Rapid transitions in viral DNA packing during infection

Top view (Left and Center) and side view (Right) schematics of encapsidated DNA transitions with increasing temperature.
Viral replication can begin only after a virus successfully ejects its genome into a host cell. To investigate how a long, tightly packed, double-stranded DNA viral genome efficiently enters a cell at rates up to 60,000 base pairs per second, Ting Liu et al. (pp. 14675–14680) studied the internal energy, mobility, and order of DNA packing within viral capsid enclosures. Within capsids, viral DNA is arranged as a hexagonal, crystalline structure with little mobility due to friction between DNA strands. The authors observed, however, that the internal energy of DNA within the capsid of bacteriophage λ transitioned abruptly at around 33 °C due to a sudden decrease in the amount of ordered DNA in the capsid. Increased disorder, along with decreased DNA density, reduced friction between strands and enhanced genome mobility, according to the authors. The results suggest that the efficiency of viral infection may depend on crystalline DNA transitioning to a fluid-like disordered state, and that the temperature at which this transition happens, near body temperature, indicates a possible adaptation by phage λ to prevent accidental loss of genetic material outside of a human host. Such a metastable DNA-ordering mechanism may provide a target for future antiviral drugs, according to the authors. — P.G.
How cells deal with endoplasmic reticulum stress
When misfolded proteins accumulate in the endoplasmic reticulum (ER), eukaryotes mount the unfolded protein response (UPR). In budding yeast, production of the Hac1 transcription factor drives the UPR by turning on target genes that increase ER protein folding capacity. In contrast, fission yeast decreases the ER protein folding load, and metazoans incorporate elements of both yeast strategies. To develop a unified model for the ER stress response in budding yeast, David Pincus et al. (pp. 14800–14805) compared the transcriptional profile of cells experiencing ER stress to that of unstressed cells expressing inducible Hac1. The authors identified 848 genes with altered transcription under stress independently of Hac1. These genes were enriched in binding sites for Msn2/4 and other transcription factors inhibited by phosphorylation by the kinase enzyme PKA. In response to increasing dosage of an ER stressor, PKA activity decreased, while expression of Msn2/4 target genes increased, with kinetics describing a second-wave stress response distinct from the Hac1-mediated UPR. This delayed PKA deactivation response also served to diminish protein synthesis, rendering it functionally analogous to aspects of the fission yeast and metazoan UPR that decrease the load of unfolded proteins in the ER. The two-pronged approach to protein misfolding enables rapid restoration of homeostasis in the ER, according to the authors. — C.B.
Evolution of mutualism

