Significance
Na+-coupled symporters use the cellular Na+ gradient to power transport of physiologically important molecules across the lipid membrane. However, the mechanism by which binding and dissociation of Na+ drive transport remains undefined. This work investigated the Na+/hydantoin transporter Mhp1, a member of the LeuT-fold class of transporters, to describe the conformations sampled during its transport cycle and elucidate the ligand-induced shifts in its conformational equilibrium. The results of this study suggest that Mhp1 isomerization between inward- and outward-facing conformations are Na+-independent and that coupling to the Na+ gradient occurs through modulation of substrate affinity by Na+ coordination. A previously unidentified model of Mhp1 transport defined by ligand-independent equilibrium fluctuations emerges from this work, offering a new perspective on Na+-coupled symport in the LeuT-fold.
Keywords: EPR, DEER, Na+-coupled symport, conformational dynamics, transport mechanism
Abstract
Ion-dependent transporters of the LeuT-fold couple the uptake of physiologically essential molecules to transmembrane ion gradients. Defined by a conserved 5-helix inverted repeat that encodes common principles of ion and substrate binding, the LeuT-fold has been captured in outward-facing, occluded, and inward-facing conformations. However, fundamental questions relating to the structural basis of alternating access and coupling to ion gradients remain unanswered. Here, we used distance measurements between pairs of spin labels to define the conformational cycle of the Na+-coupled hydantoin symporter Mhp1 from Microbacterium liquefaciens. Our results reveal that the inward-facing and outward-facing Mhp1 crystal structures represent sampled intermediate states in solution. Here, we provide a mechanistic context for these structures, mapping them into a model of transport based on ion- and substrate-dependent conformational equilibria. In contrast to the Na+/leucine transporter LeuT, our results suggest that Na+ binding at the conserved second Na+ binding site does not change the energetics of the inward- and outward-facing conformations of Mhp1. Comparative analysis of ligand-dependent alternating access in LeuT and Mhp1 lead us to propose that different coupling schemes to ion gradients may define distinct conformational mechanisms within the LeuT-fold class.
Secondary active transporters harness the energy of ion gradients to power the uphill movement of solutes across membranes. Mitchell (1) and others (2, 3) proposed and elaborated “alternating access” mechanisms wherein the transporter transitions between two conformational states that alternately expose the substrate binding site to the two sides of the membrane. The LeuT class of ion-coupled symporters consists of functionally distinct transporters that share a conserved scaffold of two sets of five transmembrane helices related by twofold symmetry around an axis nearly parallel to the membrane (4). Ions and substrates are bound near the middle of the membrane stabilized by electrostatic interactions with unwound regions of transmembrane helix (TM) 1 and often TM6 (4). The recurrence of this fold in transporters that play critical roles in fundamental physiological processes (5, 6) has spurred intense interest in defining the principles of alternating access.
Despite rapid progress in structure determination of ion-coupled LeuT-fold transporters (7–11), extrapolation of these static snapshots to a set of conformational steps underlying alternating access (4, 7, 9–12) remains incomplete, often hindered by uncertainties in the mechanistic identities of crystal structures. Typically, transporter crystal structures are classified as inward-facing, outward-facing, or occluded on the basis of the accessibility of the substrate binding site (7–11). In a recent spectroscopic analysis of LeuT, we demonstrated that detergent selection and mutations of conserved residues appeared to stabilize conformations that were not detected in the wild-type (WT) LeuT and concurrently inhibited movement of structural elements involved in ligand-dependent alternating access (13). Therefore, although crystal structures define the structural context and identify plausible pathways of substrate binding and release, development of transport models requires confirming or assigning the mechanistic identity of these structures and framing them into ligand-dependent equilibria (14).
Mhp1, an Na+-coupled symporter of benzyl-hydantoin (BH) from Microbacterium liquefaciens, was the first LeuT-fold member to be characterized by crystal structures purported to represent outward-facing, inward-facing, and outward-facing/occluded conformations of an alternating access cycle (8, 15). In these structures, solvent access to ligand-binding sites is defined by the relative orientation between a 4-helix bundle motif and a 4-helix scaffold motif (8). In Mhp1, alternating access between inward- and outward-facing conformations, was predicted from a computational analysis based on the inverted repeat symmetry of the LeuT fold and is referred to as the rocking-bundle model (16). The conservation of the inverted symmetry prompted proposal of the rocking-bundle mechanism as a general model for LeuT-fold transporters (16). Subsequent crystal structures of other LeuT-fold transporters (7, 9, 10) tempered this prediction because the diversity of the structural rearrangements implicit in these structures is seemingly inconsistent with a conserved conformational cycle.
Another outstanding question pertains to the ion-coupling mechanism and the driving force of conformational changes. The implied ion-to-substrate stoichiometry varies across LeuT-fold ion-coupled transporters. For instance, LeuT (17) and BetP (18) require two Na+ ions that bind at two distinct sites referred to as Na1 and Na2 whereas Mhp1 (15) and vSGLT (19) appear to possess only the conserved Na2 site. Molecular dynamics (MD) simulations (20, 21) and electron paramagnetic resonance (EPR) analysis (13, 22) of LeuT demonstrated that Na+ binding favors an outward-facing conformation although it is unclear which Na+ site (or both) is responsible for triggering this conformational transition. Similarly, a role for Na+ in conformational switching has been uncovered in putative human LeuT-fold transporters, including hSGLT (23). In Mhp1, the sole Na2 site has been shown to modulate substrate affinity (15); however, its proposed involvement in gating of the intracellular side (12, 21) lacks experimental validation.
