Significance
Syncytins are genes of retroviral origin that have been captured by their host as symbionts for a function in placentation. They can mediate cell–cell fusion, consistent with their ancestral retroviral envelope gene status, and are involved in fusion of mononucleate trophoblast cells to form the syncytial layer—the syncytiotrophoblast—of the feto–maternal interface. We proposed that such genes have been pivotal for the emergence of placental mammals from egg-laying animals and should be present all along the Placentalia radiation. We searched for syncytins in a superorder of eutherian mammals that emerged ancestrally during the Cretaceous terrestrial revolution and identified syncytin-Ten1, conserved over millions years of evolution of the Afrotherian tenrecs, regarded as among the most primitive of living mammals.
Keywords: endogenous retrovirus, envelope protein, syncytiotrophoblast, feto–maternal interface, placenta evolution
Abstract
Syncytins are fusogenic envelope (env) genes of retroviral origin that have been captured for a function in placentation. Syncytins have been identified in Euarchontoglires (primates, rodents, Leporidae) and Laurasiatheria (Carnivora, ruminants) placental mammals. Here, we searched for similar genes in species that retained characteristic features of primitive mammals, namely the Malagasy and mainland African Tenrecidae. They belong to the superorder Afrotheria, an early lineage that diverged from Euarchotonglires and Laurasiatheria 100 Mya, during the Cretaceous terrestrial revolution. An in silico search for env genes with full coding capacity within a Tenrecidae genome identified several candidates, with one displaying placenta-specific expression as revealed by RT-PCR analysis of a large panel of Setifer setosus tissues. Cloning of this endogenous retroviral env gene demonstrated fusogenicity in an ex vivo cell–cell fusion assay on a panel of mammalian cells. Refined analysis of placental architecture and ultrastructure combined with in situ hybridization demonstrated specific expression of the gene in multinucleate cellular masses and layers at the materno–fetal interface, consistent with a role in syncytium formation. This gene, which we named “syncytin-Ten1,” is conserved among Tenrecidae, with evidence of purifying selection and conservation of fusogenic activity. To our knowledge, it is the first syncytin identified to date within the ancestrally diverged Afrotheria superorder.
The syncytins are genes of retroviral origin that have been co-opted by their host for a function in placentation. They correspond to the envelope (env) gene of ancestral retroviruses that entered the germ line of evolutionarily distant animals and were endogenized (reviewed in refs. 1 and 2). Two such genes, syncytin-1 (3, 4) and syncytin-2 (5, 6), have been identified in simians, and distinct, unrelated ones, syncytin-A and -B (7), have been identified in muroid rodents, syncytin-Ory1 (8) in leporids, syncytin-Car1 (9) in carnivorans, and more recently, syncytin-Rum1 (10) in ruminants. Their canonical characteristic features leading to their designation as “syncytins” comprise (i) placenta-specific expression, (ii) cell–cell fusion activity, and (iii) conservation during the evolution of mammalian species for extended periods of time (e.g., >10 million years). Syncytin proteins are expected to participate in the formation of the placental syncytiotrophoblast (ST) at the maternal–fetal interface via fusion of the mononucleate cytotrophoblasts (CTs). Some of them also possess an immunosuppressive activity, as classically observed for infectious retroviral envelope glycoproteins, which may be involved in maternal–fetal tolerance (11). Recently, the direct involvement of syncytins in placentation has been demonstrated unambiguously through the generation of knockout mice for syncytin-A and -B (12, 13), whose embryonic placenta displayed defects in cell–cell fusion, resulting in decreased maternal–fetal exchanges and impaired embryo survival. Interestingly, other captured env genes have been reported to be expressed specifically in the placenta [e.g., syncytin-like env-Cav1 (14), bosenv4/fematrin-1 (10, 15), and enJSRV env (16) genes], with the last one reported to be involved in peri-implantation placental morphogenesis (17).
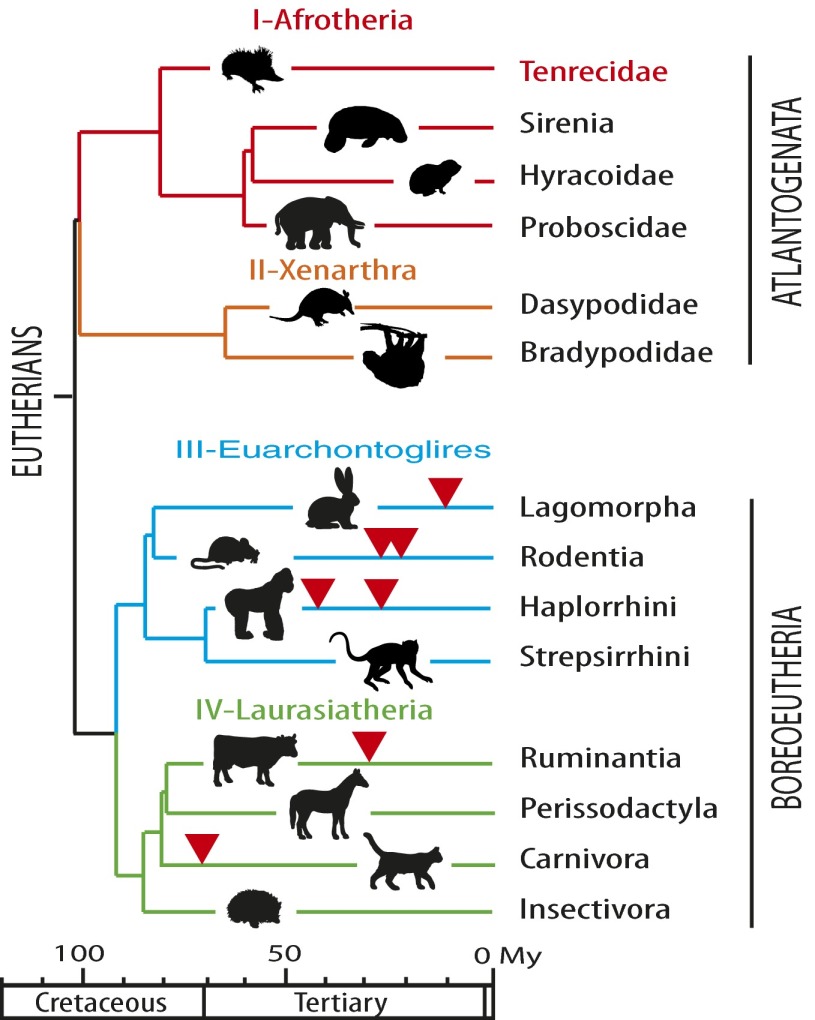
A remarkable feature of syncytins is that these retroviral env genes, which repeatedly and independently integrated into the genomes by chance in the course of evolution, are not junk DNA but are necessary for a basic function in placental mammals (reviewed in ref. 2). We therefore proposed that syncytins should be present in all placental mammals and that the capture of a founding syncytin by an oviparous ancestor was pivotal for the emergence of placentation more than 150 Mya. This founding syncytin gene would then have been replaced in the diverse emerging mammalian lineages upon successive and independent germ-line infections by new retroviruses and co-optation of the new retrovirus’s env gene, each new gene providing its host with a positive selective advantage. This process would account for the diversity in the nature and age of the captured syncytins that presently can be identified, concomitant with the diversity of placental architectures (2, 18, 19). Whatever the fine details of the proposed evolutionary scenario might be, a necessary outcome is that syncytins should be found in every mammal with a placenta. It turns out that all the syncytin genes identified to date belong either to the Laurasiatheria clade of eutherian mammals (the ruminant syncytin-Rum1 and the Carnivora syncytin-Car1) or to the Euarchontoglires (the primate syncytin-1 and -2, the rodent syncytin-A and -B, and the Lagomorpha syncytin-Ory1). These two major clades, which compose the Boreoeutheria superorder, are themselves sisters of the Afrotheria and Xenarthra clades, which constitute the Atlantogenata superorder. These two superorders, which form the eutherian mammals, diverged from one another around 100 Mya (Fig. 1).
Fig. 1.
Phylogeny of Eutheria and previously identified syncytin genes. Eutherians can be grouped into two superorders, Atlantogenata and Boreoeutheria, and four major clades, Afrotheria (I), Xenarthra (II), Euarchontoglires (III), and Laurasiatheria (IV) (adapted from ref. 40). Branch length is proportional to time (in million years, My), and the time of insertion of the different syncytins identified to date is indicated by arrowheads.
To substantiate further that syncytin capture is a general process common to all eutherian mammals, we searched for such genes in the Atlantogenata superorder. In this paper, we have focused on the Afrotherian clade, for which model species are becoming available (20), and more precisely on the Tenrecidae. We selected this taxon because (i) the genome of the lesser hedgehog tenrec, Echinops telfairi, has been sequenced and (ii) because tissues and placenta from pregnant females of a closely related species, the greater hedgehog tenrec (Setifer setosus) could be recovered. Furthermore, Tenrecidae are considered to have retained characteristic features of primitive mammals, including a small forebrain, the presence of a cloaca, nondescending testes in males, and variable body temperature (21, 22). They share an original placentation with the formation of a specialized central hemophagous region with highly invasive syncytial trophoblasts not found in related species. Finally, three additional Afrotherian genomes—the African elephant (Loxodonta africana, Proboscidae), the West Indian manatee (Trichechus manatus, Sirenia), and the rock hyrax (Procavia capensis, Hyracoidae)—also have been sequenced entirely, facilitating the dating and natural history of syncytin capture.
Results
In Silico Search for Retroviral env Genes Within the Lesser Hedgehog Tenrec (E. telfairi) Genome.
