Significance
The advance of new clinical treatment options for cancer relies heavily on the discovery of chemotherapeutic agents with new modes of action. In this paper, we describe the discovery of a potent melanoma-selective agent, seriniquinone, and elucidate its targeting of dermcidin and dermcidin-conjugated proteins within tumor cells. Early evidence indicates a direct correlation between seriniquinone activity and the levels of dermcidin within an ascribed tumor cell line, therein suggesting not only a unique target and avenue for further therapeutic exploration but also, and perhaps more importantly, revealing that dermcidin that may play a dual role as a diagnostic patient preselection biomarker and a drug target.
Keywords: mode of action, chemical biology, drug discovery, marine natural products, cancer
Abstract
Natural products continue to provide vital treatment options for cancer. Although their translation into chemotherapeutics is complex, collaborative programs continue to deliver productive pipelines for cancer chemotherapy. A new natural product, seriniquinone, isolated from a marine bacterium of the genus Serinicoccus, demonstrated potent activity over a select set of tumor cell lines with particular selectivity toward melanoma cell lines. Upon entering the cell, its journey began by localization into the endoplasmic reticulum. Within 3 h, cells treated with seriniquinone underwent cell death marked by activation of autophagocytosis and gradually terminated through a caspase-9 apoptotic pathway. Using an immunoaffinity approach followed by multipoint validation, we identified the target of seriniquinone as the small protein, dermcidin. Combined, these findings revealed a small molecule motif in parallel with its therapeutic target, whose potential in cancer therapy may be significant. This discovery defines a new pharmacophore that displayed selective activity toward a distinct set of cell lines, predominantly melanoma, within the NCI 60 panel. This selectivity, along with the ease in medicinal chemical modification, provides a key opportunity to design and evaluate new treatments for those cancers that rely on dermcidin activity. Further, the use of dermcidin as a patient preselection biomarker may accelerate the development of more effective personalized treatments.
Over the last decade, we have found that integrating a streamlined transition between marine natural products discovery (1), detailed cell and molecular biological studies (2), and medicinal chemical optimization offers a rich forum to advance leads that escape industrialized high-throughput practices (3–5). One place where drug discovery continues to be challenged lies in the discovery of new agents, which are highly selective for specific cancers (6–8).
Malignant melanoma, classified by genetic defects within pigment-producing melanocytes, is attributed to the largest number of skin-related cancer deaths (9). Although protective measures can be taken, the aggressive and rapid metastatic properties of melanoma-based tumors continue to challenge clinical practices. The recent introduction of dabrafenib (Tafinlar), trametinib (Mekinist), and ipilimumab (Yervoy) promises to provide improved melanoma treatment (9); however, selective melanoma drugs are still of great interest. In addition, the unique epidemiological concerns for the generation of melanoma (10) and the lack of curative treatment options (11) places melanoma as one of the most dangerous of cancers.
Results and Discussion
With the goal of contributing to the treatment of melanoma, we evaluated a large extract library containing novel metabolites produced by Gm+ marine bacteria (12). We identified seriniquinone (1, Fig. 1A), molecular formula C20H8O4S, from culture broths of a rare marine bacterium of the genus Serinicoccus (Fig. 1B). The structure of 1 (Fig. 1A) was deduced using chemical and spectroscopic methods (SI Appendix, Table S1). Although an undiscovered natural product, the synthesis of 1 (Fig. 1C) had been reported as part of a materials science program, allowing us to further validate our structural assignment (13). Our interest in 1 began after observing significant selectivity in melanoma (six of nine cell lines screened) along with additional cell lines (SR leukemia, HOP-62, NCI-H460 and NCI-H23 nonsmall cell lung; HCT-15 colon; SNB-75 CNS; IGROV1 and OVCAR-3 ovarian; DU-145, prostate; and MDA-MB-231 and HS 578T, breast cancers) with mean GI50 values of 758 nM and LC50 values of 13.2 μM across the NCI-60 panel (SI Appendix) (14). Given this selectivity, we turned our attention to probe its mode of action (MOA).