Here, we used site-directed spin labeling (SDSL) (24) and double electron-electron resonance (DEER) spectroscopy (25) to elucidate the conformational changes underlying alternating access in Mhp1 and define the role of ion and substrate binding in driving transition between conformations. This methodology has been successfully applied to define coupled conformational cycles for a number of transporter classes (13, 26–32). We find that patterns of distance distributions between pairs of spin labels monitoring the intra- and extracellular sides of Mhp1 are consistent with isomerization between the crystallographic inward- and outward-facing conformations. A major finding is that this transition is driven by substrate but not Na+ binding. Although the amplitudes of the observed distance changes are in overall agreement with the rocking-bundle model deduced from the crystal structures of Mhp1 (8, 15) and predicted computationally (16), we present evidence that relative movement of bundle and scaffold deviate from strict rigid body. Comparative analysis of LeuT and Mhp1 alternating access reveal how the conserved LeuT fold harnesses the energy of the Na+ gradient through two distinct coupling mechanisms and supports divergent conformational cycles to effect substrate binding and release.
Results
Approach.
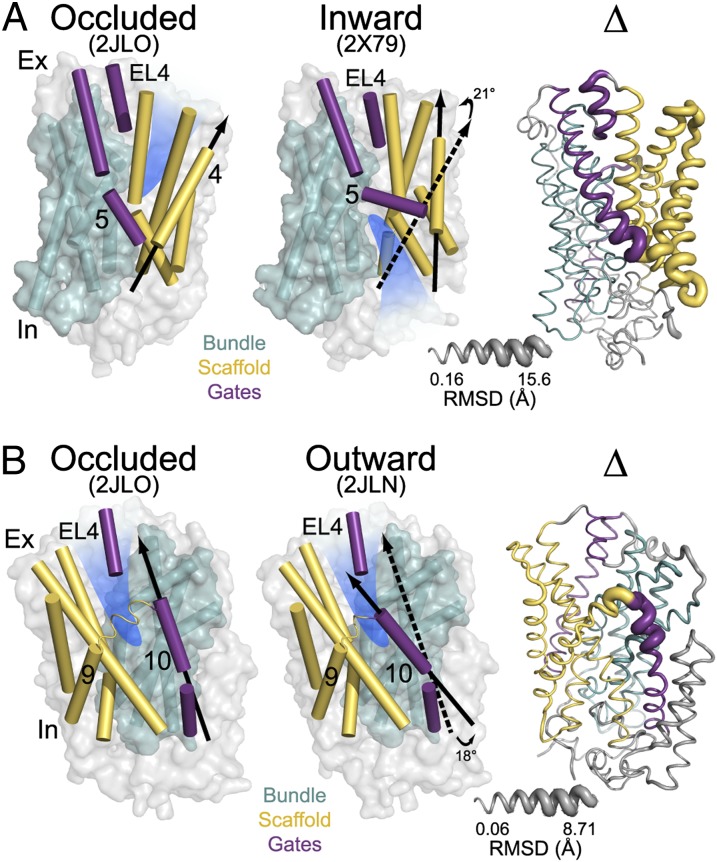
To describe the geometric transformation relating the crystallographic inward-facing, outward-facing/occluded, and outward-facing conformations, the Mhp1 structure was divided into two domains: the bundle consisting of TM1, -2, -6, and -7 (colored cyan, Fig. 1) and the scaffold (colored yellow, Fig. 1) consisting of TM3, -4, -8, and -9. The inward- and outward-facing conformations, each defined by the orientation of the substrate-binding site (highlighted in blue in Fig. 1), differ by a rigid body rotation of the bundle relative to the scaffold (Fig. 1A). The outward-facing and outward-facing/occluded (hereafter referred to as occluded) conformations are distinguished by the position of extracellular TM10 (colored purple in Fig. 1B). The closed position of TM10 partially occludes access to the extracellular side; therefore, TM10 has been referred to as a “thin gate.” The symmetrically positioned TM5 (colored purple in Fig. 1A) participates in the occlusion of the substrate-binding site from the intracellular side as an equivalent thin gate to TM10 (8). Analysis of the rmsd between the outward-facing, occluded, and inward-facing structures outlines the regions of inferred conformational changes (Fig. 1, ribbon structure labeled Δ).
Fig. 1.
(A) Comparison of outward-facing (2JL0) and inward-facing crystal structures (2X79) of Mhp1 highlighting the inferred movement of the scaffold (yellow) relative to the bundle (cyan). The rmsd between the two structures is displayed by the ribbon thickness (scale in gray) on the 2JLO backbone (Δ). The change in orientation of the scaffold between the two structures is shown by the arrows. The approximate location of the solvent accessible vestibule is indicated in blue. (B) Comparison of outward-facing conformation (2JLO) and outward-facing/occluded conformation (2JLN) highlighting the inferred movement of TM10.
To investigate the ligand-dependent conformational equilibrium of Mhp1, we measured distance distributions between spin-label pairs in detergent micelles under different ligand conditions that would be expected to promote transitions between intermediates in the transport cycle. We verified that the substitution of the native cysteines (residues 69, 234, and 327) by alanines does not compromise Na+ and BH binding with measured affinities of the cysteine-free variant in the range of those reported for the WT (15) (Fig. S1 A and B). Furthermore, all spin-labeled Mhp1 mutants reported here bind BH (Fig. S1C).
