or M. maripaludis that was obtained from SR-negative (M−) or SR-positive (M+) dilution lines from 1,000-generation–evolved H2. If loss of the trait was beneficial in the evolution environment, we would expect the mutant cocultures to grow faster and have a higher yield compared with pairings containing the strain used for mutant construction (JW710). The composition of cocultures had a significant impact on their growth rate and yield. In an ANOVA (Table S4), the effects of D. vulgaris strain [growth rate, F(6,42) = 77.3; yield, F(6,42) = 84.9], M. maripaludis strain [growth rate, F(2,42) = 548.1; yield, F(2,42) = 1,312.8], or the interaction between these variables [growth rate, F(12,42) = 43.2; yield, F(12,42) = 102.4] were all highly significant (P < 0.0001) . As expected based on the results in Fig. 1, all cocultures containing evolved D. vulgaris or M. maripaludis grew faster and had higher yield than the fully ancestral coculture (Fig. 3). The effects on coculture growth of removing sulfate reduction genes were mixed and mainly apparent in pairings with the ancestral M. maripaludis. Cocultures containing mutants with deletions of sat grew more slowly than their wild-type counterpart with both the ancestral and evolved M. maripaludis (Fig. 3A). The mutant with deleted apsAB also appeared to grow more slowly when paired with the ancestral methanogen, but this difference was not significant after a sequential Bonferroni correction. Despite these slower growth rates, all three deletion mutants paired with the ancestral M. maripaludis obtained a slightly higher yield than the wild-type D. vulgaris. The apsAB and qmoABCD mutants exhibited faster growth and a slightly higher yield when grown in monoculture on sulfite but not thiosulfate (Fig. S2).
Fig. 3.
Growth rate (A) and yield (B) of cocultures of D. vulgaris strains (aps, qmo, sat, JW710, DA, D+, D−) paired with each of three M. maripaludis populations. Results for pairings containing M+ (blue diamonds), M− (red squares), and MA (green triangles) are shown. Asterisks (*) or X (colored according to the M. maripaludis strain) indicate a significant difference (after sequential Bonferroni correction, P < 0.05 per panel for all comparisons against appropriate control) between the mean for that coculture and a control coculture in the full ANOVA model (*) or a submodel (x) containing only data with open symbols. The control coculture for D+ and D− cocultures were DA cocultures (solid symbols). The control coculture for mutants was JW710 (open symbols). M+ and D+ are evolved partners from coculture H2 dilutions that have retained the ability to respire sulfate; M− and D− from H2 dilutions that have lost that capability. MA and DA refer to ancestral strains. Error bars indicate 95% confidence interval.
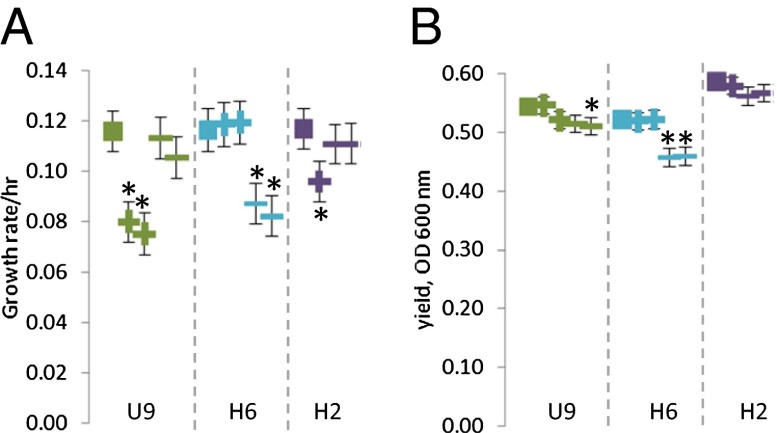
Thus, the putative beneficial effects of SR-negative mutations on fitness in syntrophy likely are dependent on other mutations in the evolved D. vulgaris population, or even on mutations in the evolved M. maripaludis. We then assessed the impact of the native SR-negative mutations in their native environment by comparing the growth of SR-positive EPD cocultures with SR-negative EPD cocultures derived from the same evolved community (Fig. 4). In every case, coculture growth rate, yield, or both, of SR-negative EPD cocultures differed significantly from SR-positive EPD cocultures (contrast; H0: mean SR-positives = mean SR-negatives; for growth rate, P < 0.0001 for U9 and H6, P = 0.0040 for H2; for yield, U9, P = 0.0066, H6, P < 0.0001, H2 = 0.6483; all except the last contrast were significant under an experiment-wide P < 0.05: see Table S5). SR-negative EPD cocultures consistently had lower yield than SR-positive EPD cocultures, although this difference was not always statistically significant. However, in two out of three evolution lines tested (U9 and H2), the SR-negative cocultures grew significantly faster than the SR-positive cocultures. In evolved line H6, the SR-positive cocultures grew faster than the SR-negative cocultures.