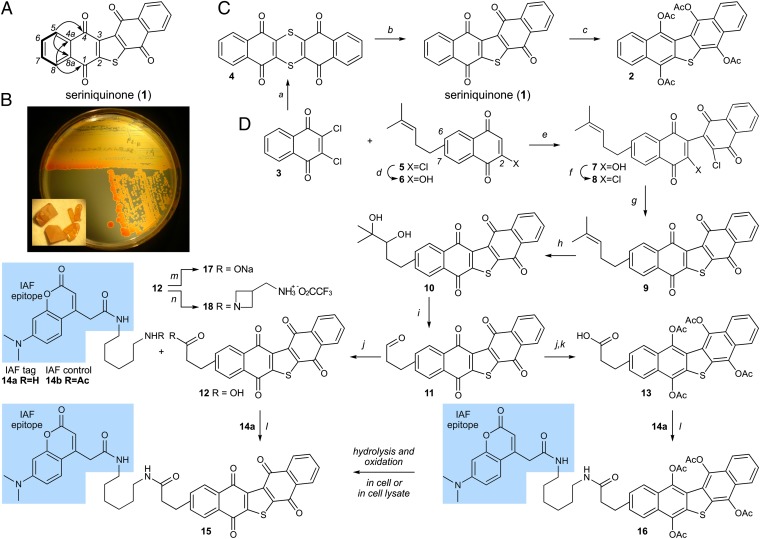
Fig. 1.
Isolation and chemical synthesis. (A) Structure of dinaphtho[2,3-b:2',3′-d]thiophene-5,7,12,13-tetraone or seriniquinone (1). Summary of salient NMR data including 1H-1H COSY (bold lines) and heteronuclear multiple-bond correlation spectroscopy correlations (arrows) observed in solutions of 1 in CDCl3. (B) Images depicting the marine bacterium Serinicoccus sp. strain CNJ927 growing on a nutrient agar surface, and crystals of 1 (Inset). Culture broths of Serinicoccus sp., strain CNJ927, provided 0.067 mg/L of 1. (C) Repetition of the published synthesis of 1 (13) confirmed the structure of the natural product. Tetra-acetate hydroquinone 2 was prepared as a prodrug motif, which when absorbed in cells underwent acetate hydrolysis followed by oxidation to 1 (SI Appendix, Fig. S2). (D) Advance of synthetic methods permitted access to IAF probes 15 and 16, as well as derivatives 10, 12, 17, and 18. Reagents and conditions: (a) dithiooxamide, Et3N, N,N-dimethylformamide (DMF), 50 °C, 10 h, 85%; (b) 30% aq. H2O2, AcOH, reflux, 3 h, 86%; (c) Zn dust, Ac2O, reflux, 2 h, 67%; (d) NaOH, MeOH, 2 h; (e) CsCO3, CH3CN, rt, 72 h; (f) (ClCO)2, DMF, CH2Cl2, 2 h, 49% (over 3 steps from 5); (g) Na2S, aq. THF, rt, 1h; (h) K2OsO4 • 2 H2O, N-methylmorpholine oxide, THF, H2O, 16 h, 67% (over 2 steps from 8); (i) NaIO4 on SiO2, CH2Cl2, 2 h, 98%; (j) oxone, DMF, 2 h, 98%; (k) Zn dust, Ac2O, reflux, 2 h, 65%; (l) 1-[bis(dimethylamino) methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU), EtNiPr2, DMF, rt, 2 h, 29% for 16; (m) NaOMe, MeOH, 16 h, 24%; (n) tert-butyl acetidine-3-ylmethylcarbamate, HATU, EtNiPr2, DMF, rt, 2 h, then TFA, CH2Cl2, 1 h, 42% (two steps). Starting material 5 was obtained as batch-dependent inseparable 5:2–6:1 mixture of 2-chloro-7-(4-methylpent-3-en-1-yl)naphthalene-1,4-dione (major) and 2-chloro-6-(4-methylpent-3-en-1-yl)naphthalene-1,4-dione (minor).
Fortunately, 1 was fluorescent and could be imaged during uptake and induction of cell death in multiple tumor cell lines. Using confocal microscopy, green–red fluorescence from 1 appeared in HCT-116 colon carcinoma cells within minutes of incubation (Fig. 2A). Comparable localization was also observed in other cell lines including the more responsive Malme-3M melanoma cells (Fig. 2B). We then combined time-course confocal imaging studies (Fig. 2C) with conventional cytotoxicity 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assays (15), which indicated an optimal activity arising in HCT-116 cells that were treated with 1 over 24 h (Fig. 3 A and B).
