Significance
Triple negative breast cancers (TNBCs) have a poor prognosis (5-y survival of 74.5%) among all breast cancer patients (5-y survival of greater than 95%) because of the aggressiveness of the disease and the lack of targeted therapeutics. We show that intercellular adhesion molecule-1 (ICAM-1) is differentially expressed in human TNBC tumor tissues by immunohistochemistry and in human TNBC cell lines via quantification of gene and protein expression. Iron oxide nanoparticles functionalized with ICAM-1 antibody (ICAM-IONP) were synthesized as MRI probes. An in vivo signal enhancement of 2.6-fold for ICAM-IONPs was measured relative to controls, demonstrating that ICAM-1 is a potential diagnostic and therapeutic target for TNBC treatment.
Abstract
Triple negative breast cancers (TNBCs) have a high mortality rate owing to aggressive proliferation and metastasis and a lack of effective therapeutic options. Herein, we describe the overexpression of intercellular adhesion molecule-1 (ICAM-1) in human TNBC cell lines and tissues, and demonstrate that ICAM-1 is a potential molecular target and biomarker for TNBC therapy and diagnosis. We synthesized ICAM-1 antibody-conjugated iron oxide nanoparticles (ICAM-IONPs) as a magnetic resonance imaging (MRI) probe to evaluate tumor targeting. Quantitative analysis of ICAM-1 surface expression predicted the targeting capability of ICAM-IONPs to TNBC cells. MRI of the TNBC xenograft tumor after systemic administration of ICAM-IONPs, coupled with iron quantification and histology, demonstrated a significant and sustained MRI contrast enhancement and probe accumulation in tumors with ICAM-1 overexpression relative to control. Identification of ICAM-1 as a TNBC target and biomarker may lead to the development of a new strategy and platform for addressing a critical gap in TNBC patient care.
Triple negative breast cancers (TNBCs) comprise a heterogeneous group of tumors with a diverse histology and genetic makeup that share the common feature of low expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) (1). TNBCs represent 15–20% of all breast cancers, occurring more frequently in women under 50 y of age, African American women, and individuals carrying the breast cancer early onset 1 (BRCA1) gene.
TNBC patients do not benefit from hormone or HER2-targeted therapies, leaving chemotherapy as a limited treatment option (1). As a result, the prognosis for TNBC patients remains poor. The 5-y survival rate of patients with TNBC is less than 74.5%, compared with 87% for patients with HER2-positive breast cancer and greater than 90% for patients with ER-positive breast cancer (2–4). Although targeted therapy using the overexpression of a specific cancer cell membrane molecule can facilitate the spatial and temporal delivery of therapeutics, the bottleneck of TNBC-targeted therapy is lacking TNBC-specific entities that can discriminate between TNBC cells and nonneoplastic cells. Several cell membrane molecules have been reported as molecular markers for TNBC, such as epidermal growth factor receptor (EGFR) and mast/stem cell growth factor receptor (c-KIT or CD117) (5–7). TNBC-targeted therapeutics, including cetuximab (anti-EGFR monoclonal antibody) (8), imatinib (c-KIT tyrosine kinase inhibitor) (9), and poly(ADP ribose) polymerase (PARP) inhibitor also are undergoing preclinical/clinical investigations (10). However, to date, trials of these targeted therapeutics have failed to demonstrate enough clinical efficacy to meet the statistical criteria (11–13). Therefore, an urgent and unmet need remains to identify new and effective molecular targets and associated therapies in TNBC.
To identify a TNBC target, we screened cell surface molecules involved in signal transmission, followed with validation using in vivo targeted imaging. Malignant cells can usurp the functions of cell surface receptors to survive and proliferate, elude the immune system, expand the blood supply, colonize tissues, and spread to other organs, making them natural candidates as therapeutic and diagnostic targets for TNBC (14, 15).
Results
Screen and Identify Intercellular Adhesion Molecule-1 as a TNBC Target and Biomarker.
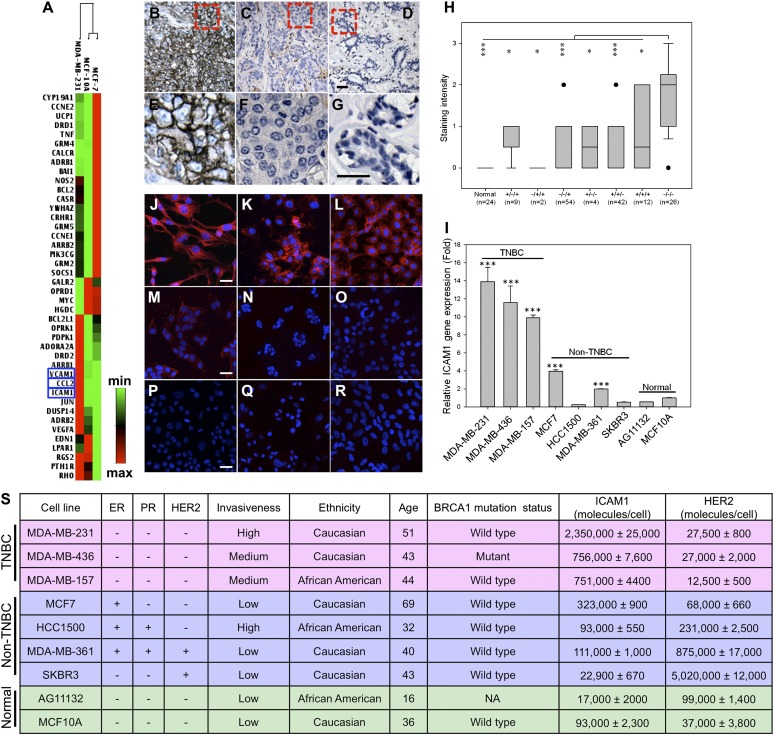
We used a real-time PCR array to obtain the expression profile of 84 genes involved in signal transduction pathways, including bioactive lipid receptors, metabotropic glutamate receptors, and proteins in the calcium signaling pathway, in three cell lines: MDA-MB-231 (TNBC), MCF7 (non-TNBC, ER+/PR−/HER2−), and MCF10A (a nonneoplastic, human mammary epithelial cell line) (as shown in Fig. S1 and Table S1). Fig. 1A demonstrates a collection of 42 differentially expressed genes in MDA-MB-231 cells compared with MCF7 and MCF10A. We narrowed the candidates down to three genes: chemokine (C-C motif) ligand 2 (CCL2), vascular cell adhesion molecule-1 (VCAM-1), and intercellular adhesion molecule-1 (ICAM-1, also called CD54) based on up-regulated levels relative to MCF7 and MCF10A cells and expression on cell membranes. CCL2 is anchored to glycosaminoglycan side chains of proteoglycans on endothelial cells and secreted by monocytes, macrophages, and dendritic cells (16–18). ICAM-1 and VCAM-1 are well-recognized biomarkers for inflammation. Unlike VCAM-1, ICAM-1 is expressed at low levels in normal tissues except the tonsil, adrenal gland, and spleen (19). ICAM-1 is present on endothelial cells at levels lower than on TNBC cells (Table S2) (19). Because of its specificity, ICAM-1 was chosen for further evaluation as a target for TNBC.