Fig. 4.
Comparison of cocultures with sulfate reduction (SR)-negative and SR-positive phenotypes. Each point is the average growth rate (A) or yield (B) of three replicates of an evolved coculture (solid squares) or an EPD coculture that was derived from evolved coculture U9 (green), H6 (aqua), or H2 (purple) and had either a SR-positive (+ symbol) or SR-negative (− symbol) phenotype. Error bars indicate 95% confidence intervals calculated from the ANOVA (see SI Methods and Table S5), and asterisks (*) indicate a significant difference between the EPD and the evolved coculture from which it was derived after a sequential Bonferroni correction (P < 0.05 for all 11 tests in the panel).
Discussion
There are numerous examples of species that have evolved to be specialized for mutualistic interactions, often to the detriment of their ability to survive independently (1, 2). However, there are few, if any, empirical observations of this happening in real time. Here, we show that, within the first 1,000 generations of adaptation to a new mutualistic interaction, D. vulgaris populations frequently became dominated by mutants that had lost their capacity to grow independently by reducing sulfate, severely restricting the conditions under which they could hypothetically survive outside of their interaction with the methanogen. This observation was based on finding repeated evolution of loss-of-function mutations in genes required for sulfate reduction, and by confirming the phenotypic effect of these mutations in evolved subpopulations. In two out of the three evolved communities tested, subcommunities populated by SR-negative genotypes grew substantially faster than those with SR-positive genotypes. Cocultures composed of D. vulgaris mutants with markerless deletions of the sat, apsAB, or qmoABC genes had slightly higher yield compared with their wild-type progenitor when paired with the ancestral M. maripaludis, but had similar or slower growth rates.
In population genetics, there are two general explanations for the loss of an unused trait during evolution. It can be lost by natural selection because of a genetically based trade-off where the same gene affects both the unused trait and another trait that is beneficial in the evolution environment. Mutations that improve the beneficial trait simultaneously cripple the unused trait, causing it to be lost because of natural selection on the genetically linked beneficial trait (26, 27). Such trade-offs explain (for example) losses in the ability to metabolize single-carbon compounds in Methylobacterium after extended evolution with succinate (28). An alternative explanation for loss of an unused trait is that it occurs by the chance accumulation of mutations in genes required for its proper functioning (27, 29). Mutations in the trait are neutral with respect to fitness and hence accumulate as a result of neutral processes, as observed in insect endosymbionts (30) and in losses in Bacillus spore production (31). Because there is no selection to increase or decrease the frequency of these mutants in a population, the timing of loss of the unused trait is determined by the mutation rate and population size (29, 31, 32). Thus, in a large population, it may take thousands of generations for any particular neutral mutation to become fixed after its emergence. In contrast, genetic trade-offs will cause a nonrandom loss of the trait that may repeatedly occur early in the process of adaptation to a new environment (27, 29).