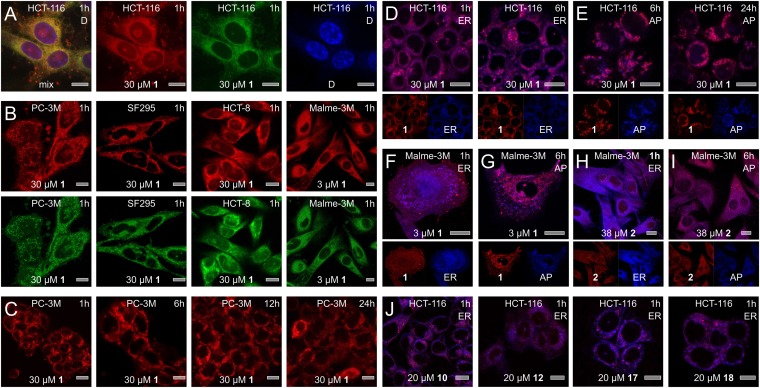
Fig. 2.
Subcellular localization studies. (A) Fluorescence from 1 was characterized by excitation maxima at 250 nm (ε = 20,800 cm−1 M−1), 289 nm (ε = 24,000 cm−1 M−1), and 342 nm (ε = 6,900 cm−1 M−1) and a broad emission from 490 to 680 nm, providing green–red fluorescence. Treatment of HCT-116 cells with 1 for 1 h results in the appearance of green–red fluorescence from 1 around the nucleus, as indicated by counterstaining with DAPI. (B) Similar subcellular localization was observed across a panel of cell lines that demonstrated sensitivity to 1. (C) Time-course analysis. Within 1 h, 1 localizes within the ER then induces autophagocytosis with 1 localizing in the forming autophagosomes (AP). (D–G) Counterstaining of the ER and AP was used to validate the subcellular localization events. Images depict staining of cells with ER stain at 1 and 6 h and AP stain at 6 and 24 h. (F–G) Similar localization was observed in Malme-3M cells albeit at lower concentrations of 1. (H–I) Treatment of Malme-3M cells with 2 also results in comparable but slower localization H, in the ER followed by I, the transition to AP. (J) Analogs 10, 12, 17, and 18 also undergo ER localization followed by transitioning into the forming AP. Each image is marked with cell line (Top Center), compound (Bottom Center), dose (Bottom Center), time (Top Right), and counterstain (Top Right). Counterstains are indicated by the following labels: ER = Tracker blue-white (ER), AP = dansylcadavarine (autophagosomes), and D = DAPI (nucleus). Bars denote 10 µm.
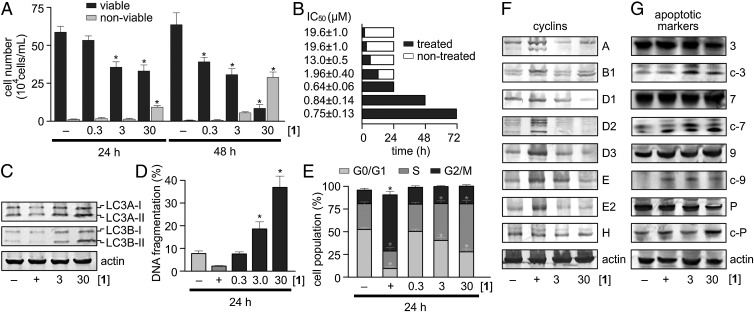
Fig. 3.