Fig. 1.
Identification of ICAM-1 as a TNBC target and biomarker. (A) Gene expression analysis of human breast cancer cell lines. Red and green represent maximum and minimum gene expression, respectively. Representative microscopic images of human TNBC tissues (B and E), non-TNBC tissues (C and F; ER+/PR+/HER2−), and normal breast epithelium (D and G) stained with an anti-human ICAM-1 antibody. (The dashed box illustrates the area subjected to increased magnification in the lower panel; scale bars represent 50 µm.) (H) Quantification of ICAM-1 staining intensities in different subtypes of breast cancer [status of ER/PR/HER2: +/−/+; −/+/+; −/−/+; +/−/−; +/+/−; +/+/+; and −/−/− (TNBC)] and normal breast tissue. Data are presented as a box-and-whisker plot. *P < 0.05; **P < 0.01; ***P < 0.001 compared with TNBC tissues. (I) ICAM-1 gene expression in TNBC, non-TNBC, and normal cells quantified by qRT-PCR. ICAM-1 fold change is relative to GAPDH (***P < 0.001). (J–R) Representative fluorescence microscope images of ICAM-1 immunofluorescent staining in MDA-MB-231 (J), MDA-MB-436 (K), MDA-MB-157 (L), MCF7 (M), HCC1500 (N), MDA-MB-361 (O), SKBR3 (P), AG11132 (Q), and MCF10A (R). DAPI was used to stain the cell nuclei; mouse anti-human ICAM-1 antibody (primary) and NL557-conjugated goat anti-mouse antibody (secondary) were used to stain ICAM-1. Scale bars represent 20 µm. (S) Collection of human TNBC, non-TNBC, and normal cell lines with their ICAM-1 and HER2 surface protein densities measured by flow cytometry.
Although its role as a TNBC target and biomarker has yet to be fully investigated, ICAM-1 expression has been shown to associate with aggressive tumor phenotypes in breast cancer, prostate cancer, and myeloma (20, 21). Evidence suggests that ICAM-1 triggers multiple cell-signaling pathways that promote cancer cell proliferation, migration, resistance to apoptosis, and development of cell adhesion molecule-induced drug resistance (20, 21).
To validate that ICAM-1 is highly overexpressed in TNBC tumors, immunohistochemistry was conducted by using 149 human breast tumor tissues representing different ER/PR/HER2 status along with 144 human normal tissues of 20 different organs. ICAM-1 staining in TNBC tissues was stronger and present in more cells (Fig. 1 B and E) compared with non-TNBC tissues (Fig. 1 C and F) and normal breast mammary epithelium (Fig. 1 D and G). TNBC exhibited a significant increase in ICAM-1 expression compared with various other subtypes of breast cancers and normal epithelium (Fig. 1H). ICAM-1 expression is absent in normal human breast, cerebrum, colon, esophagus, kidney, liver, ovary, pancreas, prostate, rectum, skin, small intestine, and uterine cervix, but positive ICAM-1 staining was observed in normal spleen, lung, lymph node, thymus, testis, and bone (Fig. S2). These findings correlate with the Human Protein Atlas database (www.proteinatlas.org), revealing that the ICAM-1 expression in normal organs is substantially less than that of EGFR, another generally accepted TNBC target. The finding that ICAM-1 is overexpressed in 26 human TNBC tissues provides clinical evidence supporting ICAM-1 as a potential molecular target for TNBC.
We further characterized and quantified ICAM-1 gene and surface protein expression in nine different human cell lines: three TNBC, four non-TNBC, and two nonneoplastic lines (Fig. 1S). Because TNBCs are more prevalent in women under 50 y of age, African American women, and individuals carrying the BRCA1 gene mutation (1), we analyzed ICAM-1 levels in seven breast cancer cell lines—derived from patients of African American and Caucasian origin, of ages spanning 32 to 69 y, and with wild-type and mutant BRCA1 gene status—relative to nonneoplastic, human mammary epithelial cells MCF10A and AG11132. As shown in Fig. 1I, TNBC cells MDA-MB-231, MDA-MB-436, and MDA-MB-157 exhibited 13.9-, 11.6-, and 9.9-fold higher ICAM-1 gene expression, respectively, than MCF10A (even higher fold relative to AG11132). In non-TNBC cells, MCF7 and MDA-MB-361 showed elevated ICAM-1 gene expression relative to nonneoplastic cells, but at markedly lower levels than TNBC cells.