The metagenome sequencing data reported here are most consistent with the hypothesis that sulfate respiration capability was lost due to natural selection. First, evolutionary losses occurred early, within less than 1,000 generations, and were highly parallel; at least 13 of 21 sequenced, independently evolved communities acquired nonsense or frameshift mutations in one or more of three genes required for the reduction of sulfate to sulfite: sat, apsA, or apsB. Moreover, in 7 of the 10 populations for which such data are available, the loss-of-function allele comprised at least 50% of the D. vulgaris population (Fig. 2). The probability of fixation of a strictly neutral mutation u, is 1/(2N), where N is the population size (32). In our experiments, this means that, if one of these sulfate reduction mutations is neutral, its probability of fixation is 1 in ∼2 × 1010 (∼5 × 108 cells per mL times 20 mL). We observed at least 19 mutations (based solely on the metagenomic data in Fig. 2) in four genes change in frequency from 1 in ∼1 × 1010 to a frequency of 1/2 or higher within 1,000 generations. The probability of observing these changes if each of these mutations is neutral and each evolutionary change is independent, is 1 in (1 × 1010)19 (mutant population size is 0.5 of total). Alternatively, these mutations could have hitchhiked repeatedly with beneficial alleles, but, unless the mutation rate in these genes is exceptional, it is unlikely that this occurs so often in these particular genes and not others (33).
Although the metagenomic data suggest a fitness benefit to losing sulfate reduction ability in the syntrophy environment, the nature of that benefit is unclear. Walker et al. (34) observed that sulfate reduction genes were highly expressed even during syntrophic growth in the absence of sulfate, suggesting that some components of the sulfate respiration pathway, such as the DsrABC system of electron transfer, are abundant during syntrophic association. Retention of all pathway components may somehow interfere with the capacity for energy acquisition through syntrophy. If so, then deleting these genes in wild-type D. vulgaris should increase absolute fitness (population growth rate and yield in absence of competitors) in syntrophy. However, in the experiments with constructed mutants reported here, deletion of genes for the Sat and the ApsAB caused similar or slower coculture growth and small but significant increases in yield with the ancestral M. maripaludis. These results are consistent with the possibility that functional ApsAB, QmoABCD, and Sat genes may impose a cost on the amount of cellular material that can be produced from a finite lactate concentration in syntrophy. However, because growth of the cocultures was slower, more experiments are necessary to determine why and how SR-negative genotypes were selected in an environment where lactate was regularly supplied in abundance.
There are several alternative explanations for repeated selection of SR-negative alleles. In the evolution experiment, the sat and apsAB genes were not deleted, but many of the nonsense and frameshift mutations occurred near the beginning of the gene. It is possible that the fitness benefit requires a piece of the nonfunctional protein, and thus the full effect was not observed with the constructed deletion mutants. Alternatively, the loss of functional sulfate reduction pathway proteins may affect competitive ability in the presence of SR-positive genotypes, with smaller effects on absolute fitness. Another possibility is that the genetic context (either in the same D. vulgaris genome, or other D. vulgaris and M. maripaludis genomes) in which these loss-of-function mutations occur affects their contribution to fitness. In support of this possibility that other mutations in the same D. vulgaris genome affect SR-negative fitness, higher absolute fitness of SR-negative compared with SR-positive evolved subpopulations was observed for two out of three evolved communities that were tested. Other data suggested that the methanogen genotype affects SR-negative fitness. The growth rate of cocultures with the SR-negative D. vulgaris subpopulation of H2 varied considerably when paired with a M. maripaludis from the same subcommunity vs. the M. maripaludis from a SR-positive subcommunity (Fig. 3). Finally, SR-positive genotypes could readily be recovered from all evolved cocultures through at least 1,300 generations of evolution. This persistence of SR-positive D. vulgaris may indicate a frequency-dependent interaction between SR-negative and SR-positive D. vulgaris in the same coculture.
