Significance
The alternative adaptive immune system of jawless vertebrates is based on three types of variable lymphocyte receptors (VLRs) that are differentially expressed by distinct B- and T-like lymphocyte lineages. Like the antibodies and T-cell receptors of jawed vertebrates, the highly variable VLR antigen receptors are generated by combinatorial assembly. However, it is not known whether VLRs are subjected to selection to mitigate the detrimental effects of self-reactivity. Here, we identify signatures for selection of the VLRC receptors that are expressed by one of the T-cell lineages of lamprey; selection concerns the length of the receptor molecules and their N-terminal sequence diversity. These findings pave the way to identifying the mechanistic basis of selection.
Keywords: adaptive immunity, molecular evolution, agnathans
Abstract
The alternative adaptive immune system of jawless vertebrates is based on different isotypes of variable lymphocyte receptors (VLRs) that are composed of leucine-rich repeats (LRRs) and expressed by distinct B- and T-like lymphocyte lineages. VLRB is expressed by B-like cells, whereas VLRA and VLRC are expressed by two T-like lineages that develop in the thymoid, a thymus-like structure in lamprey larvae. In each case, stepwise combinatorial insertions of different types of short donor LRR cassettes into incomplete germ-line genes are required to generate functional VLR gene assemblies. It is unknown, however, whether the diverse repertoires of VLRs that are expressed by peripheral blood lymphocytes are shaped by selection after their assembly. Here, we identify signatures of selection in the peripheral repertoire of VLRC antigen receptors that are clonally expressed by one of the T-like cell types in lampreys. Selection strongly favors VLRC molecules containing four internal variable leucine-rich repeat (LRRV) modules, although VLRC assemblies encoding five internal modules are initially equally frequent. In addition to the length selection, VLRC molecules in VLRC+ peripheral lymphocytes exhibit a distinct pattern of high entropy sites in the N-terminal LRR1 module, which is inserted next to the germ-line–encoded LRRNT module. This is evident in comparisons to VLRC gene assemblies found in the thymoid and to VLRC gene assemblies found in some VLRA+ cells. Our findings are the first indication to our knowledge that selection operates on a VLR repertoire and provide a framework to establish the mechanism by which this selection occurs during development of the VLRC+ lymphocyte lineage.
Jawless fishes (agnathans), comprising lampreys and hagfishes, the sister group of jawed vertebrates (gnathostomes), exhibit an alternative adaptive immune system, which uses somatically diversifying variable lymphocyte receptors (VLRs) that are composed of leucine-rich repeats (LRRs) (1) instead of the Ig domain-containing antigen receptors of jawed vertebrates (2–5). Despite these differences, the immune systems of both jawed and jawless vertebrates possess similar characteristics. For instance, recent work has indicated that lampreys have three distinct lymphocyte lineages, one of B-like cells, and two of T-like cells, and that each lymphocyte lineage of lamprey expresses a distinct type of VLRs, designated VLRA, VLRB, and VLRC, respectively (6). Hagfishes, the second clade of jawless vertebrates, also possess genes encoding orthologs of these three VLR isotypes (7, 8), suggesting that distinct lymphocyte lineages and specialized antigen receptor types were integral components of the immune system of the common ancestor of jawless vertebrates.
The vast repertoire of structurally diverse VLRs that are clonally expressed by lymphocytes is generated by a combinatorial assembly process akin to gene conversion (9–11), believed to be mediated by the activity of distinct cytosine deaminases with lineage-specific expression patterns (12): CDA1 for VLRA (13) and VLRC (6) genes, and CDA2 for VLRB (13) genes. The formation of mature VLR genes involves the insertion of tandem arrays of LRR-encoding cassettes into the incomplete VLR loci, which encode only the invariant N- and C-terminal constant regions of VLR receptors. VLRB molecules function as GPI-linked membrane-bound receptors on the B-like cells of lamprey and as soluble antibodies (10, 14–17). By contrast, VLRA and VLRC are expressed as membrane-bound receptors in a mutually exclusive fashion by the two distinct T lineages of lamprey (6). Previous work has identified in lamprey larvae a thymus equivalent (referred to as the thymoid), situated at the tips of the gill filaments; this lymphoepithelial tissue is characterized by the expression of genes such as FOXN1 and DLL4 that are also found in the thymopoietic tissues of jawed vertebrates (18). These findings, together with the observations of a high frequency of incomplete and nonfunctional VLRA (18) and VLRC (6, 19) assemblies and the expression of the CDA1 gene (18), indicate that VLRA and VLRC receptor genes are assembled in this tissue.
Two observations suggest that jawless fishes possess self-tolerant VLR repertoires. First, lampreys were found to accept skin autografts, but to reject first-set allografts slowly and second-set allografts with accelerated kinetics (20). In a mixed leukocyte reaction system of hagfish, the adherent myeloid fraction of blood cells was shown to be largely responsible for driving the alloresponse, whereas responder cells were found in the nonadherent lymphocyte fraction (21). Second, estimates of the sequence complexities of VLR assemblies (2, 4) indicate that these are comparable in magnitude to those observed in B-cell receptors (BCRs) and T-cell receptors (TCRs) of jawed vertebrates, which are known to be purged of self-reactivities. Whereas the mechanisms governing the selection of antigen receptors expressed by the lymphocytes of jawed vertebrates are already well understood, no such information exists for the antigen receptors expressed by lymphocytes of jawless vertebrates.
Here, we describe our efforts to identify any indication of selection in the VLRC-expressing lymphocyte lineage of lampreys, chosen for our studies for two reasons. First, the tissue wherein VLRC gene assembly takes place has been identified in lampreys (18). Hence, it should be possible to identify certain features of the preselection repertoire among VLRC gene sequences isolated from the thymoid in a comparison with those isolated from peripheral blood, which we assume to mainly consist of selected VLRC sequences. Second, we wished to exploit our previous observation that, whereas relatively few VLRC+ cells additionally possess assembled VLRA genes, many VLRA+ cells carry VLRC assemblies (6). Assuming that VLRC assemblies found in VLRA+ cells failed to support differentiation of VLRC+ lineage cells, their comparison with the bona fide repertoire of VLRC sequences in VLRC+ cells should provide an additional opportunity to identify signs of sequence-specific selection of the VLRC repertoire. To examine the molecular basis of possible selection processes, we also determined the crystal structure of the lamprey VLRC receptor. Our sequence analyses and structural studies provide evidence for sequence-specific selection of the VLRC repertoire.