Binding of Na+/BH Induces Large Conformational Transitions.
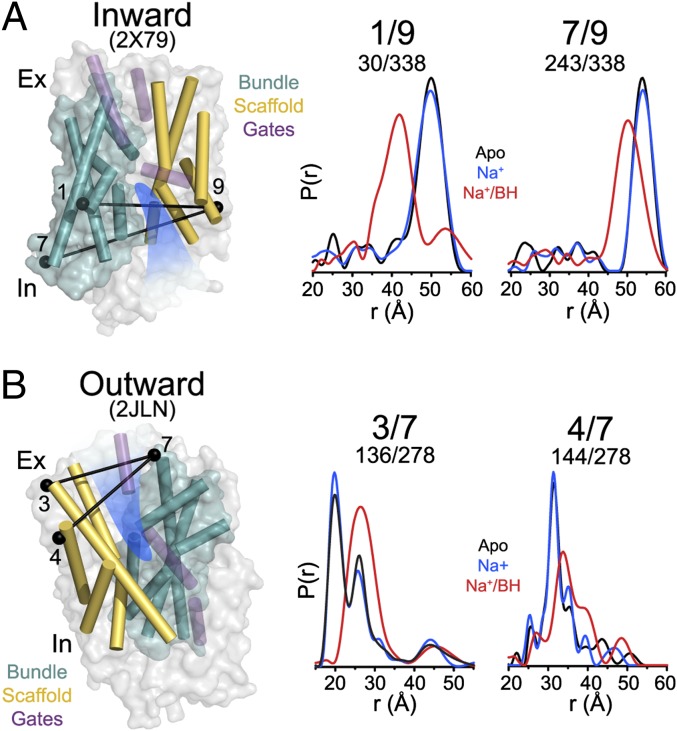
Distance distributions for spin-label pairs monitoring the TMs in the bundle relative to TMs in the scaffold reveal large-amplitude conformational changes upon concurrent Na+ and BH binding. On the intracellular side, addition of Na+ and BH to apo Mhp1 shifts the average distances between TM1 and -7 in the bundle and TM9 in the scaffold by 7–10 Å (Fig. 2A). The direction of the distance change suggests that binding of Na+/BH induces the movement of TM1 and TM7 toward TM9 consistent with closing of the intracellular vestibule. Similarly, distributions monitoring TMs on the extracellular side report changes in the relative population of distinct distance components. Specifically, binding of Na+/BH favors the distance components with larger separation between bundle and scaffold helices compared with apo (e.g., 3/7 in Fig. 2B). Thus, the pattern of distance changes reported by intracellular and extracellular pairs is consistent with an Na+/BH-induced transition from an inward-facing conformation to an outward-facing or an occluded conformation.
Fig. 2.
Monitoring ligand-dependent conformational changes of Mhp1. Distance distributions depicting the probability of a distance P(r) versus distance (r) between spin labels and reporting the ligand-dependent movement of the scaffold relative to the bundle on the intracellular (A) and extracellular (B) sides. The locations of the spin-label pairs are highlighted by black spheres connected by a line. Distance distributions for each pair were obtained in the apo, Na+-bound (Na+), and Na+- and BH-bound (Na+/BH) intermediates.
On the intracellular side, distance distributions between TMs within the bundle or TMs within the scaffold were similar in apo and Na+/BH-bound states (Fig. S2), suggesting little, if any, relative movement within these structural units. On the extracellular side, ligand-induced changes in distance distributions were observed between TMs within bundle and the scaffold suggesting deviation from strict rigid-body motion (see Fig. 4).
Fig. 4.
Conformational dynamics of the Mhp1 extracellular thin gates. (A) Distributions monitoring the distance from TM10 to bundle helices are consistent with a dynamic TM10 but do not reveal a change in its overall disposition as predicted from the occluded crystal structure. (B) Distance distributions between TM9 and helices in the scaffold (TM3) and bundle (TM9) have a broader width upon substrate binding, suggesting increased dynamic of TM9 and deviation from rigid-body movement in the scaffold. (C) Gating of the extracellular vestibule by EL4 is revealed by the substrate-dependent changes in distance distributions to the scaffold and the bundle. The asterisk denotes distance components arising from a minor aggregated population of Mhp1.
Mhp1 Substrate-Induced Conformational Changes Support the Rocking-Bundle Mechanism.
To quantitatively evaluate the relationship between experimental distance distributions and the various crystal structures, we calculated the distance distributions expected based on these structures using molecular dynamics simulations [molecular dynamics of dummy spin labels (MDDS)] (Methods) (33). For this purpose, we introduced dummy spin labels at the sites of interest and carried out MD simulations while fixing the protein backbone (33). Comparison of experimental and predicted distributions (Fig. S3) reveals that the Na+/BH-bound intermediate corresponds to the outward-facing or occluded crystal structures (15) wherein the intracellular vestibule is closed. Although the breadth of the experimental distance distributions is not consistent with a single conformation, we estimated the amplitude of the conformational changes from comparison of average distances between apo and Na+/BH distributions. The 7- to 10-Å distance change evident on the intracellular (TMs 1/9 and 1/7) and extracellular (TMs 3/7) sides is remarkably consistent with that predicted by comparing the Mhp1 inward- and outward-facing crystal structures (8, 15), supporting the conclusion that these structures are sampled in solution. This evidence alleviates the possibility of distortion associated with the crystal lattice forces and detergent selection, while leaving open the question of how the lipid bilayers and an imposed gradient shape conformational sampling.