Activity data and cell cycle studies. (A) Seriniquinone (1) induces cell death in HCT-116 cells in a concentration- and time-dependent manner. (B) Time-course studies show that the optimal activity from 1 in HCT-116 cells is obtained after constant treatment with 1 over 24 h. (C) Western blot analysis confirms compromise of autophagosome maturation by a dose-dependent increase in the levels of LC3A-II and LC3B-II (two posttranslationally modified light chain 3 [LC3] protein isoforms whose levels increase during autophagy) after treatment of HCT-116 cells with 1 for 24 h. This response was significantly greater than negative (–, DMSO) and positive (+, 17 µM etoposide) controls. (D) Cell death is accompanied by DNA fragmentation suggesting that apoptotic processes are elicited. (E) Compound 1 induces a dose-dependent arrest during DNA replication as noted by the increase in S staged cells. Western blot analyses for selected: (F) cyclins, as given by A = cyclin A, B = cyclin B1, D1 = cyclin D1, D2 = cyclin D2, D3 = cyclin D3, E = cyclin E, E2 = cyclin E2, H = cyclin H; or (G) apoptotic markers, as given by 3 = caspase 3, c-3 = cleaved caspase 3, 7 = caspase 3, c-7 = cleaved caspase 7, 9 = caspase 9, c-9 = cleaved caspase 9, P = PARP, c-P = cleaved PARP, in lysates prepared from HCT-116 cells that were treated with 1 for 24 h. Negative and positive controls are given by (–) DMSO and (+) 17 µM etoposide, respectively. Actin was used as a loading control. Unless noted otherwise, all experiments were conducted in HCT-116 cells and concentrations are provided in µM. * denotes P < 0.05.
We then applied a series of stains to identify the subcellular distribution of 1. Counterstaining with endoplasmic reticulum (ER) tracker blue–white (16) illustrated that 1 appeared in the ER rapidly after treatment. Within 2–3 h, vesicles formed bearing fluorescence from 1 (Fig. 2D). This transition was complete at 6–9 h, at which point the fluorescence from 1 correlated with an autophagosomal stain, monodansylcadaverine (Fig. 2E) (17). This observation was even more pronounced in Malme-3M melanoma cells (Fig. 2F). The fact that 1 induced autophagy was further confirmed by conversion of autophagy markers LC3A-I to LC3A-II and LC3B-I to LC3B-II, respectively (Fig. 3C) (18).
Next, we screened for the effects of 1 on the cell cycle. Flow cytometry analyses indicated an increasing number of cells with a subdiploid DNA suggesting fragmentation (Fig. 3D). The remaining cells were arrested at the S to G2 phase, indicating that treated cells were dying and not reentering the cell cycle (Fig. 3E). Analysis of cyclin expression in HCT-116 treated cells reinforced this hypothesis, showing a strong reduction in the expression of cyclins D1–D3 (Fig. 3F), proteins whose cyclin-dependent kinase complexes are essential for entry into G1 (19). We also monitored conventional apoptotic markers for further documentation of cell death. Treatment of HCT-116 cells with 1 was accompanied by increased cleavage of caspases 3, 7, and 9 and Poly (ADP-ribose) polymerase (PARP), suggesting that 1 triggers cell death through a caspase-9 dependent event, thus leading to overall observation of DNA fragmentation (Fig. 3G) (19).
With a phenotype at hand, we screened for proteins targeted by 1 using an immunoaffinity fluorescent (IAF) technique (20, 21). The first step required preparation of an IAF probe, a task that was complicated by the lack of a functional group in 1 to append an IAF tag. After evaluating multiple options, we identified a synthetic route to prepare derivatives of 1 bearing an aliphatic tail. Using the route in Fig. 1D, we were able to prepare IAF probe 15 in eight steps from 5 (22). We also prepared a second probe, 16 (Fig. 1D), which could serve to deliver 15 after hydrolysis and oxidation in tumor cells (SI Appendix, Fig. S2). As a means of further validation of this “proprobe” concept, we prepared the corresponding analog 2, a material obtained in one step from 1 (Fig. 1C, SI Appendix, Fig. S2).
We then screened these materials for their ability to mimic 1. Probe 15 maintained activity with an IC50 value of 250 ± 80 nM in HCT-116 cells, and the IAF control 14b (Fig. 1D) was inactive (IC50 value of > 50 µM). Further evidence from time-course imaging studies confirmed that 15 provided an effective mimic of 1 by its rapid uptake into the ER followed by transport to the forming autophagosomes (SI Appendix, Fig. S3). Similarly, compound 2 not only delivered comparable activity to 1 (Fig. 2), but also underwent hydrolytic release and oxidation in HCT-116 cells or cell lysates to afford 1 (SI Appendix, Fig. S2). Comparable observations were also seen for the conversion of 16 to 15 (SI Appendix, Fig. S2). Further, cytotoxicity analyses confirmed that 16 displayed comparable activity as 15 with an IC50 value of 240 nM in HCT-116 cells.