Consistent with ICAM-1 gene expression levels, TNBC cells exhibited between 8- and 25-fold higher ICAM-1 surface protein levels than non-TNBCs and normal cells (Fig. 1S). Compared with the surface protein density of HER2, a clinical target and biomarker for breast cancer, the ICAM-1 surface protein density (751,000–2,350,000 molecules/cell) on TNBC cells was comparable to the HER2 surface density (875,000– 5,020,000 molecules/cell) measured on HER2-positive breast cancer cells (MDA-MB-361 and SKBR3). Immunofluorescent staining of ICAM-1 overexpression in TNBC cells revealed greater ICAM-1 surface staining on TNBCs (Fig. 1 J–L) relative to non-TNBCs (Fig. 1 M–P) and nonneoplastic cells (Fig. 1 Q and R).
Furthermore, ICAM-1 was localized largely on the TNBC cell membranes, suggesting that it might be recognized and bound by targeted therapeutic agents. Based on the above results, we concluded that ICAM-1 expression is not ubiquitous in all breast cancers. ICAM-1 is overexpressed on the surface of TNBC cells and thus may be used as a target and biomarker for TNBC-targeted therapy.
Characterizations of TNBC-Targeted Iron Oxide Nanoparticles.
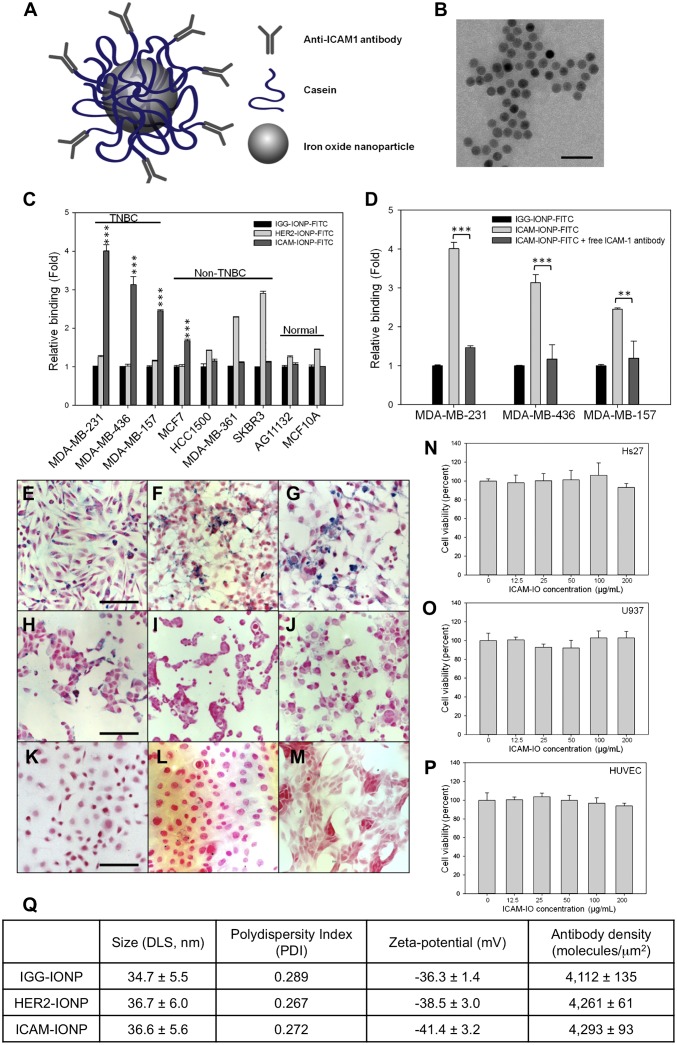
To test ICAM-1 targeting for TNBC, we synthesized a TNBC-targeted MRI probe using magnetic iron oxide nanoparticles (IONPs) conjugated with ICAM-1 antibodies (ICAM-IONPs). ICAM-1 antibodies were conjugated covalently to casein-coated IONPs via 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)/N-hydroxysuccinimide (NHS) chemistry (see schematic illustration in Fig. 2A) (22). Similarly, casein-coated IONPs were conjugated with Herceptin (humanized anti-HER2 antibody; HER2-IONPs) or nonspecific IgG (IGG-IONPs) as controls for evaluation of targeting specificity, efficiency, and delivery. The morphology and monodispersity of ICAM-IONPs were examined by transmission electron microscopy (TEM; Fig. 2B) and dynamic light scattering (DLS; Fig. 2Q). ICAM-IONPs, with a core diameter of 15 nm, had a mean hydrodynamic radius of 36.6 ± 5.6 nm and a zeta potential of −41.4 ± 3.2 mV. The antibody density for ICAM-IONP, IGG-IONP, and HER2-IONP was determined experimentally using FITC-labeled antibodies as ligands (Fig. 2Q). Approximately two or three antibody molecules were conjugated to each particle. The ICAM-IONPs obtained showed no cytotoxicity (Fig. 2 N–P).
Fig. 2.
Synthesis of ICAM-IONP and its in vitro targeting of TNBC cell lines. (A) Schematic illustration of the structure of ICAM-IONP as an MRI probe for in vivo TNBC targeting and imaging. (B) TEM image of as-synthesized ICAM-IONPs. Scale bar represents 50 nm. (C) Cellular binding of IGG-IONP, HER2-IONP, and ICAM-IONP in TNBC, non-TNBC, and normal cell lines characterized via flow cytometry. ***P < 0.001 versus resting. (D) Competition of free ICAM-1 antibody and ICAM-IONP–FITC for binding on TNBC cells. Free ICAM-1 antibodies were added to MDA-MB-231, MDA-MB-436, and MDA-MB-157 cells at a concentration of 20 μg/mL for 1 h at 4 °C before addition of the ICAM-IONP–FITC, and the fluorescence of rinsed cells was taken in a flow cytometer. **P < 0.01; ***P < 0.001. (E–M) Representative microscope images of Prussian blue staining of ICAM-IONP taken up by MDA-MB-231 (E), MDA-MB-436 (F), MDA-MB-157 (G), MCF7 (H), HCC1500 (I), MDA-MB-361 (J), SKBR3 (K), AG11132 (L), and MCF10A (M). Scale bars represent 20 µm. (N–P) ICAM-IONPs are biologically nontoxic at a test concentration of ∼200 µg/mL. Cytotoxicity effects of ICAM-IONP in HS27 (human fibroblast; N), U937 (human macrophages; O), and human umbilical vein endothelial cells (HUVEC; P). (Q) Characterization of as-synthesized IGG-IONP, HER2-IONP, and ICAM-IONP.