Transmission electron micrograph of the bacterium Desulfovibrio vulgaris. Image courtesy of Wikimedia Commons/Graham Bradley.
Some organisms that have evolved to form mutualistic relationships lack the ability to survive alone. To investigate how mutualisms can lead to a loss of function in one or both paired organisms, Kristina Hillesland et al. (pp. 14822–14827) grew a bacterium and an archaeon in coculture under selection to form a mutualistic relationship. The bacterium, Desulfovibrio vulgaris, normally obtains energy by using sulfate, but by growing D. vulgaris in a medium lacking sulfate, the authors forced the bacterium to instead obtain energy by fermentation. To complete the fermentation process and support growth of D. vulgaris, the archaeon Methanococcus maripaludis was added to help metabolize the fermentation waste product hydrogen. After 1,000 generations of growth in these conditions, the cocultures demonstrated increased stability and growth rates. Further, several replicate populations of D. vulgaris evolved mutations in one of three different genes, which resulted in an inability to use sulfate and rendered the bacterium less capable of growing in the absence of M. maripaludis. According to the authors, the results suggest that mutualistic relationships can evolve by natural selection and that the benefits gained in mutualism can outweigh the loss of gene function that confers the flexibility to survive alone. — J.P.J.
Sickle cell disease diagnostic
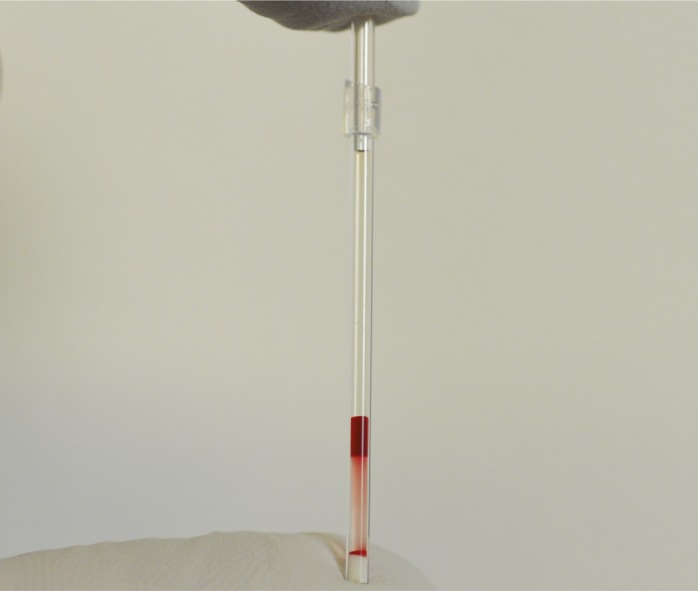
Blood cells separated by density to identify sickle cell disease.
Despite inexpensive and effective interventions for sickle cell disease, more than half of affected children in underprivileged areas die before age 5, often due to delayed diagnosis. Ashok Kumar et al. (pp. 14864–14869) developed a low-cost blood test that identifies sickle cell disease in less than 12 minutes. Sickle cell disease is currently diagnosed via the biochemical separation of blood proteins via gel electrophoresis, a technique that is not feasible in low-resource settings. Noting that blood cells that are denser than normal signify sickle cell disease, the authors used a technology known as aqueous multiphase systems to separate whole blood into self-forming fluid gradients based on differences in density that allow for the visual detection of dense, often-sickled cells. Visual inspection of a three-phase system of aqueous polymers allowed the authors to distinguish between individuals with the disease and those without it, and often delineated the two main subclasses of the disease. According to the authors, the test may help identify children with the disease prior to the onset of acute symptoms, possibly allowing for more timely and effective intervention, particularly in low-resource areas. — A.G.
Mantle updrafts and origins of intraplate volcanoes
Volcanic activity at sites within tectonic plates, such as the Hawaiian Islands or the Yellowstone Caldera, is commonly attributed to mantle plumes, described as narrow, focused, hot upwellings of material from great depth. Don Anderson and James Natland (pp. E4298–E4304) used seismic images and thermodynamic principles to challenge the typical model of mantle plumes. According to the authors, updrafts of mantle material are likely broad and slow-moving, balancing the downwelling of thin oceanic crustal slabs. The authors suggest that updrafts provide magma to both spreading midocean ridges and to intraplate volcanoes. Differences in composition and temperature between midocean ridge magma and intraplate volcanic magma may result from contamination of intraplate magma by a mélange of hot, shallow mantle material, according to the authors. Aggregated magma, trapped in a thick shear zone between the mantle and crust beneath an insulating surface boundary layer may be the source of intraplate volcanism beneath Hawaii, Samoa, and the Pitcairn Islands. The results suggest that midocean ridge magma likely originates from a source between 400 km and 650 km deep and intraplate magma originates from a thermal maximum at the base of the surface boundary layer, according to the authors. — P.G.
Unearthing a placentation gene

Young Malagasy tenrec (Tenrec ecaudatus). Image courtesy of J. F. Eisenberg (American Society of Mammalogists).
Ancient retroviral envelope genes called syncytins aid in placentation by promoting the formation of the syncytiotrophoblast, a layer of cells that enables nutrient flow between the fetus and mother. Guillaume Cornelis et al. (pp. E4332–E4341) suggest that syncytins have been pivotal in the emergence of placental mammals from egg-laying animals, postulating that the genes should therefore be present in species that fall along the Placentalia radiation, a taxonomic division apart from marsupials and monotremes. The authors conducted an in silico search for syncytin-like genes in present-day species that have retained characteristic features of primitive mammals. Analysis of genomes from living animals in the Malagasy and mainland African Tenrecidae families, members of a superorder that emerged during the Cretaceous terrestrial revolution, identified several genes with full coding capacity. The authors report that one of these genes, dubbed syncytin-Ten1, appears to have been conserved through millions of years of evolution in among the most primitive living mammals known as the Afrotherian tenrecs. Subsequent genetic and tissue analyses of the gene revealed that it is expressed in multinucleate cellular masses and layers at the maternal-fetal interface, consistent with a role in syncytiotrophoblast formation. According to the authors, the findings suggest that endogenous retroviral syncytins may have driven the evolution and diversification of placental mammals. — A.G.