Results
Number of Variable Leucine-Rich Modules in VLRC Assemblies.
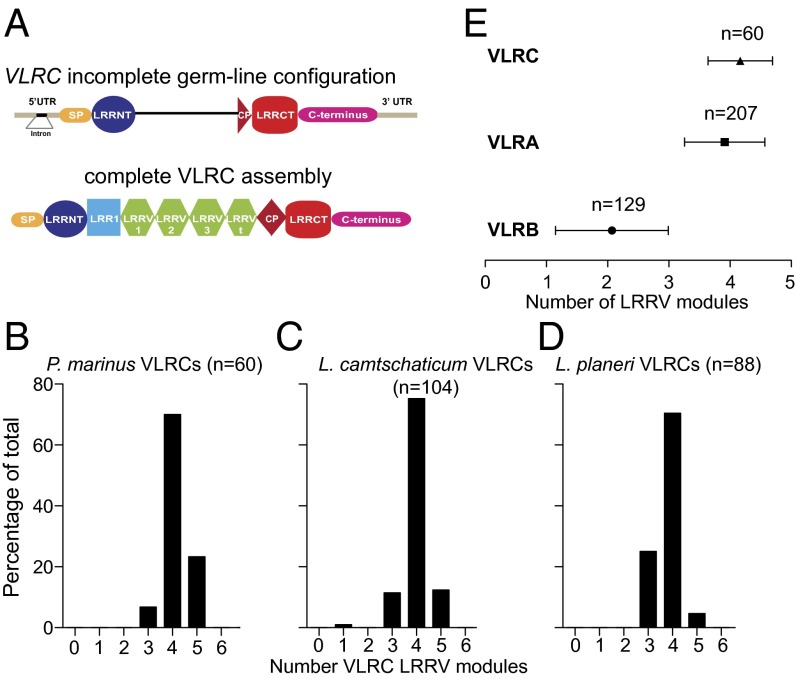
A remarkable feature of VLRC assemblies (Fig. 1A) in different lamprey species is the apparent preference for four modules of variable leucine-rich repeats (LRRVs) situated in the central part of the VLRC molecule. In blood lymphocytes of Petromyzon marinus, Lethenteron camtschaticum, and Lampetra planeri more than 70% of all functionally assembled VLRC genes exhibit this characteristic (Fig. 1 B–D). Interestingly, VLRC assemblies of hagfishes also exhibit a preference for four LRRV modules (Fig. S1A), although the size distribution of their VLRC molecules is broader than that of lampreys (Fig. S1B). A similar phenomenon, albeit less pronounced, is observed for VLRA assemblies in these species, whereas VLRB assemblies mainly possess only two LRRV elements, as exemplified for VLR assemblies of P. marinus in Fig. 1E. The reason for the remarkable homogeneity in length of VLRC assemblies is unknown. Here, we set out to examine the contributions of the following factors to repertoire formation of VLRC receptors: (i) mechanistic constraints of assembly and (ii) postassembly selection processes.
Fig. 1.
VLRC gene assemblies are restricted in length. (A) Schematic depicting the germ-line configuration of the VLRC gene in lamprey (Upper) and the organization of internal variable modules in mature proteins (Lower). CP, connecting peptide; LRR, leucine-rich repeat; LRR1, first LRR module; LRRCT, C-terminal LRR; LRRNT, N-terminal LRR; LRRV, variable LRR modules; LRRVt, terminal LRRV module; SP, signal peptide; UTR, untranslated regions. (B–D) Analysis of LRRV number in the mature VLRC blood repertoire of three lamprey species. Numbers of LRRV modules are inclusive of the terminal LRRVt module. (E) Summary of the average number of internal LRRV modules found in P. marinus peripheral blood VLRC, VLRA, and VLRB repertoires. Error bars represent SDs.
Evidence for Selection of VLRC Assemblies of a Certain Size.
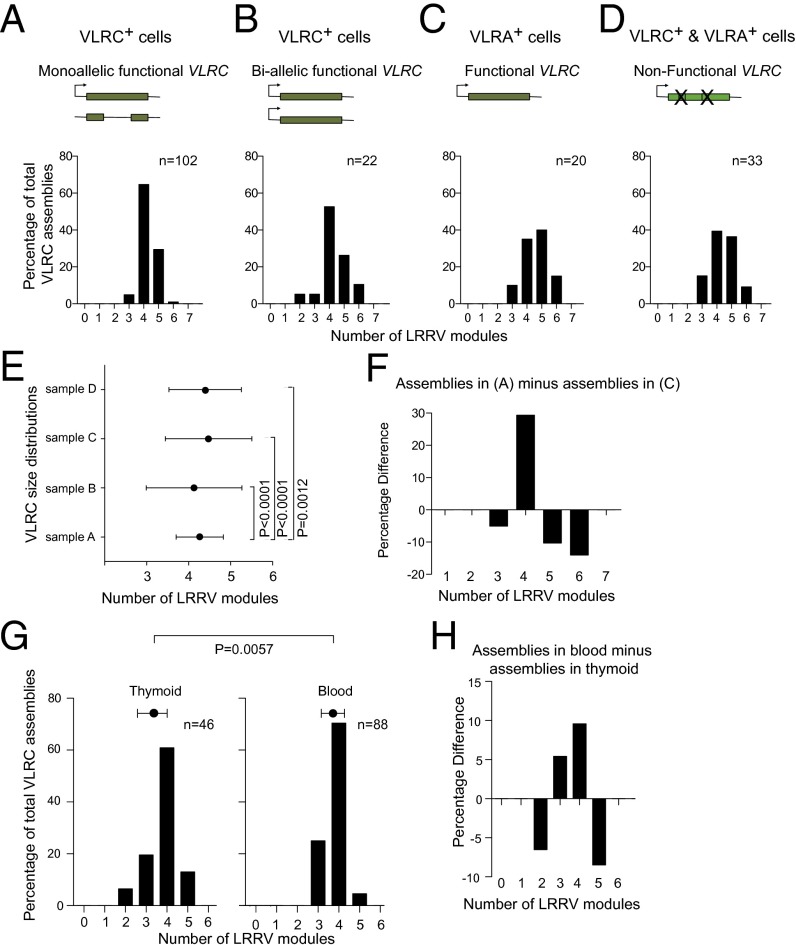
To assess whether the formation of completely assembled VLRC genes is governed entirely by the sequence peculiarities of individual modules, or whether they are subject to selection after assembly, we examined the structure of complete VLRC assemblies in individual VLRC+ and VLRA+ peripheral blood lymphocytes of P. marinus larvae. As shown previously, a certain fraction of VLRA+ cells contains complete VLRC assemblies (6). Whereas only 11% of VLRC+ cells contain VLRA gene assemblies, four times as many (43%) VLRA+ cells carry VLRC gene assemblies (Dataset S1). Thus, the analysis of VLR assemblies at the single cell level supports the earlier conclusion from population studies (6) that the assembly of VLRC genes occurs before that of VLRA genes. As expected from the analysis of VLRC assemblies of all blood lymphocytes (Fig. 1), a clear preference for four LRRV modules (about two-thirds of all assemblies) was observed in VLRC+ cells with monoallelic VLRC assemblies (Fig. 2A); in VLRC+ cells carrying biallelic VLRC assemblies, the contribution of genes with four LRRV modules to the repertoire is somewhat reduced (Fig. 2B). Most of the functional assemblies of VLRC genes in VLRA+ cells contain either four or five LRRV modules (Fig. 2C) and a similar picture emerges from complete VLRC assemblies that are rendered nonfunctional by frame-shift