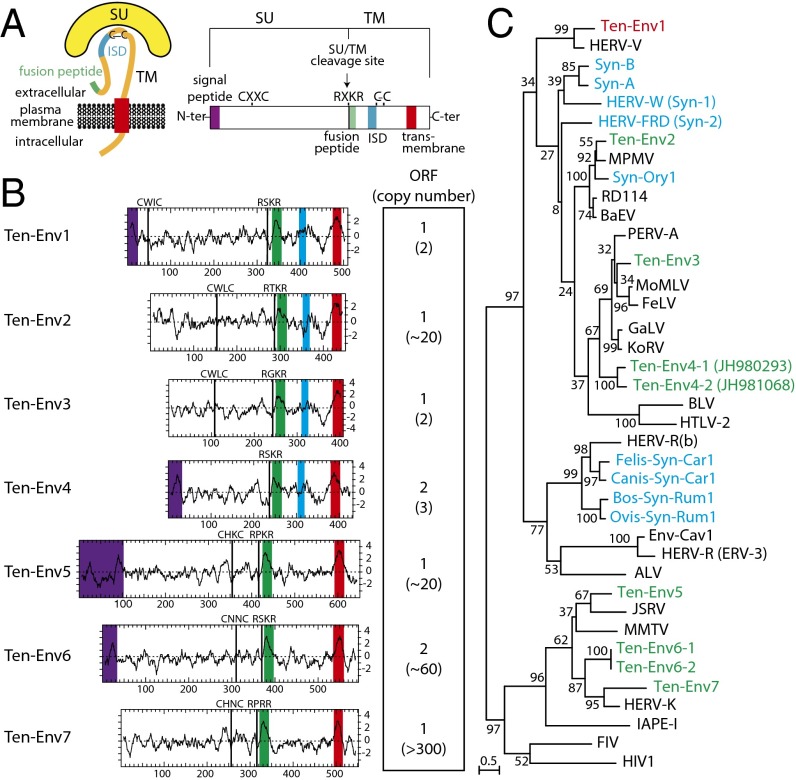
To identify putative env-derived syncytin genes in Afrotheria, we made use of the available lesser hedgehog tenrec genome sequence [2× coverage assembly of the E. telfairi genome, National Center for Biotechnology Information (NCBI) EchTel2.0, November 2012] and of the method that we previously devised to screen the cow genome for such genes (10). Basically, a BLAST search for ORFs (from the Met start codon to the stop codon) longer than 400 aa was performed using a selected series of env sequences, including all presently identified syncytins (Materials and Methods). The search resulted in a series of sequences that were further selected for the presence of a hydrophobic domain >20 aa located 3′ to a C-X5,6,7-C motif, corresponding to a highly conserved motif of retroviral envelope proteins (the C-C and transmembrane domain; see the scheme in Fig. 2A). It yielded nine sequences incorporated into the phylogenetic env tree shown in Fig. 2C (env gene coordinates are listed in Table S1). Some of the sequences can be grouped into single families, resulting in seven families that we named “Ten-Env1” to “Ten-Env7” (Fig. 2B).
Fig. 2.
Structure of a canonical retroviral env protein and characterization of the identified tenrec candidates. (A) Schematic representation of a retroviral env protein, delineating the SU and TM subunits. The furin cleavage site (consensus: R/K-X-R/K-R) between the two subunits, the C-X-X-C motif involved in SU–TM interaction, the hydrophobic signal peptide (purple), the fusion peptide (green), the transmembrane domain (red), and the putative immunosuppressive domain (ISD) (blue) along with the conserved C-X5/6/7-C motif are indicated. (B) Characterization of the candidate E. telfairi env proteins. (Left) The hydrophobicity profile for each candidate is shown with the canonical structural features highlighted in A positioned and shown in the color code used in A. (Right) Number of full-length env gene ORFs within each family of elements. The total number of genomic copies is shown in parenthesis. (C) Retroviral env protein-based phylogenetic tree with the identified Ten-Env protein candidates. The maximum likelihood tree was constructed using TM subunit amino acid sequences from syncytins and a series of endogenous and infectious retroviruses. The length of the horizontal branches is proportional to the percentage of amino acid substitutions from the node (see the scale bar at the lower left), and the percent bootstrap values obtained from 1,000 replicates are indicated at the nodes. ALV, avian leukemia virus; BaEV, baboon endogenous virus; BLV, bovine leukemia virus; Env-Cav1, syncytin-like Cavia porcellus env1 protein; FeLV, feline leukemia virus; FIV, feline immunodeficiency virus; GaLV, gibbon ape leukemia virus; HERV, human endogenous retrovirus; HIV1, HIV type 1; HTLV-2, human T-lymphotrophic virus type 2; IAPE, intracisternal A-type particle with an env gene; JSRV, Jaagsiekte retrovirus; KoRV, koala retrovirus; MMTV, murine mammary tumor virus; MoMLV, Moloney murine leukemia virus; MPMV, Mason–Pfizer monkey virus; PERV, porcine endogenous retrovirus; RD114, feline endogenous type-C retrovirus.
Analysis of the overall gene structure of the seven identified Ten-Env families (Fig. 2B) strongly suggests that they correspond to bona fide retroviral env proteins, with all or most of their characteristic features, including the presence of a predicted signal peptide sequence at the N terminus, a putative furin cleavage site delineating a surface (SU) and a transmembrane (TM) subunit (R-X-K/R-R), and a CXXC motif in the SU subunit corresponding to a binding domain between the two subunits. Hydrophobicity plots identify the hydrophobic transmembrane domain within the TM subunits required for anchoring the Env protein within the plasma membrane and a putative hydrophobic fusion peptide at the N terminus of the TM subunit. Some contain a canonical immunosuppressive domain (ISD) (11). Finally a BLAST search reveals that several families are present at a high copy number (from 20 to several hundred copies, including noncoding copies), whereas the Ten-Env1, -3, and -4 gene families display a very low copy number (i.e., fewer than three copies).
Because we had access only to tissues from the nonsequenced greater hedgehog tenrec, S. setosus, the most closely related species that diverged from E. telfairi less than 10 Mya (23), we first checked that the identified env gene families were present in S. setosus. To do so, specific primers were designed, and a highly sensitive touchdown PCR protocol was performed (Materials and Methods). Single bands of the expected size were obtained for six of the seven families but not for the Ten-Env4–encoding gene family. Full-length env genes were obtained for the genes encoding Ten-Env1, -3, -6, and -7, but only internal fragments could be amplified for Ten-Env2 and -5. Their sequencing confirmed homology to the corresponding E. telfairi env sequences. Although we cannot formally exclude the possibility that the Ten-Env4 sequences are too divergent for PCR amplification, the encoding gene probably is absent from S. setosus or is present only at a low level of sequence conservation. Hence, it was not considered further in the present study.
Identification of Placenta-Specific env Genes.
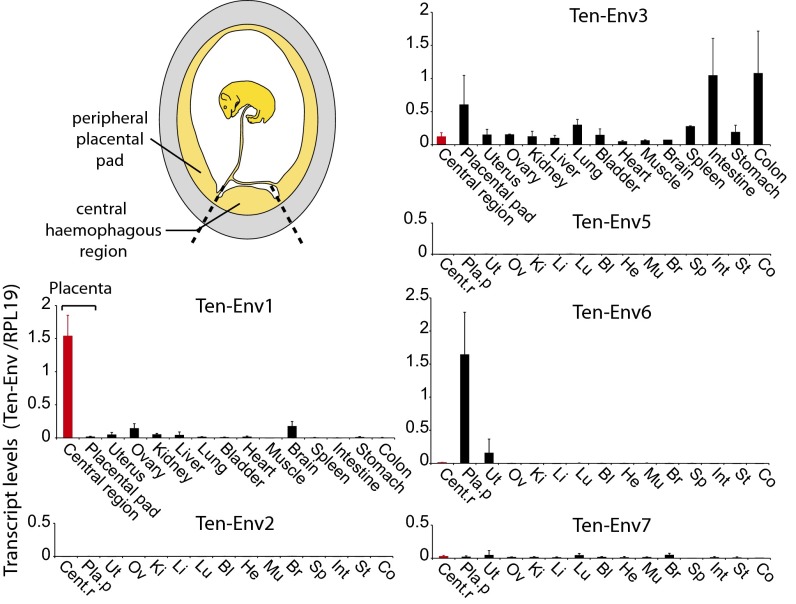
Quantitative RT-PCR (qRT-PCR) analysis of transcript levels for each candidate env gene was performed using primers that were designed to amplify each family of elements identified in S. setosus (Materials and Methods). The tenrec placenta is composed of two concentric regions, the central hemophagous region and the peripheral labyrinthine placental pad (Fig. 3) (24–26). Around midgestation, a layer of syncytial trophoblast is formed at the tip of the trophoblast villi in the central region, whereas maternal-blood spaces are delineated by nonfused mononucleated CTs in the labyrinthine placental pad (described in more detail below). Therefore we analyzed midgestation S. setosus placentas after separate dissection of the central and peripheral regions.
Fig. 3.
Real-time qRT-PCR analysis of the candidate env gene transcripts from the greater hedgehog tenrec (S. setosus) placenta and other tissues. Transcript levels are expressed as the ratio of the expression level of each env gene to that of the RPL19 control gene (Materials and Methods). Midgestation placental tissues were dissected into two regions: the central hemophagous region where syncytium can be found, and the peripheral labyrinthine placental pad, without syncytium. The results for the six env gene candidates obtained with the same series of tissues are shown (Tissues are shown in the same order in each graph; in four graphs, tissue names are abbreviated.) Values are the means of duplicates from two samples ± SEM.
As illustrated in Fig. 3, only the ten-env1 gene displays specific and high-level expression in the central region of the placenta, where cell–cell fusion occurs, and limited expression in other tissues (<10% of the placental level), as expected for a syncytin gene. Interestingly, ten-env6 also is expressed in the placenta of the greater hedgehog tenrec, at levels similar to those of ten-env1, but it is expressed only in the labyrinthine region where no syncytium formation is observed, suggesting a role other than ST formation (Fig. 3). Ten-env3 also is expressed in the labyrinthine region but at levels similar to those observed in the colon and intestine, i.e., not in a tissue-specific manner. Finally, ten-env2, -5, and -7 showed only limited expression in all the tissues tested. Altogether, the in silico analysis combined with qRT-PCR assays for the ten-env genes clearly identify ten-env1 as a putative syncytin gene, and the other genes were not considered further in the present study.