Mhp1 Is in a Conformational Equilibrium.
The experimental distributions profile a transporter in equilibrium between multiple states under all biochemical conditions (Figs. 2–4 and Fig. S4). This equilibrium is manifested by the bimodal distributions of the spin-label pairs on the extracellular side where the two components correspond to the distributions predicted from the inward- and outward-facing crystallographic conformations (Fig. 2B and Fig. S3). Similarly, distributions for pairs on the intracellular side overlap in the presence of Na+/BH, which favors an outward-facing conformation, and under apo conditions, which favor an inward-facing conformation. Thus, Mhp1 populates an open-in conformation in the Na+/BH-bound intermediate although with low probability, providing a plausible mechanism for substrate release.
Na+ Binding Does Not Shift Mhp1 Conformational Equilibrium.
A role of Na+ in driving conformational changes of Mhp1 was inferred from the crystal structures (12), supported by MD simulations and reinforced by free-energy calculations (21) suggesting that Na+ binding favors the outward-facing conformation. These conclusions are consonant with MD simulations of LeuT that linked binding of Na+ at the Na2 site to population of the outward-facing conformation (20) and release of Na+ to the population of the inward-facing conformation (34) although there is no direct data that link binding at each Na+ site to the stabilization of a particular conformation. Contrary to expectations originating from crystal structures and computational models (12, 21), distance distributions of Mhp1 bound to Na+ are superimposable on those obtained under apo conditions, suggesting that the energetics of Mhp1 equilibrium between inward-facing and outward-facing conformations are not substantially affected by Na+ binding at the Na2 site (Fig. 2 and Fig. S1). In contrast, as noted in Fig. 2, the binding of the substrate BH in the presence of Na+ shifts the equilibrium to favor the outward-facing conformation (Fig. 2).
“Thin” Dynamic Gates in the Inward- to Outward-Facing Transition.
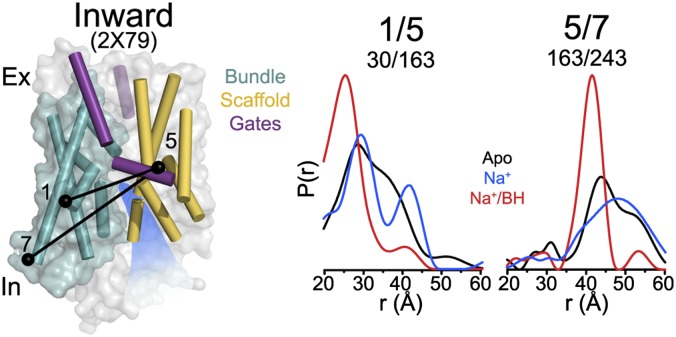
The crystal structures (8, 15) implied changes in the spatial disposition of TM5 and -10 to occlude the substrate-binding site (Fig. 1). To test this inference, we monitored the ligand-dependent movements of these TMs in the three conditions outlined in Fig. 2. At the intracellular side, distance distributions between TM5 and the bundle are broad in the apo state, suggesting a highly dynamic TM5 (Fig. 3). Binding of Na+/BH narrows the distance distributions, selecting for a subset of short distances that are consistent with the occluded and outward-facing conformations. Comparison with predicted distributions (Fig. S3) suggests that, in the apo state, TM5 samples conformations consistent with both open-in and closed-in conformations whereas its dynamics are restricted upon substrate binding. TM5 distance distributions are significantly broader than those monitoring the relationship between the bundle and scaffold motifs, indicative of a higher degree of flexibility, particularly in the inward-facing conformation.
Fig. 3.
Conformational dynamics of TM5. Distance distributions monitoring the distance between TM5 and bundle helices have broad widths indicative of a highly dynamic TM5 in the apo state. Binding of substrate restricts sampling by TM5 to a population consistent with the outward-facing conformation.