Although our results point to a complex evolutionary interplay between species during adaptive evolution, this model system now provides an experimental system to develop a fundamental mechanistic understanding of how syntrophies and other mutualisms evolve. In particular, the data address the evolutionary origins of facultative vs. obligate syntrophy in anaerobic food webs. There are several well-studied species that are considered specialized for syntrophy because they have little or no capacity for respiration (10, 12, 13). Most of these specialized syntrophs (and other facultative syntrophs) share common ancestry with sulfate-reducing bacteria (10), and some retain genes associated with sulfate respiration (11). These observations suggest a propensity for sulfate-reducing bacteria toward syntrophy, and that sometimes extended evolution in syntrophic association results in the loss of sulfate respiration ability. Here, we demonstrate the plausibility of this hypothesis by directly observing the loss of sulfate respiration capacity in model syntrophic communities that were evolving to become faster growing and more productive associations. Moreover, our observations show that SR loss can occur due to natural selection, an insight that may be difficult to obtain from natural populations alone because of uncertainty in the timing of the loss and the number of times loss did not occur. This result helps us understand the evolution of syntrophy in the natural world and also deepens our understanding of the limits of metabolic flexibility in sulfate-reducing bacteria (17).
More broadly, our results provide a rare empirical example of populations becoming more dependent on a mutualistic interaction during their adaptation to conditions requiring their cooperation. Respiration of sulfate is a key metabolic feature of Desulfovibrio and other sulfate-reducing genera that allows them to thrive in diverse environments (15, 17). Although D. vulgaris can still grow by respiring sulfite or thiosulfate, its options for growth outside syntrophy would likely be limited in many environments. This increased dependence on another species for survival is an important possible step in the process of adaptation to mutualism after the origin of the beneficial association. Species that depend on their mutualistic partner for survival are less likely to abandon the association (3, 35) and are also more vulnerable to the ecological conditions that threaten the abundance of their partner (36). Although the evolutionary history of some obligate mutualisms has been studied extensively (1, 2), the physiological, ecological, and genetic factors that affect their evolution (vs. facultative associations) are not yet well understood (3, 35, 37). We anticipate that this model laboratory system, by providing real-time observation of evolutionary processes leading to increased dependence on a mutualistic interaction, will help us to understand more generally how mutualisms evolve (37).
Methods
Strains and Culture Conditions.
The strains, culture conditions, and measurements of growth rate and yield of evolved cocultures were the same as described previously (6). Twenty-four cultures containing D. vulgaris Hildenborough clone D1 or D2 and M. maripaludis S2 clone M1 or M2 were propagated for 1,000 generations, by 152 weekly 100-fold dilutions into coculture medium A (CCMA) in Balch tubes that were either incubated upright without shaking, or in a horizontal position with constant shaking at 300 rpm (SI Methods).
Measurement of Growth Rate and Yield of 1,000-Generation Cocultures.
Freezer stocks of the ancestral cocultures and 1,000-generation–evolved cocultures were each inoculated into Balch tubes containing CCMA. D. vulgaris and M. maripaludis populations in each stationary-phase coculture were separated by transferring the coculture to one tube of medium selective for D. vulgaris (CCMA with 5 mM sulfite and 5 μg/mL puromycin to exclude M. maripaludis), and another tube selective for M. maripaludis (CCMA without lactate and 1 mg/mL spectinomycin to select against D. vulgaris). The resulting separated D. vulgaris and M. maripaludis populations were mixed in the following combinations in coculture: DAMA, DAME, DEMA, and DEME (A denotes ancestral, whereas E indicates the strain was evolved). Three days after the populations reached stationary phase, 1% of the coculture volume was transferred into fresh CCMA tubes, in duplicate, and the OD600nm was recorded every 3–6 h until the cultures reached stationary phase, as determined by at least three consecutive measurements of equivalent or declining OD. This growth experiment was repeated twice, with two replicates each time, yielding four independent measurements of growth rate and yield for each coculture pairing. Coculture growth rate was the slope of the linear portion of the curve of ln (OD600nm) vs. time, and yield was the maximum OD attained by the coculture.