mutations and/or in-frame stop codons as a consequence of errors during the assembly process (Fig. 2D). Functionally assembled VLRC genes in peripheral VLRC+ lymphocytes with monoallelic assemblies must encode receptors that successfully passed the hypothetical checkpoint(s) for inclusion into the peripheral VLRC+ pool. By contrast, functionally assembled VLRC genes in VLRA+ lymphocytes presumably encode receptors that either did not experience or failed VLRC-specific selection. Hence, the comparison of size distributions in these two types of cells provides an indication of the contribution of selection based on the length of VLRC molecules to the mature peripheral VLRC repertoire (Fig. 2E). Our analysis provides evidence for preferred admission of assemblies with four LRRV elements to the peripheral repertoire (Fig. 2F); accordingly, VLRC assemblies in VLRC+ cells with monoallelic assemblies exhibit the smallest variance of size distributions (Fig. 2E).
Fig. 2.
Functional selection of a preferred LRRV number in VLRC assemblies. VLRA and VLRC assemblies were amplified from single VLRA+ and VLRC+ blood lymphocytes of P. marinus and the number of LRRV modules present in VLRC assemblies determined (A–D). Results are presented for VLRC+ lymphocytes with a monoallelic VLRC assembly (A); VLRC+ lymphocytes with biallelic VLRC assemblies (B); VLRA+ lymphocytes with functional VLRC assemblies (C); and VLRC+ and VLRA+ lymphocytes with complete but nonfunctional VLRC assemblies (symbolized by Xs) (D) (Dataset S1). (E) Summary of the average number of LRRV modules present in the different collections of VLRC assemblies presented in A–D. Error bars represent the SDs of the means; differences of variances were assessed using the F test. (F) Percentage difference in LRRV number between VLRC assemblies in A and C shows enrichment of the preferred size of four internal LRRV modules. (G) Distributions of the number of LRRV modules in VLRC assemblies in the thymoid (Left) and peripheral blood (Right) of L. planeri. Error bars represent SDs of the means; differences of variances were assessed using the F test. (H) Percentage difference in LRRV number between blood and thymoid in G indicates enrichment of the preferred size of four internal LRRV modules.
We then sought additional evidence for size selection of VLRC receptors. The thymoid constitutes the site of development of VLRA+ and VLRC+ T-like cells in lamprey larvae. If size selection indeed occurs after assembly of VLRC genes during the formation of the peripheral repertoire, one would expect that the thymoid tissue harbors cells at both pre- and postselection stages. Assuming that the fraction of preselection cells is sufficiently large in this tissue and not too diluted by the presence of cells with the characteristics of peripheral lymphocytes, it should be possible to detect a measurable difference of the size distribution of VLRC assemblies in the thymoid to that of VLRC assemblies in the peripheral repertoire. Indeed, VLRC assemblies in the thymoid are significantly more diverse with respect to their length than in peripheral blood (Fig. 2G); the differential pattern of both distributions again demonstrates that four LRRV modules are preferred in peripheral VLRC assemblies (Fig. 2H). Collectively, our data suggest that VLRC assemblies undergo selection toward the preferred number of four LRRV elements.
Overall Structure of Lamprey VLRC.