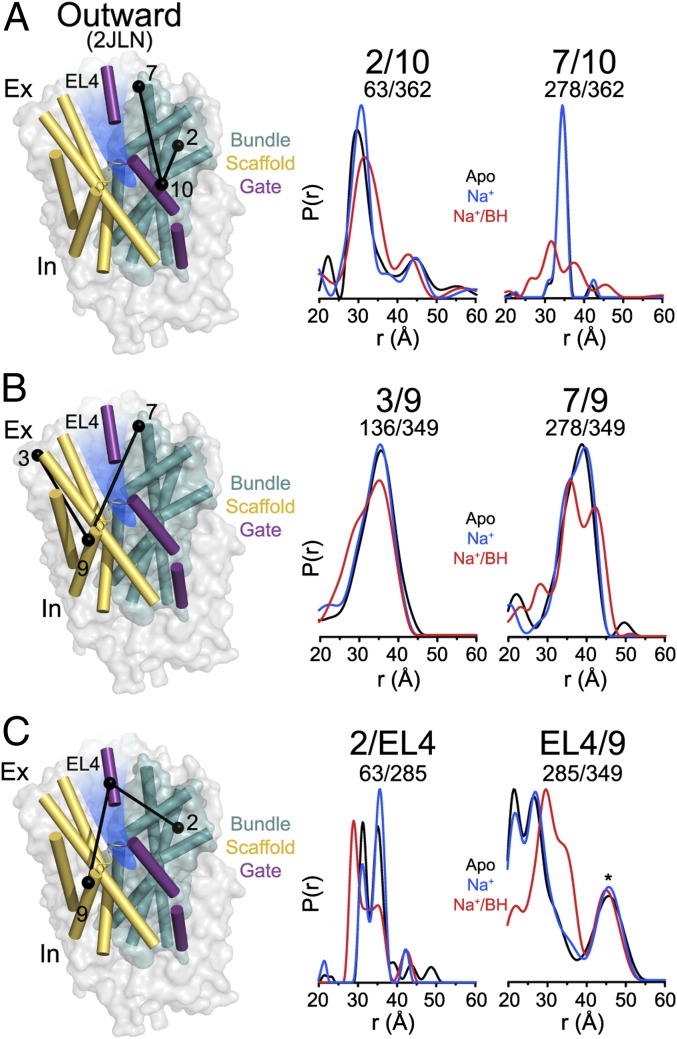
A substantial shift in the position of extracellular TM10 between the outward-facing occluded and outward-facing conformations (Fig. 1B) was interpreted as reflecting substrate-induced occlusion of the substrate-binding site (15). To test this interpretation, distance distributions relating extracellular TM10 to TMs in the scaffold and bundle were determined under different ligand conditions. We observed no evidence of the discrete change in TM10 position (Fig. 4A) predicted by comparison of the crystal structures (Fig. 1B and Fig. S3). Rather, binding of Na+ and BH increases the width of the distance distributions, suggesting an increase in flexibility of TM10 in the outward-facing conformation (Fig. 4A), similar in principle to that shown for TM5 under apo conditions (Fig. 3).
We identified a similar dynamic profile in the extracellular part of TM9, which is connected to TM10 by a short helix. Although movements in this region of TM9 were not evident from comparison of the Mhp1 crystal structures (Fig. 1B), MD simulations (8) predicted that TM9 may be coupled to TM10 fluctuations. Consistent with this prediction, distance distributions on the extracellular side describing the spatial relation of TM9 to TM3, both helices in the scaffold, show that TM9 deviates from the presumed rigid-body motion of the scaffold (Fig. 4B). In the presence of Na+ and BH, the breadth of the TM9 distributions increases, indicating an increase in conformational sampling, but does not suggest discrete alternative conformations as would be expected of a scaffold helix relative to the bundle motif (Fig. 4B). The similarity between the TM9 and TM10 profiles (specifically, TMs 2/10 and 3/9) leads us to conclude that the motions of TM9 and -10 may be coupled.
EL4, the extracellular loop between TM7 and -8 consisting of a single helix in Mhp1, was not identified as a gate element although a small-scale change in its position was evident in a comparison of the outward-facing (15) and inward-facing (8) structures (Fig. 1A). Multicomponent distance distributions for this motif suggest that conformations are in equilibrium (Fig. 4C). Relative to TM2 of the bundle, two populations are observed in the apo and Na+-bound states whereas, in the presence of Na+ and BH, a new shorter distance is observed. A similar multicomponent profile is observed in distance distributions relating EL4 to the scaffold. Na+/BH binding induces a movement of EL4 away from TM9 consistent with the distance change relative to TM2. Although the EL4/TM9 distribution is somewhat complicated by the independent dynamics of TM9 relative to the bundle, the amplitude of the distance change is substantially larger than that observed for TM9. We conclude that EL4 participates in regulation of extracellular occlusion and substrate access. We speculate that the bimodal distributions of EL4 (apo and Na+-bound) may correspond to the bimodal distributions shown in Fig. 2B reflecting the inward-facing and outward-facing positions of the bundle and scaffold. The new distance component observed in the presence of Na+ and BH may represent a distinct occluded conformation.
Discussion
Mhp1 isomerization between inward- and outward-facing conformations follows, in outline, the model deduced from comparison of the inward- and outward-facing crystal structures (8) and predicted from modeling (16, 35). The results presented here describe the ligand-dependent equilibrium between these conformations, reveal that substrate but not Na+ binding changes the energetics of these conformations, and indicate that the bundle and scaffold motifs are not strictly rigid bodies. In conjunction with a previous investigation of LeuT (13), this work illuminates commonalities and differences in the structural mechanism of alternating access in the LeuT-fold class of ion-coupled transporters.
The Distinct Role of Na+ in Mhp1 and LeuT Alternating Access.
Symported ions can drive directional movement of substrates by coordination of the substrate in the binding site and/or conformational selection to facilitate substrate access/release from the binding site. Direct coordination stabilizes the bound substrate and enables ion gradients to impose directionality on substrate transport by promoting ion binding on one side of the membrane and ion dissociation on the other side. In addition, ion binding can directly alter the energetics of the conformational equilibrium to favor conformations that provide access to the substrate-binding site. A comparative analysis of the Mhp1 and LeuT Na+-dependent conformational changes illustrates an example of these two mechanisms of ion coupling and allows us to speculate as to the roles of the Na1 and Na2 sites.