We then immunoprecipitated tumor cell lysates with either probe 15 or 16 using a mAb elicited against the IAF coumarin epitope to screen for proteins that bound to the seriniquinone motif (Fig. 1). Using established protocols (20, 21), HCT-116 cell lysates were treated for 8–12 h with 15 or 16, and immunoprecipitated with resins bearing the covalently attached anti-IAF mAb. Over multiple repetitions, we obtained a distinct series of proteins from 10 to 250 kDa, with increased levels of the immunoprecipitated proteins returned at higher concentrations of 15 (Fig. 4A) or 16 (Fig. 4B). Interestingly, the use of probe 16 provided a marked increase in the intensity of a band at 12 kDa, particularly when applied in live cells (L8–L9; Fig. 4B).
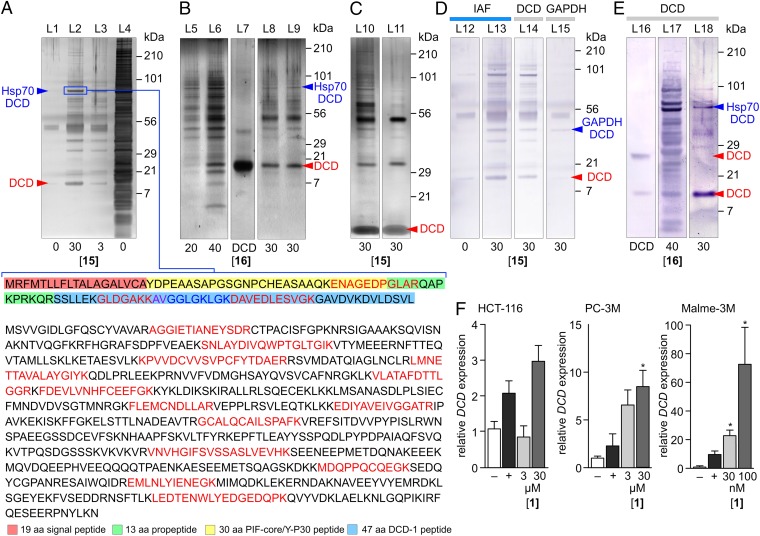
Fig. 4.
Seriniquinone (1) targets DCD. (A) Silver-stained SDS PAGE gel depicting proteins immunoprecipitated from HCT-116 cell lysates treated with 15 over 8 h at 4 °C. This response was dose dependent with an increasing return of immunoprecipitated protein at higher concentrations of 15 (L2 versus L3). Trypsin-digest LC-MS/MS analysis of the protein in a 90-kDa band returned peptides (colored in red or blue) corresponding to DCD (Upper) and Hsp70 (Lower). The DCD-derived, peptides were observed in the 30 aa PIF core, 13 aa propeptide and 47 DCD-1 peptide region (additional data provided in SI Appendix, Fig. S1). (B) Silver-stained SDS/PAGE gel depicting proteins immunoprecipitated from HCT-116 cell lysates treated with 16 at 4 °C over 12 h. DCD in L7 denotes the position of recombinant full length 110 aa DCD protein. (C) Treatment with iodoacetamide blocks the formation of disulfide linkages to DCD. A silver-stained SDS/PAGE gel provided in L10 depicts the proteins immunoprecipitated from HCT-116 cell lysates with 15 over 8 h, then treated with 5 mM iodoacetamide at room temperature for 1 h before SDS/PAGE analysis. A silver-stained SDS PAGE gel provided in L11 depicts the proteins from HCT-116 cell lysates that were treated for 1 h with 0.5 mM iodoacetamide, spin dialyzed, and then immunoprecipitated with 15 over 8 h. (D) Western blot evaluation of the immunoprecipitated fractions from cell lysates treated 15 were positive when developed with antibodies against the IAF tag in L13 and DCD in L14. A 50-kDa band (SI Appendix, Fig. S1) was also positive when stained with an antibody against GAPDH as shown in L15 suggesting that this band contained a fusion between GAPDH, 15, and DCD. L12 depicts a negative control in lysates without probe 15. (E) Western blot analysis depicting the presence of DCD fragments in the immunoprecipitated fraction from HCT-116 cell lysates treated with 16 in L17 at 4 °C over 12 h or lysates from HCT-116 cells that were cultured in media containing 16 in L18, compared with an authentic sample of the full 110 aa DCD protein in L16. (F) qPCR data showing increases in DCD mRNA in HCT-116, PC-3M, and Malme-3M cells treated with 1. The most profound increase in DCD mRNA was found in the most sensitive, Malme-3M cell line, as noted by our need to use reduced (nM) concentrations and the ∼10 fold increase in DCD mRNA expression in Malme-3M cells compared with that observed in HCT-116 and PC-3M cells. Unless noted otherwise, all experiments were conducted in HCT-116 cells and concentrations are provided in µM. Gel lanes are noted by L1-L18, respectively. *P < 0.05.