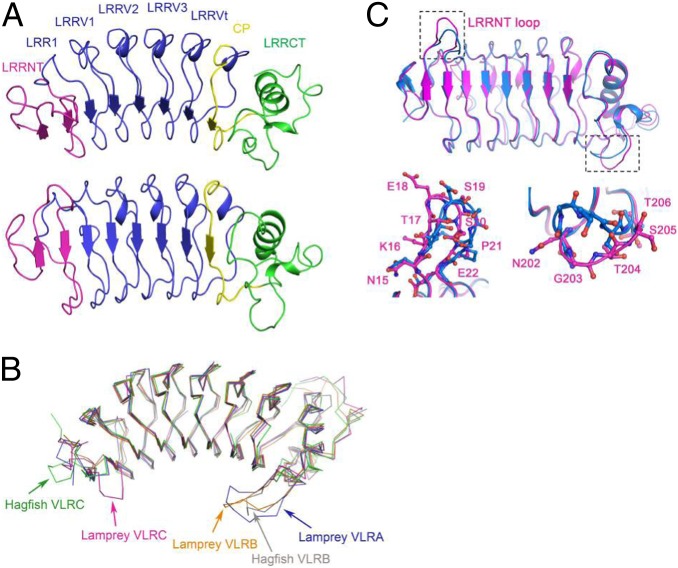
To gain insight into the molecular mechanism of the selection process, we determined the crystal structure of a VLRC receptor exhibiting the preferred number of internal LRRV elements. We determined the structure of the VLRC clone VLRC.1MP from brook lamprey (L. planeri) to 2.5-Å resolution (Dataset S2). VLRC.1MP adopts a crescent-shaped solenoid characteristic of other VLRs (Fig. 3A) (22). The structure comprises a 36-residue N-terminal LRR capping module (LRRNT), a 25-residue LRR1, four 24-residue LRRVs (LRRV1, LRRV2, LRRV3, and LRRVt), a 16-residue truncated LRR designated the connecting peptide (CP), and a 48-residue LRR C-terminal capping module (LRRCT). The inner, concave surface, through which VLRAs and VLRBs bind antigen, is formed from eight β-strands (two from LRRNT, five from LRRs, and one from CP), which assemble into a continuous β-sheet (Fig. 3A). The outer, convex surface of VLRC.1MP is composed of one α-helix from LRRCT, and an array of 310 helices and loops from the other LRR modules (Fig. 3A). In LRRNT, the characteristic four-cysteine motif of N-terminal LRR capping modules (CXnCXCXnC) gives rise to two pairs of disulfides (Cys2–Cys13 and Cys11–Cys26); a similar motif in LRRCT (CXCXnCXnC) forms disulfide pairs Cys174–Cys201 and Cys176–Cys221. These disulfide pairs are structurally equivalent to those in the LRRNT and LRRCT modules of VLRAs and VLRBs (22).
Fig. 3.
Structure of L. planeri VLRC. (A, Upper) Side view of the VLRC.1MP solenoid. LRRNT, magenta; LRR1, LRRV1–3, and LRRVt, blue; CP, yellow; LRRCT, green. (A, Lower) VLRC.1MP rotated 60° from the above orientation to show a front view of the putative antigen-binding concave surface. (B) Comparison of VLRC with VLRA and VLRB. Superposition of VLRC.1MP (magenta) onto lamprey VLRA.R2.1 (PDB accession code 3M18) (blue), lamprey VLRB.aGPA.23 (4K5U) (orange), hagfish VLRB.59 (2O6S) (gray), and hagfish VLRC.29 (2O6Q) (green). Superpositions were carried out by deleting the additional LRRV modules of lamprey VLRA.R2.1 and hagfish VLRC.29 relative to VLRC.1MP. The protruding LRRNT loop of VLRC.1MP is marked with a red arrow. The long LRRCT inserts of VLRA and VLRB are indicated with arrows in corresponding colors. (C) Superposition of VLRC.1MP (magenta) onto Japanese lamprey VLRC (3WO9) (27) (marine), highlighting conformational differences in LRRNT (Lower Left) and LRRCT (Lower Right) loops.
Unique Structural Features of VLRC Receptors.
Superposition of VLRC.1MP onto VLRA and VLRB structures gave root mean square (rms) differences in Cα positions of <2 Å for equivalent Cα atoms, indicating close similarity: 1.7 Å for lamprey VLRA.R5.1 (Protein Data Bank, PDB accession code 3M19) (23), 1.5 Å for lamprey VLRA.R2.1 (3M18) (23), 1.3 Å for lamprey VLRB.aGPA.23 (4K5U) (24), 1.4 Å for lamprey VLRB.RBC36 (3E6J) (14), 1.9 Å for lamprey VLRB.2D (3G3A) (15), 1.9 Å for lamprey VLRB.VLR4 (3TWI) (25), 1.1 Å for hagfish VLRB.59 (2O6S) (26), and 1.8 Å for hagfish VLRB.61 (2O6R) (26) (Fig. 3B). Superposition of VLRC.1MP onto two other VLRCs, hagfish VLRC.29 (2O6Q) (45% sequence identity) (26) and a VLRC from Japanese lamprey (Lethenteron japonicum) (3WO9) (78% sequence identity) (27), gave rms differences in Cα positions of 1.4 and 0.9 Å, respectively.
Compared with VLRAs and VLRBs, VLRC.1MP contains a loop insert of eight amino acids (residues 15–22) in LRRNT (Fig. S1C) that was also present in Japanese lamprey VLRC (27). This loop, which connects two antiparallel β-strands in LRRNT (β1 and β2), projects toward the concave surface of VLRC.1MP and could therefore potentially contact antigen or an antigen-presenting molecule (Fig. S1D). The LRRNT loop inserts of VLRC.1MP and Japanese lamprey VLRC have nearly identical sequences (NKTESSPD and NKTDSSPD, respectively) (Fig. S1C), but they adopt markedly different conformations in the two VLRs (Fig. 3C), thereby implying flexibility. Thus, residues Glu18 and Ser19 at the tip of the LRRNT loop of VLRC.1MP are displaced by an average of 3.4 Å in the position of their Cα atoms relative to the corresponding residues of Japanese lamprey VLRC. VLRC.1MP also differs from Japanese lamprey VLRC in the conformation of a loop in LRRCT with identical sequence in the two VLRCs that flips nearly 90° toward the concave surface of VLRC.1MP (Fig. 3C); such flexibility suggests a mechanism for antigen binding by VLRCs based on conformational selection from a dynamic equilibrium of preexisting isomers, as proposed for VLRAs and VLRBs (22, 25). According to this mechanism, the intrinsically mobile LRRCT insert samples multiple conformations, only one of which is able to bind to a particular ligand. This conformational diversity may permit a single VLR to engage multiple ligands, thereby expanding the effective size of the repertoire, as demonstrated for antibodies (28).