Characterization of the Ten-Env1 Genomic Locus.
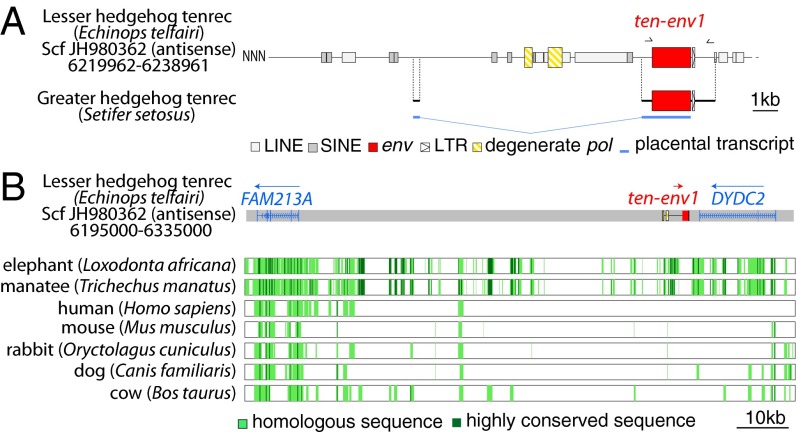
Examination of the lesser hedgehog tenrec (E. telfairi) genomic sequence reveals that ten-env1 is integrated between the predicted DYP30 domain containing 2 (DYDC2) and family with sequence similarity 213, member A (FAM213A) genes in the antisense orientation, as commonly observed for endogenous retroviruses (ERVs) (Fig. 4). The structure of the ancestral provirus is highly degenerate with only a few fragments being detected as being of retroviral origin using the RepeatMasker database (Fig. 4A). The two fragments located 5′ to the env gene show homology to retroviral pol genes, and the 130-bp fragment located 3′ to the env gene probably corresponds to part of the proviral LTR, with evidence for a polyadenylation signal. No 5′ LTR could be identified. PCR amplification of the E. telfairi genomic DNA with a forward primer located at the 5′ end of the env gene, and a reverse primer positioned ∼1 kb downstream of the 3′ LTR, in the region flanking the integration site (Fig. 4A), resulted in amplification of a fragment of the expected size and sequence. PCR carried out with the same primers on the greater hedgehog tenrec (S. setosus) DNA resulted in the amplification of the orthologous fragment (Fig. 4A), with a full-length env gene ORF (Fig. S1) followed by a 3′ LTR and a 600-bp flanking sequence with ∼95% nucleotide identity to that of E. telfairi, confirming the conservation of ten-env1 at the orthologous genomic locus in E. telfairi and S. setosus. The 5′-RACE analysis of ten-env1 placental transcripts disclosed the presence of a transcription start site about 9.5 kb 5′ to the env start codon, with evidence for a spliced subgenomic env transcript containing the entire ORF (Fig. 4A). The start site region is not predicted as a repeated element by RepeatMasker, suggesting that ten-env1 expression in the placenta is driven not by its ancestral retroviral promoter but rather by a cellular promoter neighboring the integration site.
Fig. 4.
Characterization of the lesser and greater hedgehog tenrec (E. telfairi and S. setosus) orthologous ten-env1 gene and of its proviral integration site. (A) Structure of the ten-env1 gene and evidence for orthology between the lesser and greater hedgehog tenrec ten-env1 sequences. Homologous regions common to both sequences are aligned. Repeated mobile elements as identified by the RepeatMasker web program are positioned. Of note, except for ten-env1, only a few and highly degenerated endogenous retroviral elements with only degenerate pol sequences, a fragment of the 3′ LTR, and no 5′ LTR can be identified. PCR primers used to identify the ten-env1 orthologous copy in the greater hedgehog tenrec are indicated by black half arrows. The spliced env subgenomic transcript of greater hedgehog tenrec placental RNA as determined by 5′-RACE is indicated (the sequence has been deposited in the GenBank database with the accession no. KJ934894). (B) ten-env1 is absent in the genomes of distant mammalian lineages. The genomic locus of ten-env1 (in red), along with the surrounding FAM213A and DYDC2 genes, was recovered from the UCSC Genome Browser (http://genome.ucsc.edu/) as well as the syntenic loci of the elephant, manatee, human, mouse, rabbit, dog, and cow genomes; exons (vertical lines) of the FAM213A and DYDC2 genes and the sense of transcription (arrows) are indicated. Homology of the syntenic loci was analyzed using the MultiPipMaker alignment-building tool. Homologous regions are shown as pale green boxes, and highly conserved regions (more than 100 bp without a gap displaying at least 70% identity) are shown as dark green boxes.
The syntenic genomic loci corresponding to the intergenic region between the DYDC2 and FAM213A genes were recovered from the University of California, Santa Cruz (UCSC) genome database for the elephant and manatee Afrotherian genomes and for representatives of Euarchontoglires (human, mouse, rabbit) and Laurasiatheria (cow and dog) in which syncytins have been described so far. Genomic alignment using the PipMaker synteny-building tool shows no homology to ten-env1 at the orthologous loci for any of the indicated species (Fig. 4B). However, the repeated long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs) located 5′ to the ten-env1 ORF are conserved in the Afrotherian species, indicating that these elements integrated there before the Afrotherian radiation. The presence of the conserved LINEs and SINEs between the different tenrec-specific ERV elements (pol-like and env sequences) suggests that these ERV fragments correspond to distinct retroviral integration events that occurred in the ancestor of Tenrecidae after radiation from the other Afrotherian species and that the detected pol-like elements probably do not correspond to the pol gene of the ancestral ten-env1–containing provirus. These distinct retroviruses are highly degenerate, with most of their genes being unrecognizable today, with the noticeable exception of the full-length ORF-encoding ten-env1 gene.
Ten-Env1 Is a Fusogenic Retroviral env Protein.
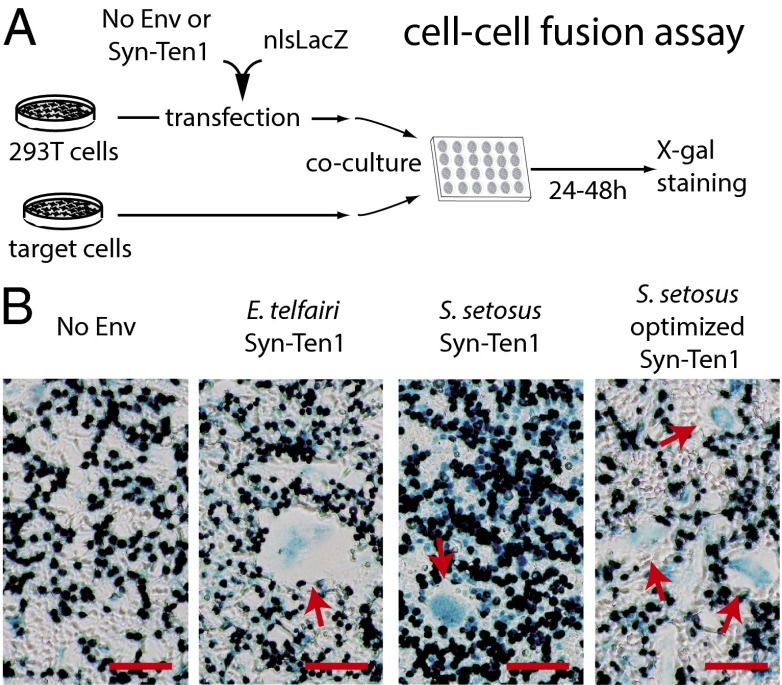
The functionality of Ten-Env1 as an ancestral, retrovirally derived, fusogenic env protein was determined by ex vivo assays in cell cultures, as described in Fig. 5. Basically, we tested whether the E. telfairi and S. setosus env proteins could induce the formation of syncytia in a cell–cell fusion assay. We transfected the highly transfectable human HEK 293T cell line with an expression vector for ten-env1 [together with a nuclear β-galactosidase (nlsLacZ) vector; Fig. 5] before coculturing the transfected cells with a panel of target cells from different species. After 24–48 h of coculture, cell–cell fusion was detected by X-Gal staining for visualization of the syncytia formed between the env-expressing and the target cells. As shown in Fig. 5, expression of Ten-Env1 resulted in cell–cell fusion, leading to the formation of large syncytial structures, when primary fibroblasts that had been derived from a tenrec species (the Tenrecidae Oryzorictes hova) were used as target cells (27). The effect is observed for both E. telfairi and S. setosus Ten-Env1 but not with an empty vector used as control. Interestingly, using the same expression vector for S. setosus Ten-Env1 but with optimized codons for expression in human cells (a synthetic gene; Materials and Methods) increased fusion efficiency, as expected (Fig. 5 and Fig. S2). Similar results were obtained using as a target another cell line classically used in this type of assay, i.e., the hamster A23 cells (Fig. S2). However, no significant syncytium formation could be detected for several other non-Afrotherian target cells (Fig. S2). A similar situation has been observed previously for other syncytins; e.g., the fusogenic mouse Syncytin-B was found to be functional in vivo (13) but was found to be fusogenic ex vivo in only one cell line (7). The restricted cell specificities for fusion suggest that the as yet unidentified receptors for the corresponding Env proteins are not evenly distributed and/or display limited conservation among species. Although a complete understanding of the fusion pattern of Ten-Env1 will require the characterization of its cognate receptor, the present experiments unambiguously establish that ten-env1 is a fusogenic gene, and we therefore named it “syncytin-Ten1.”
Fig. 5.
Syncytin-Ten1 is a fusogenic retroviral env protein. (A) Schematic representation of the coculture assay for cell–cell fusion with Syncytin-Ten1. Human 293T cells were transfected with an expression vector for syncytin-Ten1 or with an empty vector “No Env” as a control, and a plasmid expressing nlsLacZ. After transfection, the 293T cells were cocultured with primary tenrec fibroblasts (from the O. hova Tenrecidae species; see ref. 27) and were stained with X-Gal 24–48 h later. (B) Syncytium formation (arrows) with E. telfairi and S. setosus Syn-Ten1 (no syncytium formation with the “No Env” empty vector as a control). (Scale bars: 200 µm.)