In LeuT, which possesses both Na1 and Na2, we previously described Na+-dependent conformational transitions wherein binding of Na+ favors an outward-facing conformation (13) that increases water accessibility in the substrate permeation pathway (13, 22). In contrast, we find here that Na+ does not affect the equilibrium between inward- and outward-facing conformations of Mhp1. However, binding of Na+ at Na2 stabilizes substrate binding as evident in decreased KD of substrate binding (15) (Fig. S1). Thus, we speculate that the conserved Na2 site serves to couple transport of substrate to the Na+ gradient through direct stabilization of substrate binding but does not necessarily drive conformational changes between inward- and outward-facing conformations. We propose that the additional mechanism of coupling to the Na+ gradient in LeuT, which achieves conformational selection, is a consequence of the presence of the nonconserved Na1 site in addition to the Na2 site. A role of Na+ as a conformational trigger may represent a critical mechanistic divergence between classes of LeuT-fold transporters with Na+/substrate stoichiometry of 1 versus 2. Without this site, Mhp1 symport is achieved via thermodynamically coupled binding and release of Na+ and BH and equilibrium fluctuations between conformational states.
Mhp1 Transport Cycle Is Dependent on Low-Probability Transitions.
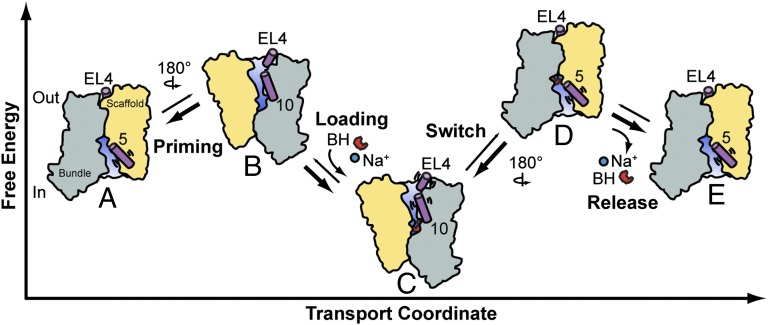
By framing Mhp1 crystal structures into a ligand-dependent conformational equilibrium, we derived a plausible model of how isomerization of the transporter between inward-facing and outward-facing conformations mediates transport (Fig. 5). Based on the dissociation constants (Fig. S1A) (15), separate binding of Na+ and substrate is unlikely to occur in a substantial fraction of transporters. Mutual stabilization of Na+ and BH implies that they concurrently bind to the minor population of outward-facing apo-Mhp1 where the substrate-binding site is accessible from the extracellular side (Fig. 5 A and B). Binding of Na+/BH favors the outward-facing conformation (Figs. 2 and 5C), but the distance distributions in the Na+/BH-bound state overlaps those of the apo state (specifically, TMs 7/9 and 4/7), consistent with Na+/BH-bound Mhp1 fluctuating to an open-in conformation from which Na+ could dissociate to the intracellular side in the presence of a gradient (Fig. 5D). Na+ release on the intracellular side lowers substrate affinity and therefore would be expected to facilitate its release (Fig. 5E). The low probability of fluctuation to the inward-facing conformation, implied by low population in the distance distribution, may define the rate-limiting step of transport (Fig. 5 C and D). After release, a new cycle of transport is initiated by the isomerization of the apo intermediate between open-in and open-out conformations (Fig. 5 A and B). This model of Mhp1 transport is distinct from previous models by the absence of an ion-dependent isomerization from inward- to outward-facing conformations and the postulated substrate-induced transition from outward-facing to inward-facing conformations (12). Consequently, it emphasizes the role of conformational sampling in the transport mechanism.
Fig. 5.
Model of Mhp1 transport. Apo-Mhp1 (A), through low probability transitions, samples outward-facing conformations (priming, B), allowing the simultaneous binding of the Na+ and substrate (loading), and resulting in a stabilization of the outward-facing conformation (C). Low-probability fluctuations allow sampling of the inward-facing conformation (switch, D) where, driven by its concentration gradient, Na+ dissociates to the intracellular solution (release). In the absence of bound Na+, BH affinity to Mhp1 is reduced, which drives dissociation of BH to the intracellular side (E). The cycle continues through the isomerization of apo Mhp1 from inward-facing (A) to outward-facing (B).
The lack of an Na+-induced shift in the equilibrium between outward- and inward-facing conformations does not exclude the possibility that Na+ induces local changes that inhibit its release to the intracellular side in the absence of substrate. Trapping of Na+ may be critical to inhibit uncoupled ion leakage that would short the gradient. Within the outward-facing or inward-facing ensembles of conformational states, there are likely to exist yet other substates in which side-chain rearrangements stabilize ligands or induce their release as was observed in BetP (10). They are not differentiated by large-amplitude conformational changes; therefore, their existence would be obscured by the inherent limitations of probe-based methods such as EPR. Previous MD simulations demonstrated that the presence of BH blocks the pathway of Na+ dissociation to the extracellular side (8). Extracellular occlusion may also be aided by a change in the position of EL4 (purple cylinder, Figs. 4C and 5).
The Divergent Transport Mechanisms of Mhp1 and LeuT.
Previous analysis of LeuT conformational changes demonstrated that the opening/closing of two extracellular motifs, consisting of TM1/TM6 and TM7/EL4, and an intracellular motif, consisting of TM6/TM7, mediate LeuT alternating access, with TM6 and -7 coupling the intracellular and extracellular sides of the transporter (13). This mechanism stands in stark contrast to the Mhp1 conformational cycle outlined in Fig. 5. Specifically, Mhp1 operates through a rocking-bundle mechanism that couples to the Na+ gradient through direct coordination of substrate by Na+. When bound to Na+/Leu, LeuT favors an occluded conformation, with all motifs in their closed conformations simultaneously whereas, in Mhp1, occlusion occurs through gates that are dynamic and, in the case of EL4, undergo ligand-dependent movement relative to both bundle and scaffold motifs.