Effective targeting of TNBC cells via the ICAM-1 antibody first was evaluated in vitro by the binding and uptake of FITC-labeled ICAM-IONPs, IGG-IONPs, and HER2-IONPs. Our normalized fluorescent intensity data demonstrated that TNBC cells exhibited 2.4- to 4-fold greater binding to ICAM-IONPs than IGG-IONPs or HER2-IONPs because of the abundance of ICAM-1 expression (Fig. 2C). Quantitative analysis of ICAM-1 surface protein expression was directly correlated with increased binding of ICAM-IONPs. In comparison, HER2-IONPs showed positive targeting to HER2-positive cells but failed to target TNBC cells because of their HER2 deficiency. Using a Prussian blue staining assay to examine the presence of iron oxide, we further confirmed the strong and specific binding of ICAM-IONPs to TNBC cells (Fig. 2 E–G and Fig. S3) compared with minimal binding to non-TNBC (Fig. 2 H–K) and nonneoplastic cell lines (Fig. 2 L and M). We observed analogous patterns of binding between ICAM-IONPs and HER2-IONPs to both ICAM-1– and HER2-overexpressing cell lines, respectively. Blocking of ICAM-1 with free ICAM-1 antibody inhibited ICAM-IONP binding and uptake by TNBC to levels commensurate with IGG-IONPs (Fig. 2D). These results indicate that ICAM-IONPs exhibit ICAM-1 targeting activity and specificity.
TNBC-Targeted MRI of TNBC Xenograft Tumor with ICAM-IONPs.
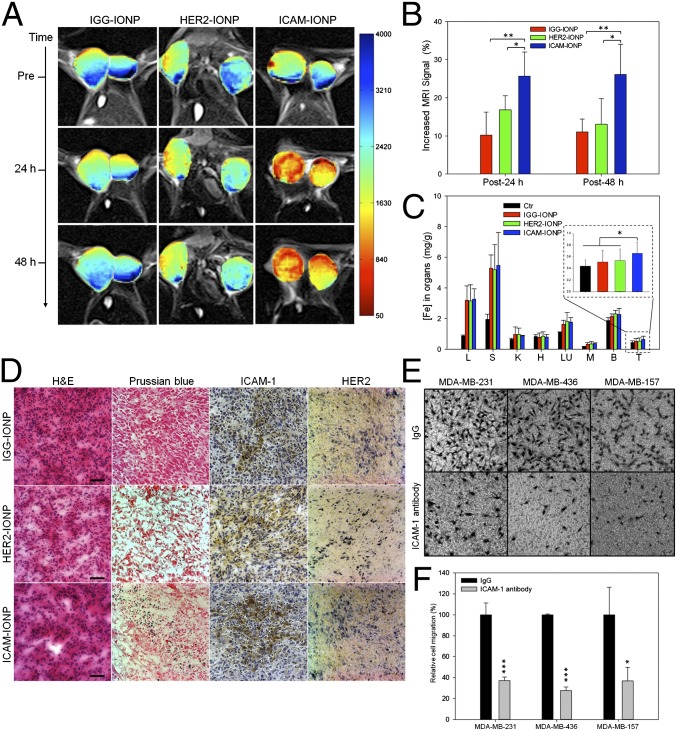
We then examined the ability of ICAM-IONPs for targeted imaging of TNBC tumors in vivo by MRI using a xenograft TNBC mouse model. MDA-MB-231 cells were implanted s.c. in immunodeficient athymic nude mice. MRI was performed on three groups of tumor-bearing mice injected i.v. with IGG-IONP, HER2-IONP, or ICAM-IONP when tumors reached 1 cm3 in volume. Each group was scanned before injection of the imaging probes (preinjection) and 24 h and 48 h post injection with a set of MRI sequences, including T1, T2-weighted spin echo imaging, and T2 relaxometry. The T2-weighted images presented in Fig. 3A show decreased signals in the regions of the tumor as the result of enhanced T2 contrast from uptake of IONP probes in tumors. Quantification of MRI signals in three groups demonstrated a 10% (IGG-IONP), 17% (HER2-IONP), and 26% (ICAM-IONP) signal drop at 24 h after administration of the probes, which lasted at least 48 h (Fig. 3B). ICAM-IONPs significantly improved MRI contrast by actively targeting the TNBC tumor via ICAM-1 binding. It is worth noting that HER2-targeted MRI probes were reported to change the MRI signal by 18–46% in HER2-positive breast tumors (23, 24), comparable to the ICAM-1–targeted MRI probe demonstrated in TNBC tumors in the current study.
Fig. 3.