Placental Structure of the Greater Hedgehog Tenrec (S. setosus) and syncytin-Ten1 Expression in Situ.
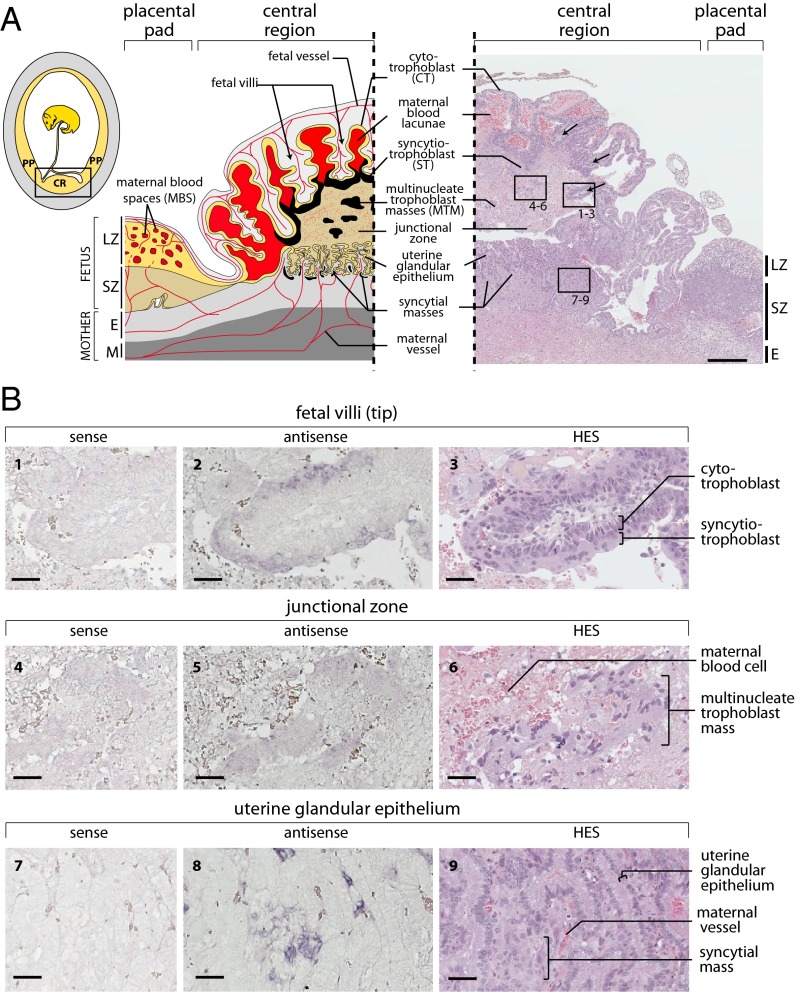
Midgestation placentas were recovered from pregnant greater hedgehog tenrec (S. setosus) females and were processed for histological, ultrastructural, and in situ hybridization analyses. As schematized in Fig. 6A, the S. setosus placenta is composed of two concentric regions, the hemophagous central region (CR) and the peripheral labyrinthine placental pad (PP) (24), as also described for the closely related lesser hedgehog tenrec (25, 26) and a few other tenrec species (22, 28). The peripheral placental pad comprises the labyrinthine zone (LZ), where the maternal blood is in direct contact with a trophoblast cell layer, which separates maternal blood spaces (MBS) from the fetal vessels. This structural organization, defining hemochorial placentation, also is observed in numerous non-Afrotherian species (e.g., mouse), where it is involved in particular in gas and nutrient exchanges (29). However, in contrast to most hemochorial placenta, in which the materno–fetal interface is constituted by a multinucleate syncytium (the syncytiotrophoblast) formed by fusion of the underlying mononucleate trophoblasts (the CTs), the labyrinthine materno–fetal interface in Malagasy tenrecs (including the greater and lesser hedgehog tenrecs) is constituted only by an unfused CT layer (ref. 25 and see below). Another uncommon feature of all tenrec placentas is the presence of an hemophagous region, a specific and complex structure found only rarely in other species, which is likely to facilitate iron uptake by phagocytosis of maternal red blood cells. In S. setosus, as in E. telfairi, in which it has been described previously, it is composed of multifolded trophoblast fetal villi enclosing the fetal vessels, with the maternal blood accumulating within the folds of the villi (Fig. 6A). As illustrated in Fig. 6, large multinucleate syncytial structures can be found at the tip of the fetal villi, with the ST layer in contact with the junctional zone, which itself is composed of a meshwork of multinucleate trophoblast masses and degenerating tissues through which maternal blood diffuses (Figs. 6A and 7A). A uterine glandular epithelium can be observed between the junctional zone and the maternal endometrium. The ST at the tip of the fetal villi contacts the uterine glandular epithelium directly only at the periphery of the central region, but it does not show signs of invasion of the maternal glandular epithelium. As revealed by electron microscopy, the ST shows signs of metabolic activity, with large nucleoli, only limited heterochromatin, a developed endoplasmic reticulum network, and numerous mitochondria (Fig. 7B). It is tightly associated with the underlying CT layer with numerous dense tight junctions at cellular contacts (Fig. 7B, 1). At the opposite side of the syncytium, only loose contacts with the junctional zone appear, with a large number of microvilli at the surface of the syncytium (Fig. 7B, 2).
Fig. 6.
Structure of the S. setosus placenta and in situ hybridization for syncytin-Ten1 expression on placental sections. (A) Schematic representation and hematoxylin eosin saffron (HES)-stained section of a midgestation S. setosus placenta. (Left, Inset) Overview of a gravid uterus displaying the hemophagous central region (CR) and the peripheral placental pad (PP) of the placenta. The yellow and gray areas represent the fetal and maternal tissues, respectively. (Left) Detailed scheme of the midgestation placental structure, with the maternal and fetal vessels schematized in red. In the placental pad tissues are, from mother to fetus, the myometrium (M), the endometrium (E), the spongy zone (SZ), and the labyrinthine zone (LZ) with cytotrophoblasts (yellow) delineating maternal blood spaces (MBS). The structure of the central region is highly divergent from that of the placental pad, with hemophagous columnar CTs (yellow) forming villi delineating large maternal blood lacunae. A thick ST layer (black) can be observed at the tip of the villi contacting a junctional zone. This junctional zone is composed of a meshwork of degenerating tissues through which maternal blood diffuses to the lacunae and in which large multinucleate trophoblast masses (MTM) are present (also represented in black). The maternal glandular epithelium in the central region is well preserved, and the ST contacts it only at the periphery. Syncytial masses also are present underneath the glandular epithelium. (Right) HES-stained section of a midgestation placenta with the positions of panels 1–9 in B indicated. Fetal villi are indicated by arrows. (Scale bar: 500 µm.) (B) In situ hybridization using digoxigenin-labeled antisense or sense riboprobes on serial sections of the HES section shown in A from the central region of the placenta revealed with an alkaline phosphatase-conjugated anti-digoxigenin antibody. (1–3) Tip of a fetal villus covered with a ST layer contacting the junctional zone. The ST is specifically labeled with the antisense probe. (4–6) Detail of the junctional zone centered on a multinucleate trophoblast mass that appears faintly labeled by the antisense probe. (7–9) Detail of the uterine glandular epithelium with the intrauterine syncytial masses, also specifically labeled. (Scale bars: 50 µm.)
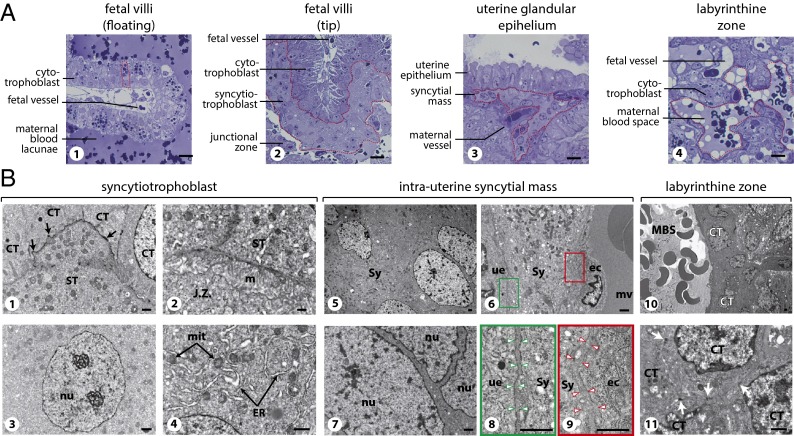
Fig. 7.
Semithin sections (A) and electron microscopy (B) of S. setosus placenta. (A, 1) Semithin sections of a floating fetal villus with hemophagous CTs (one is delineated by a dotted line). (2) The tip of a fetal villus covered by ST (delineated). (3) The maternal glandular epithelium underlaid by an intrauterine syncytial mass (delineated). (4) The labyrinthine zone with CTs separating a large maternal blood space (delineated) from fetal vessels. (Scale bars: 20 µm.) (B) Electron microscopy of the placental structures at midgestation. (1–4) Details of the ST layer. (1) Interface between ST and CTs; tight junctions are indicated by arrowheads. (2) Interface between the ST and the degenerating tissue of the junctional zone (j.z.), with microvilli (m) at the surface of the syncytiotrophoblast. (3) Syncytiotrophoblast nucleus (nu), with two large nucleolar structures and limited heterochromatin. (4) ST cytoplasm with numerous mitochondria (mit) and large endoplasmic reticulum (ER). (5–9) Details of an intrauterine syncytial mass. (5 and 6) Enlarged views of the syncytial mass (Sy) (5), with its cytoplasmic extension located between the maternal uterine glandular epithelium (ue) and the endothelial cells (ec) of a maternal vessel (mv) (6). (7) Higher magnification of the views in 5 demonstrating the absence of a cellular membrane between nuclei (nu). (8 and 9) Higher magnifications of the cells in 6 centered on the interface between the syncytium and the uterine epithelium (8) or the maternal endothelial cell (9). Note the space between the syncytium and the epithelial or the endothelial cellular membranes (indicated by arrowheads), suggesting that the basal membrane of these cells is not degraded by the syncytium at this stage. (10 and 11) Detail of the labyrinthine zone, with a maternal blood space (MBS) delineated by several CTs (10). 11 shows a higher magnification of 10 demonstrating the presence of cellular membranes between the CTs with tight junctions between the cells (white arrows). (Scale bars: 1 µm.)