The eight-residue insert loop linking strands β1 and β2 in LRRNT of VLRC.1MP is present in all known lamprey VLRC sequences, and is characterized by low sequence diversity (Fig. S1C). By contrast, the number of corresponding residues is only four in hagfish VLRC, four in lamprey VLRA, four in hagfish VLRA, and two in hagfish VLRB, which in each case are too few residues to form a protrusion in LRRNT that could possibly contact antigen (Fig. 3B). Although most (93%) lamprey VLRB also lack the LRRNT protrusion, 7% have inserts of six or eight residues that may form protruding loops similar to lamprey VLRC (Fig. S1C).
The LRRCT module of VLRAs and VLRBs contains a distinctive insert that is highly variable in both sequence and length (0–13 residues) (Fig. 3B). This insert, whose secondary structure also varies, is critical for recognition of protein and carbohydrate antigens (14, 15, 22–25). The average length of the LRRCT insert in VLRCs (2 residues) is much less than in VLRAs (10 residues) or VLRBs (8 residues), and sequence variation in the LRRCT module of VLRCs is substantially lower than in LRRCT of the other two VLR types (19, 29). In the VLRC.1MP structure, the LRRCT insert comprises only 2 residues and does not protrude from LRRCT or extend across the concave surface of the VLR. This structural feature is characteristic of both lamprey and hagfish VLRCs (26, 27) and distinguishes them from VLRAs and VLRBs.
Signatures of Phylogenetic Adaptations in VLRC Sequences.
Although the above analyses suggest the presence of selection of VLRCs according to size, other parameters must also influence the final outcome of repertoire selection; this additional requirement(s) is illustrated by the presence in VLRA+ cells of functionally assembled VLRC genes of the preferred length that must have failed to support inclusion in the VLRC+ pool (Fig. 2C). To gain insight into sequence-specific selection, we examined the presence and distribution of high entropy amino acid residue positions in VLRC molecules. In this context, entropy essentially indicates the degree of sequence diversity at a given position in the molecule; invariant sites exhibit low entropy scores and variable sites, high entropy scores.
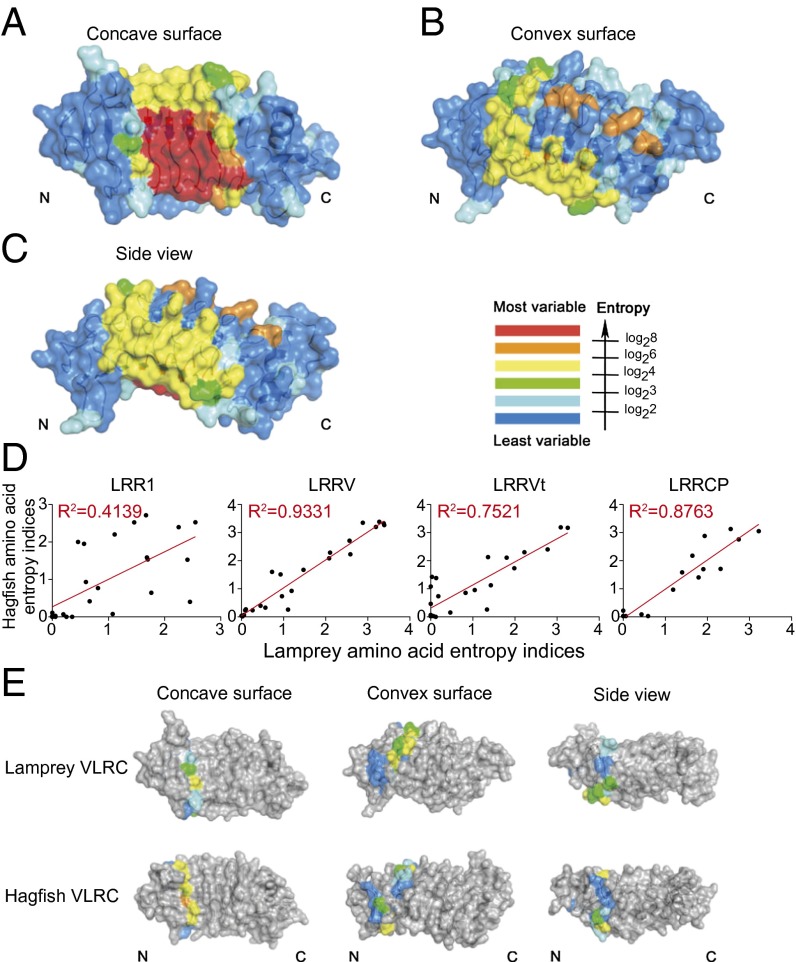
In general terms, as with VLRA and VLRB, variable aromatic residues are most frequently observed in VLRC high entropy positions. This, together with the spatial distributions of the high entropy sites in the LRRV modules suggests that the general mode of antigen engagement by the central concave region is conserved across the VLRs. As noted previously (27), VLRC molecules exhibit several high entropy sites on the convex side of the molecule, in addition to those on the concave, presumably antigen-binding surface (Fig. 4 A–C and Fig. S2C). However, sequence variations on the convex face of VLRCs are unlikely to affect the conformation of residues on the concave face, because of the structural rigidity of LRR modules. Little sequence variation is found in LRRNT and LRRCT components of the molecule, as these are constant parts derived primarily from the single incomplete VLRC genes in the three species; by contrast, substantial sequence variability is observed in the LRR1, LRRV, and LRRCP segments of the molecule. This pattern of high entropy sites across the VLRC molecule is evolutionarily conserved between lamprey species (Fig. S2A and Dataset S3). Similarly, the distribution of high entropy sites is similar for two hagfish species (Fig. S2B and Dataset S3). Accordingly, for the most variable parts of VLRC molecules (LRR1, LRRV, and LRRCP), the entropy scores of the three lamprey species and the two hagfish species are highly correlated (Fig. S3 and Dataset S3). Interclade comparisons between lampreys and hagfishes reveal highly correlated entropy scores only for LRRV and LRRCP modules, whereas those for the LRR1 module show only weak correlations (Fig. 4D, Fig. S3, and Dataset S3). The distinct sequence signatures thus suggest that among variable parts of the VLRC molecule, evolutionary divergence is particularly pronounced for the LRR1 module (Fig. 4E). The poorly conserved entropy patterns of lamprey and hagfish LRRNT modules are consistent with the clade-specific differences in the invariant part of the LRRNT region (Fig. 3 and Fig. S1). Strikingly, the LRRCT modules also showed no correlation between lamprey and hagfish VLRC. Here, in contrast to the low variability of lamprey versions, the hagfish contained several high entropy positions (Fig. S2B). These observations suggest that unlike the lamprey VLRC, the hagfish version uses LRRCT extensively to engage antigen, whereas the lamprey VLRCs exhibit a bias toward a more N-terminal engagement.