In vivo MR detection of TNBC using ICAM-1 targeting MRI probes. (A) Color maps of T2-weighted MR images of a mouse implanted with the TNBC cell line MDA-MB-231 at different time points (pre, 24 h, and 48 h) after injection of IGG-IONP, or HER2-IONP, or ICAM-IONP. (B) Quantification of the MRI signal enhanced by IGG-IONP, HER2-IONP, and ICAM-IONP at 24 and 48 h. Significant R2 signal changes were observed with time after ICAM-IONP treatment. *P < 0.05; **P < 0.01. (C) Whole-body distribution of iron accumulation of IGG-IONP, HER2-IONP, and ICAM-IONP in liver (L), spleen (S), kidney (K), heart (H), lung (LU), muscle (M), brain (B), and tumor (T). (D) Histology for MDA-MB-231 tumor accumulation of IGG-IONP, or HER2-IONP, or ICAM-IONP. Tumors were sectioned and stained with H&E, Prussian blue agent, ICAM-1 antibody, and HER2 antibody. In Prussian blue staining, the sections in blue suggest areas of IONP localization. The strongest blue staining is present in the ICAM-IONP group (bottom row, second column). Scale bar represents 50 μm. (E) Representative micrographs depicting three TNBC cells (MDA-MB-231, MDA-MB-436, and MDA-MB-157) incubated with nonspecific IgG or ICAM-1 antibody after transmigrating through 8-μm pores of a transwell membrane. Images taken were on the reverse side of the membrane facing the lower chamber. (F) ICAM-1 antibody reduces the migration of TNBC cells. Nonspecific IgG is used as control. Cells were treated with 10 μg/mL ICAM-1 antibody or nonspecific IgG. *P < 0.05, ***P < 0.001.
The biodistribution and tumor accumulation of MRI probes were evaluated by quantifying iron in collected organs and tissue. Fig. 3C shows comparative iron accumulation in seven organs harvested from mice at 48 h after a single tail vein administration of IGG-IONPs, HER2-IONPs, and ICAM-IONPs. Correlating with the in vivo MRI results, the iron accumulation of ICAM-IONPs in TNBC tumors was 3.7- and 2.1-fold higher than that of IGG-IONPs and HER2-IONPs with reference to untreated tumors, respectively (Fig. 3C, Inset). Histological analysis was performed to further confirm the targeting of ICAM-IONPs to the tumor and the observed MRI contrast change in vivo (Fig. 3D). TNBC tumor sections were stained with ICAM-1 antibody, HER2 antibody, hematoxylin and eosin (H&E), and Prussian blue. Consistent with the MRI findings, tumors from mice receiving ICAM-IONPs showed a high level of ICAM-1 expression and strong Prussian blue staining of IONPs. In contrast, Prussian blue staining was low in tumors receiving IGG-IONPs or HER2-IONPs. Low HER2 surface expression in TNBC tumors did not result in significant HER2-IONP accumulation. Thus, results from the in vivo MRI experiments suggest that the uptake of ICAM-IONPs is driven by ICAM-1 expression on TNBCs.
Discussion
It is noteworthy that our discovery of ICAM-1 as a TNBC target and biomarker also reveals promising functions of this well-characterized receptor, which may be explored for clinical applications. ICAM-1 plays an important role in inflammation. Logically, anti–ICAM-1–targeted interventions were developed for the treatment of chronic inflammatory disorders (25, 26). The role of ICAM-1 in oncology also has been under intense investigation. ICAM-1 up-regulation is observed in several types of cancers associated with advanced disease, poor survival, and resistance to chemotherapy (20, 21). Treatment with a human ICAM-1 antibody has demonstrated potent macrophage-dependent antimyeloma activity in vivo (27). ICAM-1 cross-linking leads to phosphorylation of proteins, cytoskeletal modifications, and gene regulation governing cell shape, recruitment, and migration (28).
Taken together, our finding that ICAM-1 is a promising TNBC target and biomarker may lead to an effective ICAM-1 targeting strategy for imaging and treatment of TNBC. Previous studies in wound healing, rheumatoid arthritis, and acute stroke demonstrated that enlimomab (anti–ICAM-1 antibody) was well tolerated by different patient groups, indicating that it may be safe and well tolerated in humans (29–31). Although the ICAM-1 antibody did not affect TNBC cell proliferation in vitro (Fig. S4), we observed that the ICAM-1 antibody significantly reduced TNBC cell migration (Fig. 3 E and F). Greenwood et al. (32) reported that an ICAM-1 antibody blockade resulted in ICAM-1 molecules lacking cytoplasmic tails that could not activate Rho proteins. Similar antitumor activity of the ICAM-1 antibody or siRNA was observed in several human cancers (20, 21, 27).
In summary, our results demonstrate the identification of ICAM-1 as an efficient TNBC molecular target based on the in vitro evaluation of its TNBC-specific molecular profile and preclinical in vivo ICAM-1–targeted molecular MRI in a TNBC tumor model. The findings provide a rationale for further preclinical and clinical evaluation and development of ICAM-1–targeted treatments for TNBC.
Materials and Methods
Complete details of materials are provided in SI Materials and Methods.
PCR Array.
The human signaling PathwayFinder RT2 Profiler PCR Array was used to screen possible TNBC targets in MDA-MB-231, MCF7, and MCF10A cells. First, each cell line was incubated at 5 × 105 cells per well in a six-well cell culture plate overnight. One microgram RNA of each cell line was converted to cDNA using the RT2 First Strand Kit according to the manufacturer’s instructions. Diluted cDNA was added to the RT2 SYBR Green/Fluorescein qPCR Mastermix. The human signaling PathwayFinder RT2 Profiler PCR Array was loaded with 25 μL/well of cDNA–Mastermix according to the PCR protocol provided by the manufacturer. Results were analyzed using RT2 Profiler PCR Array Data Analysis Template v3.0.
Immunohistological Staining.
One hundred forty-nine cases of human breast cancer tissue and 144 cases of human normal tissue microarray samples were evaluated for ICAM-1 expression as described previously (33, 34). Immunohistochemical staining was performed by using paraffin-embedded human breast cancer tissue microarrays (BR1503B, BR1505, and T088) and normal tissue microarrays (BN00011 and BN1002a). The individual tissue cores in the microarrays were scored by a surgical pathologist, with no knowledge of sample identity, for no staining (0), weak staining (1), moderate staining (2), or strong staining (3). Photomicrographs were taken on an Olympus BX41 microscope by using an Olympus Q-Color5 digital camera (Olympus America Inc.).