Consistent with syncytin-Ten1 having a role in ST formation, in situ hybridization using both antisense and sense (control) probes shows specific labeling of the ST layer only with the antisense probe (Fig. 6B, 1 and 2). Labeling is restricted to the ST layer and is not seen at the level of the underlying CTs. This restricted expression pattern was assessed further by examining fetal villi in regions where they are not associated with ST (dubbed “floating villi”). As illustrated in Fig. 7A, these villi are formed of hemophagous CTs which display strong hystotrophic activity, with a large number of phagocytized erythrocyte fragments at the apical regions of the cytotrophoblasts. No labeling can be detected at the level of these CTs (Fig. S3). Of note, multinucleate trophoblast masses within the junctional zone appear to be labeled only faintly, if at all (Fig. 6B, 4 and 5). Unexpectedly, specific labeling was observed with the syncytin-Ten1 antisense probe below the uterine glandular epithelium at levels similar to that observed in the fetal villi-associated ST (Fig. 6B, 7 and 8). Close examination of the corresponding region, including electron microscopy analyses, revealed the presence of intrauterine syncytial masses between the uterine epithelium and the endometrial maternal vessels (Figs. 6B, 9; Fig. 7A, 3 and 7B, 5 and 7). As observed for the ST at the tip of the placental villi, this intrauterine syncytium displays evidence of strong metabolic activity, with low heterochromatin content and well-developed endoplasmic reticulum and mitochondrion networks (Fig. 7B, 5–7). However, it does not seem to be highly histolytic; the maternal vessels (Fig. 7A, 3) and both the epithelial and endothelial basal laminae (Fig. 7B, 6, 8, and 9) are preserved.
Finally, to substantiate further the relationship between syncytin-Ten1 expression and syncytium formation, we looked at the structure of the labyrinthine region in the placental pad. As previously described for other related tenrec species (26), the materno–fetal interface is constituted by mononucleate cytotrophoblasts (Fig. 7A, 4 and 7B, 10 and 11). No syncytial structure can be found, with evidence of cellular membranes delineating the cells (see electron microscopy in Fig. 7B, 11). Consistently, no labeling for syncytin-Ten1 was detected by in situ hybridization of the corresponding region (Fig. S3).
Conclusively, the observed localization of syncytin-Ten1 labeling is consistent with a role of this fusogenic syncytin in the formation of the identified syncytial structures.
Insertion Date of Syncytin-Ten1 in Afrotherian Evolution.
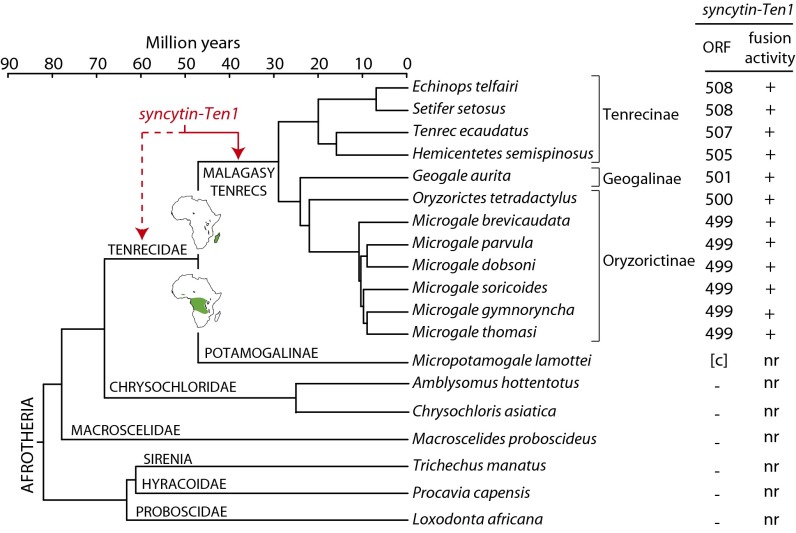
To characterize syncytin-Ten1 further and to determine its status and evolution, we searched for the orthologous gene in representative species of Malagasy tenrecs (namely, from Tenrecinae, Geogalinae, and Oryzorictinae), in the continental Tenrecidae (namely, from Potamogalinae), and in non-Tenrecidae Afrotherian species (Fig. 8). Syncytin-Ten1 was tentatively amplified from the genomic DNA of 13 Tenrecidae species using locus-specific pairs of primers (a forward primer upstream of syncytin-Ten1 and a reverse primer downstream of the provirus in the 3′-flanking sequence). In seven species of Malagasy tenrecs belonging to all three recognized subfamilies, a PCR product of the expected size could be obtained using primers designed on the E. telfairi genomic sequence, strongly suggesting the presence of the orthologous syncytin-Ten1 in the entire Malagasy tenrec lineage. This suggestion was confirmed by sequencing the PCR products, which revealed the presence of a syncytin-Ten1 gene encoding a full-length ORF (499- to 508-aa long). Orthologous syncytin-Ten1 coding copies were amplified further in the five remaining Malagasy tenrec species using primers designed in regions that were most conserved among the above-sequenced genes and with the reverse primer still located in the 3′-flanking region (Materials and Methods and Table S2). All the syncytin-Ten1 gene sequences have been deposited in the GenBank database (Materials and Methods). Using genomic DNA from Micropotamogale lamottei (belonging to the Potamogalinae, the closest outgroup of Malagasy tenrecs), locus-specific PCRs as described above and PCRs using primers bracketing the syncytin-Ten1 gene ORF were found to be negative (Fig. 8). However, PCRs using a forward primer internal to the syncytin-Ten1 ORF, placed at a position where all previously sequenced genes showed a strictly identical nucleotide sequence, and a reverse “ORF” primer located 3′ to the stop codon led to the amplification of a product of the expected size (730 bp). Sequencing of the PCR product confirmed the presence of a sequence homologous to the syncytin-Ten1 gene. Interestingly, this fragment is coding, suggesting the presence of a functional syncytin-Ten1 gene in M. lamottei, as found in other Tenrecidae species (Fig. 8). Finally, PCRs using genomic DNAs from the Chrysochloridae (Amblysomus hottentotus and Chrysochloris asiatica) and Macroscelidae (Macroscelides proboscideus) outgroups were negative for all PCR primer pairs (including primers internal to the ORF and designed to match all previously identified sequences), although an actin fragment could be amplified successfully from these DNAs as a control. Although we cannot formally exclude the possibility that sequences may be too divergent to allow primer annealing and PCR amplification, and although we could not amplify the “empty locus” in these species, the data strongly suggest that syncytin-Ten1 was inserted into the identified locus, at least in the ancestor of Malagasy tenrecs, around 30 Mya and most probably after the Tenrecidae and Chrysochloridae diverged 70–50 Mya (Fig. 8).
Fig. 8.
Entry date and conservation of syncytin-Ten1 in the Afrotheria radiation. (Left) Afrotheria phylogenetic tree (adapted from refs. 23, 40, 41). The length of the horizontal branches is proportional to time (see the scale bar above the tree). The names of the 13 Tenrecidae species tested for the presence of the syncytin-Ten1 gene are indicated, together with the names of the corresponding families. (Right) The length (in amino acids) of the syncytin-Ten1 proteins that were identified for each species is indicated. (The sequences have been deposited in the GenBank database: accession nos. KJ934881–KJ934893.) Brackets indicate that only a partial sequence could be retrieved. c, coding sequence; −, no syncytin-Ten1 homologous sequence identified by either PCR-amplification or database search, thus dating syncytin-Ten1 acquisition back 30–70 Mya, i.e., either before or after the split between the Malagasy and the mainland Africa Tenrecidae. (The African habitats of the two groups are indicated in green in the respective maps.) The fusogenic activity of each cloned gene, as determined by a fusion assay described in Fig. 5 and Fig. S2, is provided. nr, not relevant.
The presence of a syncytin-Ten1 gene also was investigated in other species of the Afrotherian clade, i.e., among Proboscidae (elephant, L. africana), Sirenia (manatee, T. manatus), and Hyracoidae (rock hyrax, P. capensis), and in other mammalian orders, namely the Euarchontoglires (primates and rodents), the Laurasiatheria (Carnivora, Ruminantia, and Perissodactyla), and the Xenarthra, by in silico search using the BLAST program among the corresponding UCSC genomic databases. No sequence with homology >50% could be found, suggesting that syncytin-Ten1 capture took place only once in the course of eutherian mammal evolution (Fig. 8).
Purifying Selection and Functional Conservation of Syncytin-Ten1 in Tenrecidae.
Sequence analysis of the syncytin-Ten1 genes identified as described above demonstrates high similarities, ranging from 73–99% identity (Fig. 9, Right), as expected for a bona fide cellular gene. Interestingly, the phylogenetic tree generated from an alignment of these sequences (Fig. 9, Left) is congruent with the Tenrecidae phylogenetic tree shown in Fig. 8, with only minor differences for some poorly resolved nodes.
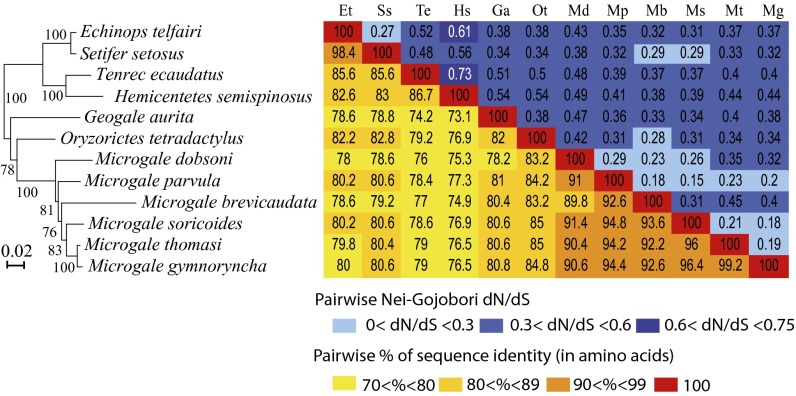
Fig. 9.