Trypsin digestion coupled with LC-MS/MS analyses identified Hsp70 (Fig. 4A) and GAPDH (SI Appendix, Fig. S1) within two of the more abundant bands. In each analysis, we also found peptide fragments that corresponded to the 30 aa proteolysis-inducing factor (PIF) core, 13 aa propeptide, and 47 aa DCD-1 peptide region of the poorly characterized dermcidin protein (DCD) (Fig. 4A) (23). LC-MS/MS analyses also identified a 12-kDa band as DCD itself (SI Appendix, Fig. S1). This suggested that the immunoprecipitated proteins in Fig. 4 A and B arose from a common linkage to DCD, which in turn was targeted by the seriniquinone pharmacophore.
The fact that peptides derived from DCD were observed in each of the bands [L2–L3 (Fig. 4A) and L5, L6, L7, L8 (Fig. 4B)], but not in gel controls or IP experiments without 15 (L1; Fig. 4A) was supported by a recent study that identified DCD peptides during the analysis of Hsp70 (24). In these studies, DCD was observed linked via a disulfide bond to Hsp70, which was released upon treatment with iodoacetamide. Interested if this were the case in our experimentation, we treated our immunoprecipitated fractions with 5 mM iodoacetamide before gel analysis. As indicated (L10; Fig. 4C), a low molecular weight band corresponding to DCD appeared. We further probed this observation by repeating the immunoprecipitation after treating the cell lysates with iodoacetamide (L11; Fig. 4C) and observed the isolation of DCD, thereby further validating the targeting of DCD by 1.
We were also able to confirm this observation by Western blot. As shown in Fig. 4D, we found that the immunoprecipitated GAPDH band (SI Appendix, Fig. S1) contained not only the IAF tag from 15, as shown by a positive blot using an anti-IAF antibody (L13; Fig. 4D), but also DCD (L14; Fig. 4D) and GAPDH (L15; Fig. 4D) compared with control (L12; Fig. 4C). Comparable results were also obtained when using 16, thus providing further evidence for the presence of DCD in the immunoprecipitated fraction (L17–L18; Fig. 4E). This was supported by a comparison with recombinant full-length DCD protein (L16; Fig. 4E).
Further validation was also obtained by examining gene expression data. qPCR analysis identified a statistically relevant increase in DCD mRNA in cells treated with 1 (Fig. 4F). Most importantly, an increasing level of DCD expression directly correlated with cell line selectivity, as illustrated by the modulation of the DCD levels in Malme-3M (Fig. 4F). Combined, these data strongly indicated that DCD plays a role in the activity and likely the melanoma selectivity of 1.
Finally, we sought to further confirm that the melanoma selectivity arose from the seriniquinone pharmacophore. The cell line selectivity observed for 1 was consistently maintained by other analogs, including 10, 12, 17, or 18 (SI Appendix, Table S2, as well as NCI-60 data provided within the SI Appendix). Time-course imaging studies on these derivatives further confirmed that they underwent comparable subcellular localization as 1, and were directed into the forming autophagosomes (Fig. 2J).