Cell Culture.
Three human TNBC cell lines (MDA-MB-231, MDA-MB-436, and MDA-MB-157), four human non-TNBC cell lines (MCF7, HCC1500, SKBR3, and MDA-MB-361), and two nonneoplastic mammary epithelial cell lines (AG11132 and MCF10A) were studied. MDA-MB-231, MDA-MB-436, MDA-MB-157, MCF7, HCC1500, SKBR3, MDA-MB-361, and MCF10A were available through the American Type Culture Collection; AG11132 was obtained from Coriell Institute. MDA-MB-231, MDA-MB-436, MDA-MB-157, MCF7, and MDA-MB-361 were cultured in DMEM, and HCC1500 was cultured in Roswell Park Memorial Institute (RPMI)-1640, SKBR3 in McCoy-5A, AG11132 in Mammary Epithelial Cell Basal Medium, and MCF10A in DMEM/F12 (1:1) medium, with all recommended supplements, respectively. All cells were maintained at 37 °C in a humidified incubator with 5% (vol/vol) CO2.
Quantification of ICAM-1 Gene Expression.
The gene expression level of ICAM-1 of breast cancer cell lines was characterized by using quantitative RT-PCR (qRT-PCR). MDA-MB-231, MDA-MB-436, MDA-MB-157, MCF7, HCC1500, MDA-MB-361, SKBR3, AG11132, and MCF10A cells were cultured at 5 × 105 cells per well in a six-well cell culture plate overnight. Cells then were removed from each well by incubating with a trypsin/EDTA solution for 3 min. The cells were washed with PBS three times. RNA was extracted, purified using the Qiagen RNeasy Mini Kit, and quantified using a SpectraMax Plus 384 UV-visible spectrophotometer (Molecular Devices). Reverse transcription was conducted using the Applied Biosystems TaqMan RT protocol. Detection and quantification of mRNA were performed with the StepOnePlus Real-Time PCR System (Applied Biosystems). All PCR samples were referenced to the gene expression of GAPDH.
Quantification of ICAM-1 Surface Expression.
Breast cancer cell ICAM-1 surface protein expression was evaluated by a BD FACSCalibur flow cytometer (BD Biosciences) as described previously (35, 36). Quantification of the ICAM-1 density on the cell surface was determined with reference to Quantum Simply Cellular microbeads, using the protocol as provided by the manufacturer. Briefly, 106 cells were collected and rinsed twice through suspension–spin cycles. Cells were blocked by 1% BSA in PBS for 30 min in an ice bath. After BSA blockage, cells were incubated with phycoerythrin/anti–ICAM-1 antibody for 1 h at room temperature (RT). Cells were rinsed with 1% BSA in PBS three times, resuspended in PBS, and evaluated by flow cytometry.
ICAM-1 Immunofluorescent Staining.
MDA-MB-231, MDA-MB-436, MDA-MB-157, MCF7, HCC1500, MDA-MB-361, SKBR3, AG11132, and MCF10A (2 × 105 cells) were seeded in a Lab-Tek II Chamber Slide System separately with 1 mL medium overnight at 37 °C. After medium was removed, cells were rinsed with PBS three times and fixed with 4% formaldehyde in PBS at RT for 10 min, followed by washing with PBS. Samples were blocked with 1% BSA in PBS for 30 min in an ice bath. After BSA blocking, samples were stained with mouse anti-human ICAM-1 antibody (primary antibody) for 1 h and rinsed with PBS. Samples were incubated with NorthernLights 557 conjugated goat anti-mouse secondary antibody (NL557 Abs) for another 1 h, followed by washing with PBS. DAPI was used to stain the cell nucleus. Immunofluorescent stained samples were dried overnight in the dark and used for fluorescent microscopic imaging. Samples were examined under a Leica TCS SP5 confocal fluorescent microscope (Leica Microsystems). Digital images were captured with AxioVision digital image-processing software.
Synthesis of ICAM-IONP, HER2-IONP, and IGG-IONP.
Casein-coated IONPs were prepared as described previously (22) and stocked at a concentration of 5 mg/mL in PBS. A 200-μL stock IONP solution (1 mg) was mixed with 200 μL Activation Buffer (Ocean Nanotech), 50 μg EDC, and 25 μg NHS for 20 min at RT. Next, 100 μg ICAM-1 antibody or HER2 antibody or IgG and 400 μL Coupling Buffer (Ocean Nanotech) were added to the IONP solution and reacted for 2 h at RT with continuous mixing. As-synthesized ICAM-IONPs or HER2-IONPs or IGG-IONPs were purified by ultracentrifugation using a Nanosep 300K Omega centrifugal device.
Characterization of ICAM-IONPs.
The morphology and size of ICAM-IONP nanoparticles were studied by using TEM (Hitachi H-7500; accelerating voltage, 75 kV). The TEM samples were prepared by dropping diluted nanoparticle solutions on a carbon-coated copper grid and were air dried. The hydrodynamic size and surface charges of IONPs in aqueous solution were evaluated using a DLS instrument (Malvern Zetasizer Nano S-90) equipped with a 22-mW He-Ne laser operating at 632.8 nm. FITC-conjugated ICAM-IONPs, or IGG-IONPs, or HER2-IONPs (ICAM-IONP-FITC, or IGG-IONP-FITC, or HER2-IONP-FITC) also were prepared to evaluate the antibody densities on obtained MRI probes. FITC-conjugated IgG, or HER2 antibody, or ICAM-1 antibody was used in the synthesis by replacing their non–fluorophore-tagged forms. Other conditions were kept the same during the synthesis. Antibody density on each type of MRI probe was calculated by using a FITC standard concentration curve.
In Vitro Nanoparticle Probe Binding.