Sequence conservation and evidence for purifying selection of syncytin-Ten1 in Tenrecidae. (Left) Syncytin-Ten1–based maximum-likelihood phylogenetic tree determined using nucleotide alignment of the syncytin-Ten1 genes identified in Fig. 8. The length of the horizontal branches and scale indicate the percentage of nucleotide substitutions. The percent bootstrap values obtained from 1,000 replicates (RAxML method) are indicated at the nodes. An identical tree topology is obtained using the MetaPiga maximum-likelihood method with posterior probabilities >0.9 for each nodes. (Right) Double-entry table for the pairwise percentage of amino acid sequence identity between the syncytin-Ten1 proteins among the indicated species (lower triangle) and the pairwise Nei–Gojobori ratio of nonsynonymous to synonymous mutation rate (dN/dS) (upper triangle). A color code for both series of values is provided below the table.
To characterize further the conservation/evolution of the syncytin-Ten1 gene, analysis of the nonsynonymous (dN) to synonymous (dS) mutation ratio (dN/dS) between all pairs of species was performed, using the Nei–Gojobori method (30). Accordingly, the entire env gene shows purifying selection between all pairs of species, with dN/dS ratios being lower than unity (0.15–0.73) (Fig. 9, Right). This pattern of dN/dS ratio is classically observed for cellular genes with a physiological function, in which nonsynonymous mutations are strongly selected against. Finally, we performed a more refined analysis of the sequences using methods allowing differences in selection pressure between different domains of the proteins (site-specific selection) to be revealed. Such an analysis, using the phylogenetic analysis by maximum likelihood (PAML) package (31), provided support for a model (model M7) in which 70% of the codons are under purifying selection (dN/dS <0.75) and 30% are under nearly neutral selection (0.9 ≤ dN/dS < 1). There is no significant support for a positive selection model (model M8 versus M7: χ2 = 5.2, df = 2, P = 0.076), suggesting that no site is under positive selection (Fig. S4). Analyses using the HyPhy package (32) with a slightly different site-specific model, random effect likelihood (REL), led to similar conclusions. Conclusively, the syncytin-Ten1 genes are under strong purifying selection, as expected for a cellular gene with a physiological function.
To determine whether the strong selective pressure exerted on the syncytin-Ten1 gene correlates with the conservation of its functional properties, an ex vivo assay for its fusogenic activity, such as that illustrated in Fig. 5 and Fig. S2 for the S. setosus and Echinops representatives, was performed. PCR-amplified syncytin-Ten1 genes from the Malagasy tenrec species listed in Fig. 8 were cloned into the same eukaryotic expression vector, and cell–cell fusion assays were performed. As shown in Fig. 8, all 12 species tested were found positive, demonstrating functional conservation of syncytin-Ten1 cell–cell fusion activity in the entire Malagasy tenrec clade. Taken together, the data suggest that syncytin-Ten1 is a bona fide cellular gene co-opted for a physiological role in placentation.
Discussion
We have identified syncytin-ten1, the env gene from an endogenous retrovirus that has integrated into the genome of a common ancestor of Tenrecidae. This gene has been maintained as a functional retroviral env gene since its integration, being conserved in a large series of species selected among the Tenrecidae major lineages. This gene displays all the canonical characteristics of a syncytin gene: (i) it exhibits fusogenic activity, in an ex vivo cell–cell fusion assay; (ii) it has been subject to purifying selection in the course of evolution, displaying low rates of nonsynonymous to synonymous substitutions and full conservation of its fusogenic property; (iii) it is expressed specifically in the placenta, as evidenced by both RT-PCR analyses and in situ hybridization of placental tissue sections. In situ hybridization experiments using syncytin-Ten1 sequences as a probe clearly show that specific expression takes place at the level of the multinucleate syncytial structures generated at the feto–maternal interface of the placenta, consistent with this fusogenic syncytin gene having a direct role in syncytium formation. Syncytin-Ten1 adds to the syncytin genes previously identified in the Euarchontoglires and Laurasiatheria clades, with the two primate syncytin-1 and -2 genes (3–5), the two Muroidea syncytin-A and -B genes (7), the Leporidae syncytin-Ory1 gene (8), and the syncytin-Car1 and syncytin-Rum1 genes recently identified in the Carnivora and Ruminantia, respectively (9, 10). Identification of syncytin-Ten1 clearly establishes that syncytin capture extends beyond the Boreoeutheria superorder within eutherian mammals, with syncytin-Ten1 being, to our knowledge, the first syncytin gene identified to date in the ancestrally diverged Atlantogenata superorder. Importantly, all these syncytins are unrelated and correspond to independent captures, in separate mammalian lineages, of genes of retroviral origin. Experiments in knockout mice unambiguously demonstrated that the murine syncytin-A and -B genes are absolutely required for placentation, with evidence that their absence results in a defect in ST formation resulting in decreased maternal–fetal exchange and impaired embryo survival (12, 13). Therefore it can be proposed that all the identified syncytins, including the newly identified syncytin-Ten1, are likely to play a similar role in placentation by being involved in ST formation.
At a refined structural level, as observed in E. telfairi (26), we demonstrated the presence of an ST layer at the surface of placental villi in the central hemophagous region of the S. setosus placenta (also see ref. 24) with significant expression of syncytin-Ten1. In addition to this well-characterized ST layer, the in situ hybridization experiments allowed us to unravel multinucleate syncytial masses within the uterine tissues, in which syncytin-Ten1 also is highly expressed. The origin of these syncytial masses remains elusive, but syncytin-Ten1 expression strongly suggests that they are formed by the fusion of mononucleate cells rather than by endomitosis. We cannot exclude the possibility that these syncytia are of fetal origin, with either nonfused fetal trophoblast cells or already-formed syncytial structures (e.g., from the ST layer at the tip of the fetal villi or from the multinucleate trophoblast masses of the junctional zone) penetrating the maternal endometrium by migrating between epithelial cells without disrupting the glandular epithelium, but this origin is unlikely. Indeed, in the S. setosus placenta, the maternal glandular epithelium at midgestation appears to be well preserved in the central region, and no invasion of this epithelial layer can be observed at the periphery of the central region, where the ST layer and the maternal epithelium are in direct contact. In addition, the syncytial masses do not seem to be highly invasive at this stage of gestation, with basal membranes of both maternal epithelial and endothelial cells being preserved when contacting these syncytial structures. Accordingly, these syncytial masses are likely to be of maternal origin. If so, this instance would be the first example, to our knowledge, of a concerted expression of a syncytin gene in both maternal and fetal tissues during gestation, and it would be of interest to study the molecular mechanisms responsible for this concerted expression and determine whether they share a common hormonal regulation. Only few cases of uterine syncytia formation have been described to date in other mammalian species, and they usually are formed by the fusion of uterine epithelial cells during implantation, as reported in the rabbit (33, 34). These syncytia usually are involved in the degradation of the maternal epithelium, favoring placental invasion of the maternal tissues. Specific syncytial structures also have been described in ruminants, in which the uterine epithelium is replaced by hybrid syncytial structures, to variable degrees depending on the species, and which are formed by the heterologous fusion of fetal trophoblast cells with uterine epithelial cells (35). Here, the observed syncytium is within the maternal tissues late in pregnancy and is located underneath the maternal epithelium, suggesting that these syncytial masses are not derived from uterine epithelial cells and that their function might be different from that of the previously described uterine syncytia observed at implantation.
At an evolutionary level, we have shown that syncytin-Ten1 is present at least in the entire Malagasy tenrec lineage, being conserved in a functional, fusogenic state for at least 30 million years in all species tested. However, we could not demonstrate the presence of a full-length coding gene in M. lamottei, which belongs to the Potamogalinae subfamily, most probably because sequence divergences between the two lineages, which separated >50 Mya, prevented complete gene PCR amplification. However, we could amplify a large coding fragment, thus strongly suggesting that syncytin-Ten1 is conserved in the entire Tenrecidae lineage. This finding, furthermore, would be consistent with the reported placental structure of Potamogalinae (28), which is closely related to that of Malagasy tenrecs and also displays two distinct regions, a peripheral labyrinthine region and a central hemophagous region. The hemophagous region of Potamogalinae also possesses an ST layer at the tip of fetal villi, where fetal cells are in contact with the maternal tissues (28). Interestingly, the placental structure of a representative species of the closest Tenrecidae outgroup, the A. hottentotus species belonging to the Chrysochloridae, also has been described, and the absence of an hemophagous region has been demonstrated in this species (36). Again, this absence would be consistent with our observation that syncytin-Ten1 could not be found in the two tested Chrysochloridae species. Taken together, these data strongly suggest that syncytin-Ten1 has been captured and conserved in a functional state in the entire Tenrecidae lineage, where it is involved in ST formation within the hemophagous region.
Another interesting correlation can be drawn from the present investigation, based on the structure of the other major placental domain, the labyrinthine region of the Tenrecidae placenta. Actually, in the Malagasy tenrecs, the labyrinthine region is of the hemochorial type but, unlike most hemochorial placentas (e.g., human, mouse, rabbit), the mononucleate cytotrophoblasts are in direct contact with the maternal blood, without an intervening syncytiotrophoblast. Again, this peculiar structural organization is consistent with our in situ hybridization results, which show no evidence of syncytin-Ten1 expression in this tissue. Conversely, the labyrinthine region of Potamogalinae is of the endotheliochorial type, with an ST layer surrounding the preserved maternal vessels (28). Thus it would be of interest to analyze the syncytin-Ten1 expression pattern in a Potamogalinae species and to determine if it is expressed in the labyrinthine region, where it could account for ST formation at this feto–maternal interface in addition to the hemophagous region. If this prediction is correct, it would reinforce the notion that syncytin diversity and the regulation of syncytin expression have profoundly affected the differences in placental structure among species, emphasizing the intimate link between syncytin genes and mammalian placenta evolution.