Fig. 4.
Sequence variability of VLRCs. (A–C) Sequence variability mapped onto the VLRC.1MP structure indicating the degree of sequence diversity in color code. (D) Correlation of entropy values between hagfish and lamprey VLRC sequences. Shannon entropy scores were generated for each amino acid position in the aligned VLRC sequences of hagfish and lamprey species and individually shown as scatterplots for LRR1, LRRV, LRRVt, and LRRCP modules (Dataset S3 for data on LRRNT and LRRCT); the coefficients of determination (R2) were calculated from linear regression to provide a measure of the relationship between the sequences of the two clades in terms of module diversity. (E) Sequence variability of LRR1 modules of lamprey and hagfish VLRCs. Color code indicates the degree of sequence diversity as in A–C.
When entropy correlations were determined between lamprey VLRA and VLRC isotypes, LRRNT, LRR1, LRRCP, and LRRCT modules exhibited weak correlations, consistent with isotype-specific adaptions; as expected, the entropy patterns are very similar for LRRV elements (Fig. S4 and Dataset S3).
Collectively, the module-specific entropy differences revealed by intra- and interclade comparisons of VLRA and VLRC isotypes described above suggest that adaptive changes are particularly pronounced in the LRR1 module of VLRC molecules over evolutionary time. This led us to hypothesize that the LRR1 module would also be the target of sequence-specific selection of functional VLRC elements during the elaboration of the peripheral VLRC repertoire. An alternative explanation for the high entropy of LRR1 could be that this module is simply less constrained than other VLRC elements; however, this alternative is not supported by the structural features of the VLRC molecule.
Sequence-Related Selection Process of VLRC Receptors.
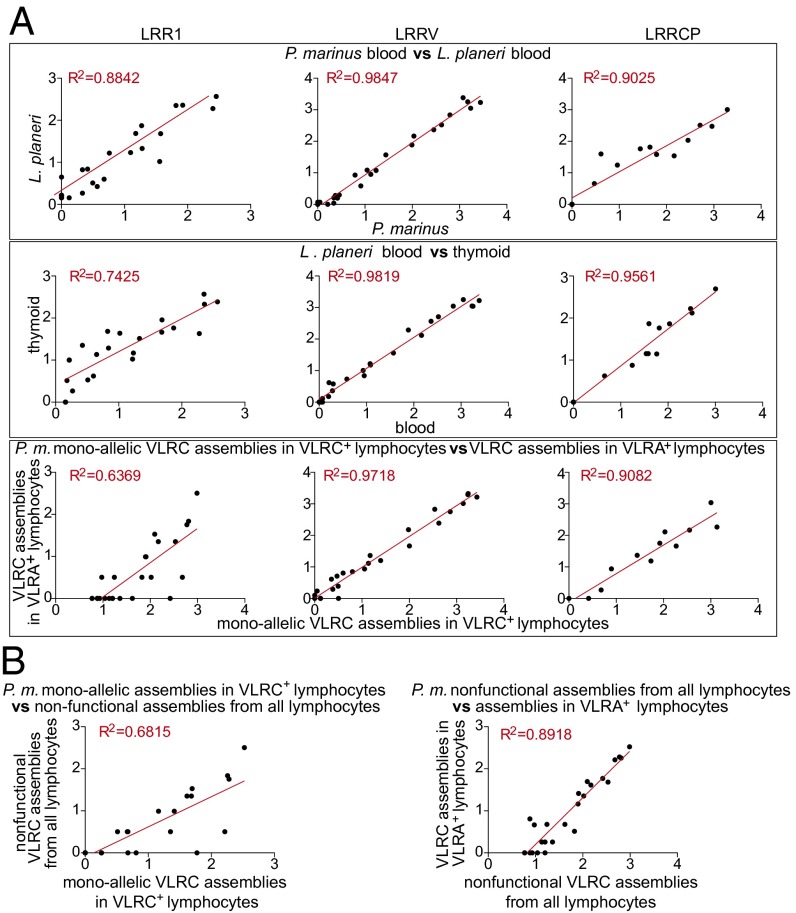
To further explore the possibility of LRR1-specific selection, we examined correlations of entropy values of LRR1 modules for different types of VLRC assemblies (Fig. 5A). As expected from clade-specific comparisons, LRR1 entropy values of VLRC assemblies in peripheral blood of L. planeri and P. marinus—consisting mostly of functionally selected assemblies—are very similar (R2 = 0.88); however, this degree of similarity is less pronounced than that for LRRV modules (P = 0.0006; Dataset S4). Entropy patterns of LRRCP elements of L. planeri and P. marinus are also very similar (R2 = 0.90), yet the degree of similarity is again less than that of LRRV modules (P = 0.0068). These results are compatible with the notion that among LRR1, LRRV, and LRRCP elements, species-specific differences occur primarily in LRR1 and LRRCP elements. A comparison between VLRC assemblies in the thymoid (consisting of a mixture of pre- and postselection assemblies) and those in the peripheral blood of L. planeri reveals a different picture. The correlation of entropy patterns is again weak for LRR1 (R2 = 0.74), but is substantially higher for both LRRV (R2 = 0.98) and LRRCP (R2 = 0.96) elements. Accordingly, the entropy patterns of LRR1 modules differ significantly from those of LRRV (P = 4 × 10−6) and LRRCP (P = 0.007) modules, whereas the entropy patterns of LRRV and LRRCP do not differ significantly (P = 0.2). Thus, whereas an interspecific comparison between L. planeri and P. marinus suggests that the differences in entropy patterns of LRRCP modules are the result of species-specific adaptations, an intraspecific comparison implicates the LRR1 module as a primary target of selection. This conclusion is supported by a comparison of the entropy patterns of functional VLRC assemblies observed in VLRC+ and VLRA+ cells of P. marinus. Whereas the correlation for LRR1 modules is weak (R2 = 0.64), those of LRRV and LRRCP modules are much stronger (R2 = 0.97 and R2 = 0.91, respectively) (Fig. 5A, Bottom), again emphasizing that only the entropy patterns of LRR1 modules are significantly different in these two groups of VLRC assemblies (Dataset S4). Next, we examined the correlation of entropy values for LRR1 modules between monoallelic VLRC assemblies of VLRC+ cells and LRR1 modules of completely assembled but nonfunctional VLRC genes; this comparison was possible because the majority of the frame shifts and in-frame stop codons in nonfunctional assemblies occur downstream of the LRR1 module. As expected, only a weak correlation was observed (R2 = 0.68); interestingly, the entropy values are highly correlated between nonfunctional VLRC assemblies and functional assemblies found in VLRA+ cells (R2 = 0.89), supporting the notion that both types of assemblies exhibit features of the preselection repertoire (Fig. 5B).