Quantitative analysis of ICAM-IONP–FITC binding to TNBCs (MDA-MB-231, MDA-MB-436, MDA-MB-157) was conducted using flow cytometry. Non-TNBCs (MCF7, HCC1500, MDA-MB-361, and SKBR3) and nonneoplastic cells (AG11132 and MCF10A) were selected as controls. Cells were seeded in six-well plates (3 × 105 cells per well) and allowed to adhere overnight. Then, cells were incubated for 4 h at 37 °C with (i) IGG-IONP–FITC, (ii) HER2-IONP–FITC, and (iii) ICAM1-IONP–FITC. The nanoparticle concentration used was 100 μg/mL All nanoparticle-treated cells were washed with PBS, harvested using a 0.25% trypsin/2.6 mM EDTA solution, and washed with PBS (pH 7.4) three times. Binding data were acquired with a BD FACSCalibur flow cytometer and analyzed using FlowJo software. The increase in binding value was calculated by dividing the mean fluorescence intensity of HER2-IONP–FITC– or ICAM1-IONP–FITC–stained cells by that of the nonspecific IGG-IONP–FITC–stained cells.
Prussian Blue Staining.
The nine cell lines (2 × 105 cells) in Fig. 1S were seeded separately in a Lab-Tek II Chamber Slide System with 1 mL medium overnight at 37 °C. After medium was removed, cells were rinsed with PBS three times and fixed with 4% formaldehyde in PBS at RT for 10 min, followed by washing with PBS then soaking in working solution composed of 10% potassium ferrocyanide (II) trihydrate and 20% HCl solution (vol:vol = 1:1) at 37 °C for 4 h. After being washed with PBS, slices were counterstained with nuclear fast red for 5 min. Blue dots representing the remaining IONPs in organs were investigated with a Leica TCS SP5 confocal fluorescent microscope (Leica Microsystems).
In Vivo MRI.
Animal experiments were performed according to the protocol approved by the Institutional Animal Care and Use Committee of Emory University. Breast tumors were established s.c. by injecting 5 × 106 MDA-MB-231 cells into the fourth mammary fat pad of athymic nude mice (Charles River; n = 5 for each group). Tumors were developed for 5–7 wks until they were at least 1 cm3 in volume. In vivo MRI was performed on the tumor-bearing mice in three groups, which were injected i.v. with IGG-IONP, HER2-IONP, and ICAM-IONP (at the dosage of 20 mg Fe/kg mouse weight), respectively. Images were obtained at pre-, 24 h, and 48 h post injection with a 3 T MRI scanner (Siemens Healthcare) with fast spin echo and multiecho time (TE) sequence for T2-weighted MRI. The imaging parameters were as follows: repetition time (TR) of 3,200 ms, TE of 86 ms, 320 × 128 matrix, 120 × 60-mm2 field of view, 150° flip angle, and 1.00-mm slice thickness for T2-weighted imaging; TR of 3,710 ms and 20 different TEs, starting at 12 ms with 12-ms increments for multi-TE imaging. To quantify the signal intensity for tumor, regions of interest (ROIs) were drawn around the whole tumor at the same slice with the same imaging depth. The pixel intensity was calculated and normalized to the area of ROIs by ImageJ software.
Histology.
The organs (liver, spleen, kidney, lung, heart, and muscle) and tumor samples were collected at 48 h after injection. The phenanthroline colorimetric method was used to determine the iron concentration in organs after they were digested in concentrated HNO3. Pathologies of MDA-MB-231 tumors with IGG-IONP, or HER2-IONP, or ICAM-IONP were investigated by H&E staining, Prussian blue staining, and ICAM-1 and HER2 immunohistological staining. All staining was performed for the tumor slices following the standard protocol.
Statistical Analysis.
Quantitative data are presented as means ± SD. Differences were compared using an unpaired t test. P values ≤0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Kristin Johnson for assistance with the schematic illustration. D.T.A. acknowledges the support of the National Institutes of Health (NIH; National Cancer Institute Grant 1DP2CA174495). M.A.M. acknowledges the support of the Breast Cancer Research Foundation. H.M. acknowledges support from the NIH (5R01CA154846-02 and 1P50CA128301-01A1).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408556111/-/DCSupplemental.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Ovcaricek T, Frkovic S, Matos E, Mozina B, Borstnar S. Triple negative breast cancer—prognostic factors and survival. Radiol Oncol. 2011;45(1):46–52. doi: 10.2478/v10019-010-0054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slamon D, et al. Breast Cancer International Research Group Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu K-D, Wu J, Shen Z-Z, Shao Z-M. Hazard of breast cancer-specific mortality among women with estrogen receptor-positive breast cancer after five years from diagnosis: Implication for extended endocrine therapy. J Clin Endocrinol Metab. 2012;97(12):E2201–E2209. doi: 10.1210/jc.2012-2423. [DOI] [PubMed] [Google Scholar]
- 5.Bilici A, Arslan C, Altundag K. Promising therapeutic options in triple-negative breast cancer. J BUON. 2012;17(2):209–222. [PubMed] [Google Scholar]
- 6.Gucalp A, Traina TA. Triple-negative breast cancer: Adjuvant therapeutic options. Chemother Res Pract. 2011;2011:696208. doi: 10.1155/2011/696208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: Therapeutic options. Lancet Oncol. 2007;8(3):235–244. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 8.Carey LA, et al. TBCRC 001: Randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol. 2012;30(21):2615–2623. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cristofanilli M, et al. Imatinib mesylate (Gleevec) in advanced breast cancer-expressing C-Kit or PDGFR-beta: Clinical activity and biological correlations. Ann Oncol. 2008;19(10):1713–1719. doi: 10.1093/annonc/mdn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Shaughnessy J, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364(3):205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 11.Modi S, et al. A phase II trial of imatinib mesylate monotherapy in patients with metastatic breast cancer. Breast Cancer Res Treat. 2005;90(2):157–163. doi: 10.1007/s10549-004-3974-0. [DOI] [PubMed] [Google Scholar]