Conclusion
Combined, these studies define the discovery of 1 and associated derivatives as highly selective anticancer agents. Our data suggest that 1 targets DCD resulting in a profound overexpression of DCD mRNA in melanoma cell lines (Fig. 4F). Although a clear link has yet to be forged between DCD and its peptides as they function during autophagy and apoptotic cell death, these studies show that 1 serves to induce autophagocytosis, a process that may result from the interaction of 1 with DCD or 1 with DCD disulfide-linked protein complexes, as observed with immunoprecipitated Hsp70 (24). Although the mechanism of these events requires further investigation, the fact that 1 and its synthetic derivatives offer potent activity, while being readily accessed synthetically, offers a robust potential to further explore the interplay between autophagy and apoptosis and targeting of dermcidin (25).
There is a growing interest in dermcidin since its discovery in 2001 (23). The role of DCD in cancer proliferation is of particular interest as the DCD-derived peptide Y-P30 has been shown to provide a cancer-associated function in the prostate (26). Furthermore, it is known that dermcidin is overexpressed in melanoma (27). It has recently been reported that DCD induces proliferation in neuronal, breast cancer and prostate cancer cells with the derived peptide Y-P30 acting as the key mediator (28). Hence, it appears that the role of dermcidin in selected tumor cell lines is an important determinant in cancer proliferation.
This study provides the first evidence, to our knowledge, that the targeting of dermcidin, a protein with a growing role in stabilizing cancer, by small molecules may offer a cell-selective means to initiate autophagy and apoptotic cell death. Finally, the recognition that dermcidin may be overexpressed in specific cancers provides an opportunity to use this protein as a patient preselection biomarker to enhance personalized cancer treatments.
Supplementary Material
Acknowledgments
We thank E. Chapman (University of Arizona) and J. Tillotson (University of Arizona) for preparation of the full length 110 aa DCD protein; A. Mrse (University of California at San Diego) for assistance with NMR studies; C. Kauffman [Scripps Institution for Oceanography (SIO)] for assistance with bacterial cultivation; L. Farnaes, L. Paul, and J. Busch (SIO) for assistance with the HCT-116 cytotoxicity bioassays; D. D. Rocha [Universidade Federal do Ceará (UFC)] for assistance with the confocal imaging; and D. Pascoalino (UFC) for assistance with qPCR analyses. We also thank Erin Gontang for assistance in the isolation of bacterial strain CNJ927. This work was supported by National Cancer Institute Grant R37-CA044848 (to W.F.) and Brazilian National Council for Scientific and Technological Development CNPq (to L.V.C.-L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The 16S RNA gene sequence for this strain has been deposited in GenBank (accession number DQ448704.1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410932111/-/DCSupplemental.
References
- 1.Hughes CC, Fenical W. Antibacterials from the sea. Chemistry. 2010;16(42):12512–12525. doi: 10.1002/chem.201001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.La Clair JJ. Natural product mode of action (MOA) studies: A link between natural and synthetic worlds. Nat Prod Rep. 2010;27(7):969–995. doi: 10.1039/b909989c. [DOI] [PubMed] [Google Scholar]
- 3.Hughes CC, MacMillan JB, Gaudêncio SP, Fenical W, La Clair JJ. Ammosamides A and B target myosin. Angew Chem Int Ed Engl. 2009;48(4):728–732. doi: 10.1002/anie.200804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes CC, et al. Marinopyrrole A target elucidation by acyl dye transfer. J Am Chem Soc. 2009;131(34):12094–12096. doi: 10.1021/ja903149u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farnaes L, La Clair JJ, Fenical W. Napyradiomycins CNQ525.510B and A80915C target the Hsp90 paralogue Grp94. Org Biomol Chem. 2014;12(3):418–423. doi: 10.1039/c3ob41355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li JW, Vederas J. Drug discovery and natural products: End of an era or an endless frontier? Science. 2009;325(5937):161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- 8.Butler MS, Newman DJ. Mother Nature’s gifts to diseases of man: The impact of natural products on anti-infective, anticholestemics and anticancer drug discovery. Prog Drug Res. 2008;65(1):1, 3–44. doi: 10.1007/978-3-7643-8117-2_1. [DOI] [PubMed] [Google Scholar]