Finally, it also is noteworthy that the existence of a hemophagous region is not a common/obligatory feature of the mammalian placenta, although it is observed in several species where it is thought to facilitate iron uptake by direct phagocytosis of maternal red blood cells. The Malagasy tenrecs, together with the hyena in the Carnivora order, are among the very few species possessing a hemophagous region in the context of a hemochorial placenta (where maternal blood is in direct contact with the fetal syncytiotrophoblast); usually, the hemophagous region is associated with less invasive placentas of the endotheliochorial type (where the maternal vessels are preserved). Interestingly, for both the hemochorial tenrecs and hyena, the most closely related species with a hemophagous region, within the Potamogalinae and Felidae, respectively, all possess an endotheliochorial placenta, suggesting that the hemophagous region of Malagasy tenrecs and of the hyena might be reminiscent of—and a remnant structure from—an ancestral placenta of the endotheliochorial type for both lineages.
In conclusion, the molecular and structural data altogether strongly suggest that syncytin gene capture is a widespread process that took place in several widely separate lineages in the course of eutherian evolution and that the remarkable variability in placental structures might result simply from the diversity of the syncytin genes that have been captured stochastically in the course of mammalian evolution. The data argue for endogenous retroviral syncytins being a major driving force in the evolution and diversification of eutherian mammals.
Materials and Methods
Materials.
Greater hedgehog tenrec (S. setosus) females used in this study were bred at the Laboratory of Artificial and Natural Evolution, University of Geneva, Geneva. Maintenance of and experiments on animals were approved by the Geneva Canton ethical regulation authority (authorization 1008/3421/1R) and were performed in accordance with Swiss law. Gestation length of S. setosus females in our colony is 55 ± 3 d, and the males and females were put in contact for 5 d. Placental and uterine tissues were collected from three pregnant females killed at midgestation (32 d after the first contact with the male) and either were stored in liquid nitrogen or were processed for in situ hybridization. Other tissues (ovary, kidney, liver, lung, bladder, heart, muscle, brain, spleen, intestine, stomach, and colon) were collected from the same females. Total RNA was extracted from the frozen organs using the RNeasy RNA isolation kit (Qiagen) or after tissue lysis in TRIzol Reagent (Invitrogen) for fibrous muscle tissues (muscle and heart). Origins of tissues used for genomic DNA extraction are listed in Table S3. Genomic DNAs were purified by phenol-chloroform extraction.
Database Screening and Sequence Analyses.
Retroviral endogenous env gene sequences were searched for by BLAST on the lesser hedgehog tenrec genome (2× coverage assembly of the E. telfairi genome, NCBI EchTel2.0, November 2012). Sequences containing an ORF longer than 400 aa from start to stop codons were extracted from the EchTel2.0 genomic database using the getorf program of the EMBOSS package (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html) and were translated into amino acid sequences. These sequences were BLASTed against the TM subunit amino acid sequences of 35 retroviral env glycoproteins (from representative ERVs, among which are known syncytins, and infectious retroviruses), using the BLASTP program of the NCBI (www.ncbi.nlm.nih.gov/BLAST). Putative env protein sequences then were selected based on the presence of a hydrophobic transmembrane domain located 3′ to a highly conserved C-X5,6,7-C motif. The identified env-encoding sequence coordinates are listed in Table S1.
The lesser hedgehog tenrec genome was secondarily screened with the identified env glycoprotein sequences using the BLAST programs from the NCBI (www.ncbi.nlm.nih.gov/BLAST). Multiple alignments of amino acid sequences were carried out using the Seaview program in ClustalW. Maximum likelihood phylogenetic trees were constructed with RaxML 7.3.2 (37), with bootstrap percentages computed after 1,000 replicates using the GAMMA + general time reversible (GTR) model for the rapid bootstrapping algorithm. We also computed branch-support posterior probabilities using the MetaPIGAv3.0 (38) after selection of the best-fitting maximum likelihood substitution model (GAMMA + GTR) on the basis of the Akaike Information Criterion. PAML4 (31) was used to run site-specific selection tests and obtain dN/dS ratios for all syncytin-Ten1 sequences. PAML models analyzed assumed no molecular clock (clock = 0) and a single dN/dS for all tree branches (model = 0), and we used likelihood ratio tests to compare the improvement in likelihood for a model allowing positive selection (M8) with a model (NS site = 7–8) that does not (M7). Each analysis ran until convergence (Small_Diff = 0.5e−6). The control file is available upon request. HyPhy (32) was used on the datamonkey web server (www.datamonkey.org) to run site-specific REL tests.
The genomes of the elephant (Loxodonta africana, UCSC/Broad loxAfr3, 2009, 7× coverage), manatee (Trichechus manatus, UCSC/Broad/Harvard triMan1, 2011), rock hyrax (Procavia capensis, UCSC/Broad proCap1, 2008), human (Homo sapiens, UCSC GRCh37/hg19, 2009), mouse (Mus musculus, UCSC GRCm38, 2011), rabbit (Oryctolagus cuniculus, UCSC/Broad oryCun2.0, 2009, 7.48× coverage), dog (Canis lupus familiaris, UCSC/Broad CanFam3.1, 2011), and cow (Bos taurus, UCSC/Baylor bosTau7, 2011) also were screened for the presence of the identified syncytin-Ten1–containing provirus sequence using syntenic genomic regions from the UCSC genome browser (http://genome.ucsc.edu/). Analyses of the syntenic conserved sequences in each genome were performed using the MultiPipMaker alignment tool (http://pipmaker.bx.psu.edu) with the E. telfairi genome sequence as a reference.
Search for a Conserved env Gene in the S. setosus Genome.
Homologous genes of the ten-env families identified in the E. telfairi genomic database were searched for in the S. setosus genome. A highly sensitive touchdown PCR was performed with 100 ng of genomic DNA (10 PCR cycles with an annealing temperature decreasing from 60 °C to 50 °C by 1 °C per cycle followed by 40 PCR cycles with a low annealing temperature of 55 °C). Homologous genes were searched using two different sets of primers (Table S2) designed externally to amplify the entire env gene (before the start and after the stop codons) or designed internally in the most conserved regions of retroviral env genes (namely the furin cleavage site and the C-X5,6,7-C motif).
Real-Time RT-PCR.
Ten-env mRNA expression was determined by qRT-PCR. Reverse transcription was performed with 500 ng of DNase-treated RNA as in ref. 39. PCR was carried out with 5 μL of cDNA diluted (1:20) in a final volume of 25 μL using the FastSYBR Green PCR Master Mix (Qiagen) in an ABI PRISM 7000 sequence detection system. Primers are listed in Table S2. Transcript levels were normalized relative to the amount of the housekeeping gene ribosomal protein L19 (RPL19). Samples (two per organ) were assayed in duplicate. We performed 5′ RACE with 100 ng of DNase-treated RNA using the SMARTer RACE cDNA Amplification Kit (Clontech).
In Situ Hybridization.
Freshly collected tenrec (S. setosus) midgestation placentae were fixed in 4% (wt/vol) paraformaldehyde at 4 °C and were embedded in paraffin, and serial 7-μm sections either werestained with H&E or were used for in situ hybridization. For syncytin-Ten1, three PCR-amplified fragments of 444 bp, 422 bp, and 442 bp (primers are listed in Table S2) were cloned into pGEM-T Easy (Promega) for in vitro synthesis of the antisense and sense riboprobes generated with SP6 RNA polymerase and digoxigenin 11-UTP (Roche Applied Science) after cDNA template amplification. Sections were processed, hybridized at 42 °C overnight with the pooled riboprobes, and incubated further overnight at 4 °C with alkaline phosphatase-conjugated anti-digoxigenin antibody Fab fragments (Roche Applied Science). Labeling was revealed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indoyl phosphate phosphatase alkaline substrates, as indicated by the manufacturer (Roche Applied Science).
Electron Microscopy.
Tissues were fixed at least 1 h at 4 °C in 2.5% (vol/vol) glutaraldehyde in 0.1 M Sörensen phosphate buffer (pH 7.3), postfixed 2 h in aqueous 2% (wt/vol) osmium tetroxide, stained en bloc in 2% (wt/vol) uranyl acetate in 30% (vol/vol) methanol, dehydrated, and finally embedded in Epon. Analysis of the cellular organization of the tissue at the optical level was performed with 1-µm semithin sections. Mounted on a microscope slide, the sections were de-embedded for 20 min at room temperature in sodium hydroxide-saturated ethanol and stained for 30 min at 56 °C with a 1% aqueous solution of thionine blue. After washings in absolute ethanol and xylene, the sections were mounted with Eukitt mounting medium. For ultrastructural observation, 70-nm ultrathin sections were stained with uranyl acetate and lead citrate and were examined with a FEI Tecnai 12 microscope at 80Kv.
Search for Syncytin-Ten1 in Other Species.
PCRs were performed on 100 ng of genomic DNA using Accuprime Taq DNA Polymerase (Invitrogen). A highly sensitive touchdown PCR protocol was performed (elongation time at 68 °C, with 30-s hybridization at temperatures decreasing from 60 °C to 50 °C by 1 °C per cycle for 10 cycles, followed by 40 cycles at 55 °C). Genomic DNAs were tentatively amplified as indicated in Results with primers (listed in Table S2) that were external to the provirus (locus primers), close to the start and stop codons of the syncytin-Ten1 ORF (ORF primers), or conserved among all sequenced syncytin-Ten1 either internal to the ORF (internal primers) or close to the stop codon (ORF-R2). Syncytin-Ten1 orthologous copies could be amplified directly in seven Malagasy tenrec species (E. telfairi, S. setosus, Tenrec ecaudatus, Hemicentetes semispinosus, Geogale aurita, Oryzorictes tetradactylus, and Microgale dobsoni) using primers designed on the E. telfairi genomic sequence. The orthologous copies of the remaining Microgale species could be amplified using a forward primer located 5′ to the syncytin-Ten1 start codon (locus-F2) and a degenerate primer in the 3′-flanking sequence (locus-R2) in regions most conserved among the sequenced orthologous syncytin-Ten1 genes named above. PCR products were sequenced directly without cloning to avoid low-level mutations introduced by PCR. The sequences were deposited in the GenBank database, accession nos. KJ934881–KJ934893.