- 12.Baselga J, et al. Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J Clin Oncol. 2013;31(20):2586–2592. doi: 10.1200/JCO.2012.46.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu JF, et al. A Phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur J Cancer. 2013;49(14):2972–2978. doi: 10.1016/j.ejca.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7(2):79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 15.Müller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 16.Soria G, Ben-Baruch A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008;267(2):271–285. doi: 10.1016/j.canlet.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Xia M, Sui Z. Recent developments in CCR2 antagonists. Expert Opin Ther Pat. 2009;19(3):295–303. doi: 10.1517/13543770902755129. [DOI] [PubMed] [Google Scholar]
- 18.Chui R, Dorovini-Zis K. Regulation of CCL2 and CCL3 expression in human brain endothelial cells by cytokines and lipopolysaccharide. J Neuroinflammation. 2010;7:1. doi: 10.1186/1742-2094-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes SH, Seigel GM. Immunoreactivity of ICAM-1 in human tumors, metastases and normal tissues. Int J Clin Exp Pathol. 2009;2(6):553–560. [PMC free article] [PubMed] [Google Scholar]
- 20.Roland CL, Harken AH, Sarr MG, Barnett CC., Jr ICAM-1 expression determines malignant potential of cancer. Surgery. 2007;141(6):705–707. doi: 10.1016/j.surg.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 21.Rosette C, et al. Role of ICAM1 in invasion of human breast cancer cells. Carcinogenesis. 2005;26(5):943–950. doi: 10.1093/carcin/bgi070. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, et al. Casein-coated iron oxide nanoparticles for high MRI contrast enhancement and efficient cell targeting. ACS Appl Mater Interfaces. 2013;5(11):4632–4639. doi: 10.1021/am400713j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen T-J, et al. Targeted Herceptin-dextran iron oxide nanoparticles for noninvasive imaging of HER2/neu receptors using MRI. J Biol Inorg Chem. 2009;14(2):253–260. doi: 10.1007/s00775-008-0445-9. [DOI] [PubMed] [Google Scholar]
- 24.Huh Y-M, et al. In vivo magnetic resonance detection of cancer by using multifunctional magnetic nanocrystals. J Am Chem Soc. 2005;127(35):12387–12391. doi: 10.1021/ja052337c. [DOI] [PubMed] [Google Scholar]
- 25.Burns RC, et al. Antibody blockade of ICAM-1 and VCAM-1 ameliorates inflammation in the SAMP-1/Yit adoptive transfer model of Crohn’s disease in mice. Gastroenterology. 2001;121(6):1428–1436. doi: 10.1053/gast.2001.29568. [DOI] [PubMed] [Google Scholar]
- 26.Yusuf-Makagiansar H, Anderson ME, Yakovleva TV, Murray JS, Siahaan TJ. Inhibition of LFA-1/ICAM-1 and VLA-4/VCAM-1 as a therapeutic approach to inflammation and autoimmune diseases. Med Res Rev. 2002;22(2):146–167. doi: 10.1002/med.10001. [DOI] [PubMed] [Google Scholar]
- 27.Veitonmäki N, et al. A human ICAM-1 antibody isolated by a function-first approach has potent macrophage-dependent antimyeloma activity in vivo. Cancer Cell. 2013;23(4):502–515. doi: 10.1016/j.ccr.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Lawson C, Wolf S. ICAM-1 signaling in endothelial cells. Pharmacol Rep. 2009;61(1):22–32. doi: 10.1016/s1734-1140(09)70004-0. [DOI] [PubMed] [Google Scholar]
- 29.Mileski WJ, et al. Clinical effects of inhibiting leukocyte adhesion with monoclonal antibody to intercellular adhesion molecule-1 (enlimomab) in the treatment of partial-thickness burn injury. J Trauma. 2003;54(5):950–958. doi: 10.1097/01.TA.0000030626.84680.11. [DOI] [PubMed] [Google Scholar]
- 30.Kavanaugh AF, Schulze-Koops H, Davis LS, Lipsky PE. Repeat treatment of rheumatoid arthritis patients with a murine anti-intercellular adhesion molecule 1 monoclonal antibody. Arthritis Rheum. 1997;40(5):849–853. doi: 10.1002/art.1780400511. [DOI] [PubMed] [Google Scholar]
- 31.Schneider D, et al. Safety, pharmacokinetics and biological activity of enlimomab (anti-ICAM-1 antibody): An open-label, dose escalation study in patients hospitalized for acute stroke. Eur Neurol. 1998;40(2):78–83. doi: 10.1159/000007962. [DOI] [PubMed] [Google Scholar]
- 32.Greenwood J, et al. Intracellular domain of brain endothelial intercellular adhesion molecule-1 is essential for T lymphocyte-mediated signaling and migration. J Immunol. 2003;171(4):2099–2108. doi: 10.4049/jimmunol.171.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, et al. Lipocalin 2 promotes breast cancer progression. Proc Natl Acad Sci USA. 2009;106(10):3913–3918. doi: 10.1073/pnas.0810617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy R, Wewer UM, Zurakowski D, Pories SE, Moses MA. ADAM 12 cleaves extracellular matrix proteins and correlates with cancer status and stage. J Biol Chem. 2004;279(49):51323–51330. doi: 10.1074/jbc.M409565200. [DOI] [PubMed] [Google Scholar]
- 35.Guo P, et al. Inhibiting metastatic breast cancer cell migration via the synergy of targeted, pH-triggered siRNA delivery and chemokine axis blockade. Mol Pharm. 2014;11(3):755–765. doi: 10.1021/mp4004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo P, You J-O, Yang J, Moses MA, Auguste DT. Using breast cancer cell CXCR4 surface expression to predict liposome binding and cytotoxicity. Biomaterials. 2012;33(32):8104–8110. doi: 10.1016/j.biomaterials.2012.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.