A novel perspective on the functional diversity of the LeuT fold emerges from comparative analysis of LeuT and Mhp1. Even within a fold, transport of diverse substrates executed by different coupling modes, number and identity of the cotransported ion, coupled with high sequence heterogeneity among transporters, entails distinct structural schemes of alternating access. Furthermore, the work illuminates a critical mechanistic element that has been missing from the analysis thus far: namely, how ion binding shapes the energy landscape of conformations. Although we speculate, based on our results, that the role of the conserved Na2 site is to stabilize substrate binding without conformational selection, analysis of other Na+-coupled transporters is needed to test this conjecture. Kinetic and conformational investigations of putative LeuT-fold members, hSGLT and PutP, have begun to tease out such details (23, 36, 37). Such analysis will also test a similarly tantalizing notion that subclasses of ion-coupled LeuT-fold transporters, defined by their transport modes and/or type and number of symported ions, share commonalities in their structural mechanics of alternating access.
Methods
Mutagenesis, Expression, Purification, and Labeling of Mhp1.
The Mhp1 construct was engineered to be cysteine-free (C69A, C234A, and C327A) with a C-terminal decahistidine tag. Cysteine residues were introduced into the cysless construct using site-directed mutagenesis (24) and confirmed by DNA sequencing. Mhp1 mutants were expressed in Escherichia coli C43(DE3) by induction with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and shaken at 25 °C for 16 h. Mhp1 was extracted from native membranes in 2% (wt/vol) n-dodecyl-β-d-maltoside (β-DDM) in 50 mM Tris⋅HCl, 200 mM NaCl, and 20% (vol/vol) glycerol at pH 7.5 before purification by Ni2+ affinity chromatography. The protein was spin-labeled and purified using the same protocol as previously described for LeuT (13) in a buffer consisting of 50 mM Tris-Mes, 0.05% (wt/vol) β-DDM, and 20% (vol/vol) glycerol at pH 7.2. Protein concentration was determined using an extinction coefficient of 1.84 cm2⋅mg−1 at 280 nm. Purified Mhp1 were concentrated with Amicon Ultra columns (100 kDa; Millipore). Samples for DEER spectroscopy were prepared in the 50–200 μM protein concentration range. A final concentration of glycerol of 30% (wt/vol) was used in all samples as a cryoprotectant. The Na+ state was obtained by addition of 200 mM NaCl. The Na+/BH state was obtained through addition of 5 mM 5-BH (Toronto Research Chemicals) and 200 mM NaCl.
Mhp1 Functional Analysis.
Binding of Na+ and BH to purified and spin-labeled mutant Mhp1 protein was monitored using a Trp fluorescence quenching assay (15). Measurements were conducted using 2.5 µM Mhp1 in 50 mM Tris-Mes, pH 7.2, 0.05% (wt/vol) β-DDM, and 20% (vol/vol) glycerol containing buffer at room temperature. Complete Na+ and BH binding curves for the cysless mutant were also obtained using varying concentrations of NaCl between 0 mM and 10 mM and BH between 0 mM and 2 mM. Curves were fitted, and KD was determined using the nonlinear curve fit, one site binding function with no weights in Origin 8 (OriginLab). Static concentrations of 15 mM NaCl and 2 mM BH were used in additional binding curves to measure mutual ligand affinity stabilization and in ligand-binding experiments for all mutants. Samples were excited at 285 nm, and fluorescence intensity was collected at 348 nm before and after addition of BH. The decrease in fluorescence is expressed as a percentage of peak height.
DEER Spectroscopy.
Distance measurements were conducted on a Bruker 580 pulsed electron paramagnetic resonance (EPR) spectrometer operating at Q-band frequency (33.9 GHz) using a standard four-pulse DEER sequence as previously described (38). All DEER experiments were performed at 83 K. Dipolar evolution times were designed to allow us to identify background slope, when possible. For long distances (>50 Å), dipolar evolution times can be insufficient, resulting in some uncertainty in the widths of the distance distributions. Echo decays were background-corrected and fit with the DEER Analysis 2011 program (39) using Tikhonov regularization (40) to obtain distance distributions. Aggregated protein, resulting from concentration and validated by gel electrophoresis, appears in some samples as a nonspecific peak near 50 Å.
Molecular Dynamics of Dummy Spin Label Attached to Mhp1.
All molecular dynamic simulations of the spin-labeled Mhp1 were carried out with the CHARMM (41) program package, using the all-atom CHARMM27 protein force field (42) with the CMAP corrections and the dummy nitroxide (ON) spin-label force field parameters (33). Three crystal structures of Mhp1 [2JLN (15), 2JLO (15), and 2X79 (8)] were used to construct the geometries of the Mhp1 systems for simulation. The dummy ON spin-labels were linked directly to the Cα atoms of the protein backbone at all residues included in the DEER mutant dataset. All of the ON dummy spin labels were introduced simultaneously into a single protein for a long molecular-dynamics simulation. However, long side-chain residues were truncated after the Cβ atom to avoid steric clashes with the ON labels. All of the simulations were performed under vacuum at 300 K using the Langevin (43) thermostat with a collision frequency of 10.0 ps−1. To begin, an Adopted Basis Newton-Raphson (ABNR) energy minimization (100 steps) and a short (10 ps) molecular dynamics simulation of the dummy ON spin labels were performed by fixing the coordinates of all other atoms of the protein to its X-ray crystallographic structure. Using the same simulation criteria, side-chain atoms were allowed freedom of motion in a second simulation. Finally, a 1-ns equilibration simulation and a 4-ns production MD simulation were performed fixing the protein and using a timestep of 2 fs from which the spin-pair distance distributions were calculated.