Fig. 5.
LRR1 diversity shows poor correlation between different groups of VLRC assemblies. The relationships of entropy values are displayed as scatterplots (see Fig. 4D) (A and B) for groups of VLRC assemblies described in Fig. 2 A–D and G. The coefficients of determination (R2) are indicated. Significant differences between the different correlation coefficients (R) were assessed using Fisher’s z transformation and are tabulated in Dataset S4. Entropy correlations for LRRV modules did not include LRRVt sequences.
In conclusion, our data suggest that the formation of the peripheral repertoire is the result of both length- and sequence-specific selection processes targeting the preselection repertoire.
Discussion
This study identifies unique characteristics of peripheral repertoires of VLRC receptors that are indicative of selection. Two levels of selection have become apparent, the first pertaining to the number of internal LRR (LRRV) modules, the second affecting protein sequence composition of individual modules. During the formation of complete VLRC genes, the assembly process appears to incorporate four or five LRRV modules with approximately similar probability, as indicated by the size distributions of nonfunctional VLRC genes in VLRC+ and VLRA+ cells, functional VLRC assemblies in VLRA+ cells, and VLRC assemblies in the thymoid. By contrast, assemblies with four LRRV elements predominate in peripheral lymphocytes. This suggests selection of VLRCs on the basis of their length after completion of assembly in the thymoid. A possible mechanism to explain such selection would be a requirement for VLRCs to interact with another protein on the lymphocyte surface to form a signaling complex that is able to deliver a maturation signal to developing VLRC+ lymphocytes. If formation of this putative signaling complex requires protein(s) to simultaneously engage both N- and C-terminal sites on the VLRC, this protein (or proteins) could act as a kind of “molecular ruler” to select for VLRCs with lengths (i.e., number of LRRVs) that fall within a certain range. Because the accessory components of the VLRC signaling complex are likely structurally invariant, they probably interact with those parts of the VLRC molecule that exhibit minimal sequence variability, such as LRRNT and LRRCT, in analogy to the interaction of the CD3 complex with the constant regions of the T-cell receptor chains (30). Alternatively, length selection could be imposed by a molecule(s) functionally analogous to MHC expressed on thymoid stromal cells with which VLRC lymphocytes interact during development, if that molecule(s) also binds VLRCs at both N- and C-terminal regions. However, such molecules, if they exist, are not expected to be structurally related to MHC molecules, because VLRCs and TCRs do not share any structural homology.
It is unclear whether the selection of VLRC molecules based on the number of LRRV modules as observed here is related to their mode of antigen recognition. Nonetheless, length selection of the antigen binding surface appears to be unique to antigen receptors of jawless fishes, because CDR3 length distributions of α and β TCR chains, which are the most constrained among the antigen receptors of jawed vertebrates, are unaltered by thymic selection (31).
A second layer of selection of VLRCs became apparent when we determined the Shannon entropy indices for each amino acid position in VLRC molecules of different origins. Significant deviations from the peripheral repertoire of functional VLRC sequences in VLRC+ cells were found in comparisons to collections of VLRC molecules reflecting preselection stages, such as those of thymoid and those in VLRA+ cells, the latter probably resulting from nonproductive VLRC selection. Comparison of different parts of VLRC molecules indicated that the most prominent selection signal is detectable in the first LRR element (LRR1) of VLRC assemblies. Interestingly, this region also features high entropy sites situated on the convex side of the molecule, indicating that any mechanism explaining sequence-specific selection of VLRC molecules must take into account possible interaction(s) with the back side of the VLRC molecule. Whereas the selection based on length of VLRC molecules likely involves structurally invariant interaction partners, selection based on sequence could also involve structurally diverse molecules. It is unclear at present whether the hypothetical molecular complex mediating this interaction also involves self-antigen.
In conclusion, our findings to our knowledge provide the first indication of selection impacting on the primary repertoire of a lamprey antigen receptor and pave the way for the identification of presumptive interaction molecules mediating this process.
Materials and Methods
The diversity region of VLRC.1MP was expressed by in vitro folding from bacterial inclusion bodies. The crystal structure of VLRC.1MP was determined by molecular replacement. Details on the determination of VLRC repertoires, sequence and statistical analyses, protein production and purification, and crystallization and structure determination can be found in SI Materials and Methods. Animal experiments were approved by the Institutional Animal Care and Use Committee at Emory University and the Review Committee of the Max Planck Institute.
Supplementary Material
Acknowledgments
This work was supported by the Max Planck Society and has received funding from the European Research Council (ERC) under the European Union's Seventh Framework Programme (FP7/2007-2013), ERC Grant Agreement ERC-2012-AdG–323126. R.A.M. was supported by National Institutes of Health (NIH) Grant AI036900. M.L. was supported by the Joint Supervision PhD Project of the China Scholarship Council. Work by L.M.I. and L.A. is funded by the Intramural Research Program of the NIH, Department of Health and Human Services, and that of M.D.C. and M.H. by NIH Grants AI072435 and GM100151.