Sequencing and Identification of Mutations in Evolved Cocultures.
Metagenomic sequences of 13 of the 22 evolved cocultures were determined using the Illumina platform. DNA was extracted with the Epicentre Masterpure kit. Illumina libraries were prepared with the Nextera DNA library preparation kit (Illumina) according to the protocol of the manufacturer. Sample libraries for sequencing were prepared according to the MiSeq Reagent Kit Preparation Guide (Illumina). The resulting raw Illumina sequences were aligned to the reference D. vulgaris (NC_002937, NC_005863) and M. maripaludis (NC_005791) published genomes using the breseq pipeline (38). See SI Methods for more details about the sequencing methods and alignment.
Construction of Markerless Deletion Mutants.
The sat (DVU1295) deletion mutant (JW9271; see Table S6 for strains and plasmids used in this study) was constructed at the University of Missouri in a similar manner as described previously (39, 40). A detailed description of mutant construction can be found in SI Methods. In short, two plasmids were used for construction: pMO9268 (the “marker-exchange plasmid”) and pMO9270 (the “markerless deletion plasmid”). Transformed cells were cultured in MOYLS3 medium (25). One of the marker-exchange isolates (JW9269) was electroporated with pMO9270, recovered, and plated in the presence of 5-fluorouracil, which selects for cells that no longer have the marker. Strains were confirmed by Southern blot verification hybridization. Construction of the apsBA (DVU0846-7) deletion mutant (JW9259) and the qmoABC (DVU0848-50) deletion mutant (JW9263) was accomplished in a similar manner to the sat deletion mutant. The plasmids pMO9256 and pMO9260 (marker-exchange apsBA and qmoABC, respectively), pMO9258 and pMO9262 (markerless-deletion apsBA and qmoABC, respectively), and intermediate strains JW9257 and JW9261 (marker-exchange apsBA and qmoABC, respectively) were also made.
All PCR products were amplified (see Table S2 for primers) with Herculase II (Life Technologies) and assembled together by sequence and ligation-independent cloning (41) using cells supplied by Bioline. Putative constructs were sequenced (DNA Core, University of Missouri) to confirm the correct sequences for regions necessary for homologous recombination.
Mutant Experiment Methods.
Freezer stocks of the following cultures were inoculated into Balch tubes containing the appropriate medium and allowed to grow to stationary phase: apsAB, qmoABC, sat deletion mutants and strain JW710, which was used to make the deletion mutants; two EPDs of evolved coculture line H2 after 1,000 generations; the D. vulgaris ancestor for the H2 line; and the M. maripaludis ancestor for the H2 line. Once in stationary phase, cocultures were separated into their constituent partners, as described above. One EPD coculture was SR-negative (its D. vulgaris and M. maripaludis populations are denoted D− or M−), whereas the other was SR-positive (D+ or M+). The following combinations of cocultures were made: DmutMA, DmutM−, DmutM+, DA MA, DE−MA, D+MA, D−M−, D+M−, D−M+, and D+M+. Dmut referred to any of the deletion mutants or background strain JW710. MA or DA refer to the ancestral D. vulgaris or M. maripaludis populations. Once the coculture combinations reached stationary phase, they were transferred into three tubes of fresh CCMA, and their OD at 600 nm was recorded every few hours until they once again reached stationary phase. Coculture growth rate and yield were calculated as above.
Measurement of Growth Rate and Yield of SR-Positive and SR-Negative Cocultures.
Triplicate growth experiments were set up in 20-mL CCMA Balch tubes from glycerol freezer stocks for three EPDs from H2 and four EPDs from H6 and U9 lines, as well as the evolved and ancestral cocultures from which they were derived. These lines were transferred twice after growing up to stationary phase to allow for stable growth, and then their optical density was measured at 600 nm every 2 h.