- 9.Korman JB, Fisher DE. Developing melanoma therapeutics: Overview and update. Wiley Interdiscip Rev Syst Biol Med. 2013;5(3):257–271. doi: 10.1002/wsbm.1210. [DOI] [PubMed] [Google Scholar]
- 10.Battie C, Gohara M, Verschoore M, Roberts W. Skin cancer in skin of color: An update on current facts, trends, and misconceptions. J Drugs Dermatol. 2013;12(2):194–198. [PubMed] [Google Scholar]
- 11.Hartman KG, McKnight LE, Liriano MA, Weber DJ. The evolution of S100B inhibitors for the treatment of malignant melanoma. Future Med Chem. 2013;5(1):97–109. doi: 10.4155/fmc.12.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gontang EA, Fenical W, Jensen PR. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol. 2007;73(10):3272–3282. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuoka M, Iwamoto A, Kitao T. Reaction of 2,3-dichloro-1,4-naphthoquinone with dithiooxamide. Synthesis of dibenzo[b,i]thianthrene-5,7,12,14-tetrone. J Heterocycl Chem. 1991;28(5):1445–1447. [Google Scholar]
- 14.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6(10):813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 15.Sumantran VN. Cellular chemosensitivity assays: An overview. Methods Mol Biol. 2011;731:219–236. doi: 10.1007/978-1-61779-080-5_19. [DOI] [PubMed] [Google Scholar]
- 16.Cole L, Davies D, Hyde GJ, Ashford AE. ER-tracker dye and BODIPY-brefeldin A differentiate the endoplasmic reticulum and golgi bodies from the tubular-vacuole system in living hyphae of Pisolithus tinctorius. J Microsc. 2000;197(Pt 3):239–249. doi: 10.1046/j.1365-2818.2000.00664.x. [DOI] [PubMed] [Google Scholar]
- 17.Biederbick A, Kern HF, Elsässer HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995;66(1):3–14. [PubMed] [Google Scholar]
- 18.Barth S, Glick D, Macleod KF. Autophagy: Assays and artifacts. J Pathol. 2010;221(2):117–124. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11(8):558–572. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 20.Alexander MD, et al. A central strategy for converting natural products into fluorescent probes. ChemBioChem. 2006;7(3):409–416. doi: 10.1002/cbic.200500466. [DOI] [PubMed] [Google Scholar]
- 21.Yu WL, Guizzunti G, Foley TL, Burkart MD, La Clair JJ. An optimized immunoaffinity fluorescent method for natural product target elucidation. J Nat Prod. 2010;73(10):1659–1666. doi: 10.1021/np100371k. [DOI] [PubMed] [Google Scholar]
- 22.Miguel del Corral JM, et al. Synthesis and biological evaluation of cytotoxic 6(7)-alkyl-2-hydroxy-1, 4-naphthoquinones. Arch Pharm (Weinheim) 2002;335(9):427–437. doi: 10.1002/1521-4184(200212)335:9<427::AID-ARDP427>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 23.Schittek B. The multiple facets of dermcidin in cell survival and host defense. J Innate Immun. 2012;4(4):349–360. doi: 10.1159/000336844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stocki P, Wang XN, Morris NJ, Dickinson AM. HSP70 natively and specifically associates with an N-terminal dermcidin-derived peptide that contains an HLA-A*03 antigenic epitope. J Biol Chem. 2011;286(14):12803–12811. doi: 10.1074/jbc.M110.179630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: The interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15(2):81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith AP, Hoek K, Becker D. Whole-genome expression profiling of the melanoma progression pathway reveals marked molecular differences between nevi/melanoma in situ and advanced-stage melanomas. Cancer Biol Ther. 2005;4(9):1018–1029. doi: 10.4161/cbt.4.9.2165. [DOI] [PubMed] [Google Scholar]
- 27.Stewart GD, et al. Dermcidin expression confers a survival advantage inprostate cancer cells subjected to oxidative stress or hypoxia. Prostate. 2007;67(12):1308–1317. doi: 10.1002/pros.20618. [DOI] [PubMed] [Google Scholar]
- 28.Lowrie AG, et al. Proteolysis-inducing factor core peptide mediates dermcidin-induced proliferation of hepatic cells through multiple signalling networks. Int J Oncol. 2011;39(3):709–718. doi: 10.3892/ijo.2011.1064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.