Syncytin-Ten1 Expression Vector and Fusion Assay.
The syncytin-Ten1 fragments PCR-amplified from the genomic DNA of each Malagasy tenrec species using the syncytin-Ten1 ORF primers (Table S2) were digested with XhoI and MluI, and the PCR products were cloned into the phCMV-G vector (GenBank accession no. AJ318514, a gift from F.-L. Cosset, INSERM, Lyon, France). The syncytin-Ten1 gene sequence also was synthesized and optimized for expression in human cells without modifying its amino acid sequence (GenCust, France). In particular, codon use was corrected for the human bias, a consensus Kozak sequence was added 5′ to the start codon (GCCACCATG), and AT/GC-rich (<80%) or GC-poor (<30%) regions were eliminated, together with cryptic splice donor and acceptor sites, repeated sequences, and RNA secondary structures. The sequence has been deposited in the GenBank database (accession no. KJ934895). Cell–cell fusion assays were performed by cotransfecting cells (1–2 × 105 cells per well) with syncytin-Ten1-expressing vectors along with a vector expressing an nlsLacZ gene (0.5–1 µg at a ratio of 1:1) using the Lipofectamine LTX and Plus reagent transfection kit (Invitrogen) before coculturing them (1:1 ratio) with a panel of target cells. At 24–48 h after co-culture, cells were fixed and stained with X-Gal. The fusion index was calculated as [(N − S)/T] × 100, where N is the number of nuclei in the syncytia, S is the number of syncytia, and T is the total number of nuclei counted. All cell lines are described in refs. 7 and 9, except for the O. hova fibroblast tenrec cells, which were established from animal tissues collected in Madagascar (27). Cells were grown in DMEM supplemented with 10% (vol/vol) FCS (Invitrogen), 100 mg/mL streptomycin, and 100 U/mL penicillin, except for Oryzorictes tenrec cells, which were grown in supplemented AmnioMax (Invitrogen) rich medium with antibiotics.
Supplementary Material
Acknowledgments
We thank J. Decher for the gift of M. lamottei tissues, U. Zeller for the gift of M. proboscideus tissues, V. Soarimalala for samples of different Malagasy Tenrecidae, O. Bawa for her contribution to the histological analyses, A. Enders for helpful comments, C. Lavialle for discussion and critical reading of the manuscript, and M. Funk for bibliographic contributions. This work was supported by the Centre National de la Recherche Scientifique and by grants from the Ligue Nationale contre Le Cancer (Equipe Labellisée) and the Agence Nationale de la Recherche (Retro-Placenta) (to T.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KJ934881–KJ934895).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412268111/-/DCSupplemental.
References
- 1.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta. 2012;33(9):663–671. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 2.Lavialle C, et al. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci. 2013;368(1626):20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blond JL, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74(7):3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mi S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403(6771):785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 5.Blaise S, de Parseval N, Bénit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci USA. 2003;100(22):13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaise S, Ruggieri A, Dewannieux M, Cosset F-L, Heidmann T. Identification of an envelope protein from the FRD family of human endogenous retroviruses (HERV-FRD) conferring infectivity and functional conservation among simians. J Virol. 2004;78(2):1050–1054. doi: 10.1128/JVI.78.2.1050-1054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupressoir A, et al. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci USA. 2005;102(3):725–730. doi: 10.1073/pnas.0406509102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: A new “syncytin” in a third order of mammals. Retrovirology. 2009;6(1):107. doi: 10.1186/1742-4690-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis G, et al. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc Natl Acad Sci USA. 2012;109(7):E432–E441. doi: 10.1073/pnas.1115346109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelis G, et al. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc Natl Acad Sci USA. 2013;110(9):E828–E837. doi: 10.1073/pnas.1215787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangeney M, et al. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci USA. 2007;104(51):20534–20539. doi: 10.1073/pnas.0707873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupressoir A, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci USA. 2009;106(29):12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupressoir A, et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci USA. 2011;108(46):E1164–E1173. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernochet C, et al. A syncytin-like endogenous retrovirus envelope gene of the guinea pig specifically expressed in the placenta junctional zone and conserved in Caviomorpha. Placenta. 2011;32(11):885–892. doi: 10.1016/j.placenta.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 15.Nakaya Y, Koshi K, Nakagawa S, Hashizume K, Miyazawa T. Fematrin-1 is involved in fetomaternal cell-to-cell fusion in Bovinae placenta and has contributed to diversity of ruminant placentation. J Virol. 2013;87(19):10563–10572. doi: 10.1128/JVI.01398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Black SG, Arnaud F, Palmarini M, Spencer TE. Endogenous retroviruses in trophoblast differentiation and placental development. Am J Reprod Immunol. 2010;64(4):255–264. doi: 10.1111/j.1600-0897.2010.00860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunlap KA, et al. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc Natl Acad Sci USA. 2006;103(39):14390–14395. doi: 10.1073/pnas.0603836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leiser R, Kaufmann P. Placental structure: In a comparative aspect. Exp Clin Endocrinol. 1994;102(3):122–134. doi: 10.1055/s-0029-1211275. [DOI] [PubMed] [Google Scholar]
- 19.Enders AC, Carter AM. 2004. What can comparative studies of placental structure tell us? A review. Placenta 25 (Suppl A):S3–S9.
- 20.Tzika AC, Milinkovitch MM. A Pragmatic Approach for Selecting Evo-Devo Model Species in Amniotes. In: Minelli A, Fusco G, editors. Evolving Pathways; Key Themes in Evolutionary Developmental Biology. Cambridge, UK: Cambridge Univ Press; 2008. pp. 119–140. [Google Scholar]
- 21.Eisenberg JF, Gould E. 1970. The tenrecs: A study in mammalian behavior and evolution. Smithson Contrib Zool 27:1e138.
- 22.Enders AC, Blankenship TN, Goodman SM, Soarimalala V, Carter AM. Placental diversity in malagasy tenrecs: Placentation in shrew tenrecs (Microgale spp.), the mole-like rice tenrec (Oryzorictes hova) and the web-footed tenrec (Limnogale mergulus) Placenta. 2007;28(7):748–759. doi: 10.1016/j.placenta.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Poux C, Madsen O, Glos J, de Jong WW, Vences M. Molecular phylogeny and divergence times of Malagasy tenrecs: Influence of data partitioning and taxon sampling on dating analyses. BMC Evol Biol. 2008;8(1):102. doi: 10.1186/1471-2148-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strauss F. Die Placentation von Ericulus setosus. Rev Suisse Zool. 1943;50(2):17–87. [Google Scholar]
- 25.Carter AM, Blankenship TN, Künzle H, Enders AC. Structure of the definitive placenta of the tenrec, Echinops telfairi. Placenta. 2004;25(2–3):218–232. doi: 10.1016/j.placenta.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Carter AM, Blankenship TN, Künzle H, Enders AC. Development of the haemophagous region and labyrinth of the placenta of the tenrec, Echinops telfairi. Placenta. 2005;26(2-3):251–261. doi: 10.1016/j.placenta.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Gilbert C, et al. Chromosomal evolution in tenrecs (Microgale and Oryzorictes, Tenrecidae) from the Central Highlands of Madagascar. Chromosome Res. 2007;15(8):1075–1091. doi: 10.1007/s10577-007-1182-6. [DOI] [PubMed] [Google Scholar]
- 28.Carter AM, Blankenship TN, Enders AC, Vogel P. The fetal membranes of the otter shrews and a synapomorphy for afrotheria. Placenta. 2006;27(2-3):258–268. doi: 10.1016/j.placenta.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Enders AC, Carter AM. Review: The evolving placenta: Different developmental paths to a hemochorial relationship. Placenta. 2012;33(S):S92–S98. doi: 10.1016/j.placenta.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 31.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 32.Kosakovsky Pond SL, Frost SDW. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22(5):1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 33.Enders AC, Schlafke S. Penetration of the uterine epithelium during implantation in the rabbit. Am J Anat. 1971;132(2):219–230. doi: 10.1002/aja.1001320208. [DOI] [PubMed] [Google Scholar]
- 34.Larsen JF. Electron microscopy of the implantation site in the rabbit. Am J Anat. 1961;109:319–334. doi: 10.1002/aja.1001090307. [DOI] [PubMed] [Google Scholar]
- 35.Wooding FB. Current topic: The synepitheliochorial placenta of ruminants: Binucleate cell fusions and hormone production. Placenta. 1992;13(2):101–113. doi: 10.1016/0143-4004(92)90025-o. [DOI] [PubMed] [Google Scholar]
- 36.Jones CJP, Carter AM, Bennett NC, Blankenship TN, Enders AC. Placentation in the Hottentot golden mole, Amblysomus hottentotus (Afrosoricida: Chrysochloridae) Placenta. 2009;30(7):571–578. doi: 10.1016/j.placenta.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 38.Helaers R, Milinkovitch MC. MetaPIGA v2.0: Maximum likelihood large phylogeny estimation using the metapopulation genetic algorithm and other stochastic heuristics. BMC Bioinformatics. 2010;11:379. doi: 10.1186/1471-2105-11-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: Identification and transcriptome of the genes with coding capacity for complete envelope proteins. J Virol. 2003;77(19):10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meredith RW, et al. Impacts of the Cretaceous Terrestrial Revolution and KPg extinction on mammal diversification. Science. 2011;334(6055):521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- 41.Olson L, Goodman S. Phylogeny and biogeography of tenrecs. In: Goodman SM, Benstead JP, editors. The Natural History of Madagascar. Univ of Chicago Press; Chicago: 2003. pp. 1235–1242. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.