Supplementary Material
Acknowledgments
We thank Marc Vrana for assistance in growth experiments. This material is based upon work supported by the National Science Foundation (NSF) under Grant 1257525 (awarded to K.L.H.), NSF-1262637 and NSF-1330912 (to N.S.B.), and by the Office of the Vice President for Research at the University of Oklahoma, and the Collaborative Innovation Center for Regional Environmental Quality. Research conducted as part of Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA) was supported by the Office of Science, Office of Biological Research, of the US Department of Energy under Contract DE-AC02-05CH1123.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All genome sequence data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (BioProject ID PRJNA248017; Title: “Distribution of mutations in evolved syntrophic cocultures of sulfate-reducing bacteria Desulfovibrio vulgaris Hildenborough and methanogenic archaea Methanococcus maripaludis S2”).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407986111/-/DCSupplemental.
References
- 1.Pellmyr O, Thompson JN, Brown JM, Harrison RG. Evolution of pollination and mutualism in the yucca moth lineage. Am Nat. 1996;148(5):827–847. [Google Scholar]
- 2.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 3.Sachs JL, Simms EL. Pathways to mutualism breakdown. Trends Ecol Evol. 2006;21(10):585–592. doi: 10.1016/j.tree.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 4.Losos JB. Seeing the forest for the trees: The limitations of phylogenies in comparative biology. (American Society of Naturalists Address) Am Nat. 2011;177(6):709–727. doi: 10.1086/660020. [DOI] [PubMed] [Google Scholar]
- 5.Shou W, Ram S, Vilar JMG. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci USA. 2007;104(6):1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci USA. 2010;107(5):2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harcombe W. Novel cooperation experimentally evolved between species. Evolution. 2010;64(7):2166–2172. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 8.Summers ZM, et al. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science. 2010;330(6009):1413–1415. doi: 10.1126/science.1196526. [DOI] [PubMed] [Google Scholar]
- 9.Schink B. Syntrophic associations in methanogenic degradation. In: Overmann J, editor. Molecular Basis of Symbiosis. Vol 41. Springer; Berlin: 2006. pp. 1–19. [DOI] [PubMed] [Google Scholar]
- 10.Sieber JR, McInerney MJ, Gunsalus RP. Genomic insights into syntrophy: The paradigm for anaerobic metabolic cooperation. Annu Rev Microbiol. 2012;66:429–452. doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 11.Imachi H, et al. Non-sulfate-reducing, syntrophic bacteria affiliated with desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl Environ Microbiol. 2006;72(3):2080–2091. doi: 10.1128/AEM.72.3.2080-2091.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosaka T, et al. The genome of Pelotomaculum thermopropionicum reveals niche-associated evolution in anaerobic microbiota. Genome Res. 2008;18(3):442–448. doi: 10.1101/gr.7136508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McInerney MJ, et al. The genome of Syntrophus aciditrophicus: Life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci USA. 2007;104(18):7600–7605. doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McInerney MJ, et al. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann N Y Acad Sci. 2008;1125:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, et al. How sulphate-reducing microorganisms cope with stress: Lessons from systems biology. Nat Rev Microbiol. 2011;9(6):452–466. doi: 10.1038/nrmicro2575. [DOI] [PubMed] [Google Scholar]
- 16.Bryant MP, Campbell LL, Reddy CA, Crabill MR. Growth of Desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl Environ Microbiol. 1977;33(5):1162–1169. doi: 10.1128/aem.33.5.1162-1169.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plugge CM, Zhang W, Scholten JC, Stams AJ. Metabolic flexibility of sulfate-reducing bacteria. Front Microbiol. 2011;2:81. doi: 10.3389/fmicb.2011.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steger D, et al. Microorganisms with novel dissimilatory (bi)sulfite reductase genes are widespread and part of the core microbiota in low-sulfate peatlands. Appl Environ Microbiol. 2011;77(4):1231–1242. doi: 10.1128/AEM.01352-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raskin L, Rittmann BE, Stahl DA. Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl Environ Microbiol. 1996;62(10):3847–3857. doi: 10.1128/aem.62.10.3847-3857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muyzer G, Stams AJM. The ecology and biotechnology of sulphate-reducing bacteria. Nat Rev Microbiol. 2008;6(6):441–454. doi: 10.1038/nrmicro1892. [DOI] [PubMed] [Google Scholar]