Footnotes
The authors declare no conflict of interest.
Reviewers: J.P.R., Sunnybrook Research Institute/University of Toronto; J.W., Dana-Farber Cancer Institute, Harvard Medical School.
Data deposition: The sequences reported in this paper have been deposited in GenBank database (accession nos. KJ734027–KJ734078, KJ751404–KJ751454, KJ649525–KJ649607, KJ670495, and KC732806–KC733164). Atomic coordinates and structure factors for VLRC.1MP have been deposited in the Protein Data Bank (PDB) (PDB ID code 4PO4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415655111/-/DCSupplemental.
References
- 1.Pancer Z, et al. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430(6996):174–180. doi: 10.1038/nature02740. [DOI] [PubMed] [Google Scholar]
- 2.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124(4):815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Herrin BR, Cooper MD. Alternative adaptive immunity in jawless vertebrates. J Immunol. 2010;185(3):1367–1374. doi: 10.4049/jimmunol.0903128. [DOI] [PubMed] [Google Scholar]
- 4.Boehm T, et al. VLR-based adaptive immunity. Annu Rev Immunol. 2012;30:203–220. doi: 10.1146/annurev-immunol-020711-075038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasahara M, Sutoh Y. Two forms of adaptive immunity in vertebrates: Similarities and differences. Adv Immunol. 2014;122:59–90. doi: 10.1016/B978-0-12-800267-4.00002-X. [DOI] [PubMed] [Google Scholar]
- 6.Hirano M, et al. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature. 2013;501(7467):435–438. doi: 10.1038/nature12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pancer Z, et al. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005;102(26):9224–9229. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Das S, Herrin BR, Hirano M, Cooper MD. Definition of a third VLR gene in hagfish. Proc Natl Acad Sci USA. 2013;110(37):15013–15018. doi: 10.1073/pnas.1314540110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alder MN, et al. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310(5756):1970–1973. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- 10.Alder MN, et al. Antibody responses of variable lymphocyte receptors in the lamprey. Nat Immunol. 2008;9(3):319–327. doi: 10.1038/ni1562. [DOI] [PubMed] [Google Scholar]
- 11.Nagawa F, et al. Antigen-receptor genes of the agnathan lamprey are assembled by a process involving copy choice. Nat Immunol. 2007;8(2):206–213. doi: 10.1038/ni1419. [DOI] [PubMed] [Google Scholar]
- 12.Rogozin IB, et al. Evolution and diversification of lamprey antigen receptors: Evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8(6):647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- 13.Guo P, et al. Dual nature of the adaptive immune system in lampreys. Nature. 2009;459(7248):796–801. doi: 10.1038/nature08068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321(5897):1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velikovsky CA, et al. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat Struct Mol Biol. 2009;16(7):725–730. doi: 10.1038/nsmb.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrin BR, et al. Structure and specificity of lamprey monoclonal antibodies. Proc Natl Acad Sci USA. 2008;105(6):2040–2045. doi: 10.1073/pnas.0711619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tasumi S, et al. High-affinity lamprey VLRA and VLRB monoclonal antibodies. Proc Natl Acad Sci USA. 2009;106(31):12891–12896. doi: 10.1073/pnas.0904443106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajoghli B, et al. A thymus candidate in lampreys. Nature. 2011;470(7332):90–94. doi: 10.1038/nature09655. [DOI] [PubMed] [Google Scholar]
- 19.Das S, et al. Organization of lamprey variable lymphocyte receptor C locus and repertoire development. Proc Natl Acad Sci USA. 2013;110(15):6043–6048. doi: 10.1073/pnas.1302500110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perey DYE, Finstad J, Pollara B, Good RA. Evolution of the immune response. VI. First and second set skin homograft rejections in primitive fishes. Lab Invest. 1968;19(6):591–597. [PubMed] [Google Scholar]
- 21.Raison RL, Gilbertson P, Wotherspoon J. Cellular requirements for mixed leucocyte reactivity in the cyclostome, Eptatretus stoutii. Immunol Cell Biol. 1987;65(Pt 2):183–188. doi: 10.1038/icb.1987.20. [DOI] [PubMed] [Google Scholar]
- 22.Deng L, Luo M, Velikovsky A, Mariuzza RA. Structural insights into the evolution of the adaptive immune system. Annu Rev Biophys. 2013;42:191–215. doi: 10.1146/annurev-biophys-083012-130422. [DOI] [PubMed] [Google Scholar]
- 23.Deng L, et al. A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc Natl Acad Sci USA. 2010;107(30):13408–13413. doi: 10.1073/pnas.1005475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo M, et al. Recognition of the Thomsen-Friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor. J Biol Chem. 2013;288(32):23597–23606. doi: 10.1074/jbc.M113.480467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchdoerfer RN, et al. Variable lymphocyte receptor recognition of the immunodominant glycoprotein of Bacillus anthracis spores. Structure. 2012;20(3):479–486. doi: 10.1016/j.str.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim HM, et al. Structural diversity of the hagfish variable lymphocyte receptors. J Biol Chem. 2007;282(9):6726–6732. doi: 10.1074/jbc.M608471200. [DOI] [PubMed] [Google Scholar]
- 27.Kanda R, et al. Crystal structure of the lamprey variable lymphocyte receptor C reveals an unusual feature in its N-terminal capping module. PLoS ONE. 2014;9(1):e85875. doi: 10.1371/journal.pone.0085875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299(5611):1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 29.Kasamatsu J, et al. Identification of a third variable lymphocyte receptor in the lamprey. Proc Natl Acad Sci USA. 2010;107(32):14304–14308. doi: 10.1073/pnas.1001910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J-H, Reinherz EL. The structural basis of αβ T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol Rev. 2012;250(1):102–119. doi: 10.1111/j.1600-065X.2012.01161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rock EP, Sibbald PR, Davis MM, Chien Y-H. CDR3 length in antigen-specific immune receptors. J Exp Med. 1994;179(1):323–328. doi: 10.1084/jem.179.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.