- 21.Mussmann M, et al. Clustered genes related to sulfate respiration in uncultured prokaryotes support the theory of their concomitant horizontal transfer. J Bacteriol. 2005;187(20):7126–7137. doi: 10.1128/JB.187.20.7126-7137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein M, et al. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J Bacteriol. 2001;183(20):6028–6035. doi: 10.1128/JB.183.20.6028-6035.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim SS, Stolyar SS, Hillesland KL. Culturing anaerobes to use as a model system for studying the evolution of syntrophic mutualism. In: Sun L, Shou W, editors. Engineeering and Analyzing Multicellular Systems: Methods and Protocols, Methods in Molecular Biology. Vol 1151. Springer; New York: 2014. pp. 103–115. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira TF, et al. The crystal structure of Desulfovibrio vulgaris dissimilatory sulfite reductase bound to DsrC provides novel insights into the mechanism of sulfate respiration. J Biol Chem. 2008;283(49):34141–34149. doi: 10.1074/jbc.M805643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zane GM, Yen HC, Wall JD. Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl Environ Microbiol. 2010;76(16):5500–5509. doi: 10.1128/AEM.00691-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stearns SC. Trade-offs in life-history evolution. Funct Ecol. 1989;3(3):259–268. [Google Scholar]
- 27.Fong DW, Kane TC, Culver DC. Vestigialization and loss of nonfunctional characters. Ann Rev Ecol Syst. 1995;26:249–268. [Google Scholar]
- 28.Lee M-C, Chou H-H, Marx CJ. Asymmetric, bimodal trade-offs during adaptation of Methylobacterium to distinct growth substrates. Evolution. 2009;63(11):2816–2830. doi: 10.1111/j.1558-5646.2009.00757.x. [DOI] [PubMed] [Google Scholar]
- 29.Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407(6805):736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- 30.Moran NA. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr Opin Microbiol. 2003;6(5):512–518. doi: 10.1016/j.mib.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Maughan H, Masel J, Birky CW, Jr, Nicholson WL. The roles of mutation accumulation and selection in loss of sporulation in experimental populations of Bacillus subtilis. Genetics. 2007;177(2):937–948. doi: 10.1534/genetics.107.075663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; New York: 1983. [Google Scholar]
- 33.Cooper VS, Schneider D, Blot M, Lenski RE. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J Bacteriol. 2001;183(9):2834–2841. doi: 10.1128/JB.183.9.2834-2841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker CB, et al. The electron transfer system of syntrophically grown Desulfovibrio vulgaris. J Bacteriol. 2009;191(18):5793–5801. doi: 10.1128/JB.00356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mondo SJ, Toomer KH, Morton JB, Lekberg Y, Pawlowska TE. Evolutionary stability in a 400-million-year-old heritable facultative mutualism. Evolution. 2012;66(8):2564–2576. doi: 10.1111/j.1558-5646.2012.01611.x. [DOI] [PubMed] [Google Scholar]
- 36.Wernegreen JJ. Mutualism meltdown in insects: Bacteria constrain thermal adaptation. Curr Opin Microbiol. 2012;15(3):255–262. doi: 10.1016/j.mib.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461(7268):1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 39.Keller KL, Bender KS, Wall JD. Development of a markerless genetic exchange system for Desulfovibrio vulgaris Hildenborough and its use in generating a strain with increased transformation efficiency. Appl Environ Microbiol. 2009;75(24):7682–7691. doi: 10.1128/AEM.01839-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks JM, et al. The genetic basis for bacterial mercury methylation. Science. 2013;339(6125):1332–1335. doi: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 41.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4(3):251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.