Supplementary Material
Acknowledgments
We thank Richard Stein for assistance with EPR data collection and distance analysis, and Jamie Wenke for contributions to collection of preliminary experimental data. We thank P. Ryan Steed and Derek Claxton for critical reading and editing of the manuscript. We thank Extreme Science and Engineering Discovery Environment for computer time. This work was supported by National Institutes of Health Grants U54-GM087519 (to H.S.M., S.M.I., and B.R.) and S10RR027091 (to H.S.M.). K.K. was supported by predoctoral National Research Service Award F31-MH095383-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1410431111/-/DCSupplemental.
References
- 1.Mitchell P. A general theory of membrane transport from studies of bacteria. Nature. 1957;180(4577):134–136. doi: 10.1038/180134a0. [DOI] [PubMed] [Google Scholar]
- 2.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211(5052):969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 3.Patlak CS. Contributions to the theory of active transport. II. The gate type non-carrier mechanism and generalizations concerning tracer flow, efficiency, and measurement of energy expenditure. Bull Math Biophys. 1957;19(3):209–235. [Google Scholar]
- 4.Krishnamurthy H, Piscitelli CL, Gouaux E. Unlocking the molecular secrets of sodium-coupled transporters. Nature. 2009;459(7245):347–355. doi: 10.1038/nature08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh SK. LeuT: A prokaryotic stepping stone on the way to a eukaryotic neurotransmitter transporter structure. Channels (Austin) 2008;2(5):380–389. doi: 10.4161/chan.2.5.6904. [DOI] [PubMed] [Google Scholar]
- 6.Wright EM, Loo DD, Hirayama BA. Biology of human sodium glucose transporters. Physiol Rev. 2011;91(2):733–794. doi: 10.1152/physrev.00055.2009. [DOI] [PubMed] [Google Scholar]
- 7.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481(7382):469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimamura T, et al. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science. 2010;328(5977):470–473. doi: 10.1126/science.1186303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe A, et al. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature. 2010;468(7326):988–991. doi: 10.1038/nature09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez C, Koshy C, Yildiz O, Ziegler C. Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature. 2012;490(7418):126–130. doi: 10.1038/nature11403. [DOI] [PubMed] [Google Scholar]
- 11.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325(5943):1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weyand S, et al. The alternating access mechanism of transport as observed in the sodium-hydantoin transporter Mhp1. J Synchrotron Radiat. 2011;18(1):20–23. doi: 10.1107/S0909049510032449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kazmier K, et al. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat Struct Mol Biol. 2014;21(5):472–479. doi: 10.1038/nsmb.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McHaourab HS, Steed PR, Kazmier K. Toward the fourth dimension of membrane protein structure: Insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19(11):1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weyand S, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322(5902):709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forrest LR, Rudnick G. The rocking bundle: A mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 2009;24:377–386. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl-dependent neurotransmitter transporters. Nature. 2005;437(7056):215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 18.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature. 2009;458(7234):47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 19.Faham S, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321(5890):810–814. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao C, et al. Ion-controlled conformational dynamics in the outward-open transition from an occluded state of LeuT. Biophys J. 2012;103(5):878–888. doi: 10.1016/j.bpj.2012.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao C, Noskov SY. The molecular mechanism of ion-dependent gating in secondary transporters. PLOS Comput Biol. 2013;9(10):e1003296. doi: 10.1371/journal.pcbi.1003296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claxton DP, et al. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat Struct Mol Biol. 2010;17(7):822–829. doi: 10.1038/nsmb.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sala-Rabanal M, et al. Bridging the gap between structure and kinetics of human SGLT1. Am J Physiol Cell Physiol. 2012;302(9):C1293–C1305. doi: 10.1152/ajpcell.00397.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hubbell WL, Mchaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4(7):779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 25.Jeschke G, Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys. 2007;9(16):1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 26.Masureel M, et al. Protonation drives the conformational switch in the multidrug transporter LmrP. Nat Chem Biol. 2014;10(2):149–155. doi: 10.1038/nchembio.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smirnova I, et al. Sugar binding induces an outward facing conformation of LacY. Proc Natl Acad Sci USA. 2007;104(42):16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishra S, et al. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife. 2014;3:e02740. doi: 10.7554/eLife.02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph B, Korkhov VM, Yulikov M, Jeschke G, Bordignon E. Conformational cycle of the vitamin B12 ABC importer in liposomes detected by double electron-electron resonance (DEER) J Biol Chem. 2014;289(6):3176–3185. doi: 10.1074/jbc.M113.512178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hellmich UA, et al. Probing the ATP hydrolysis cycle of the ABC multidrug transporter LmrA by pulsed EPR spectroscopy. J Am Chem Soc. 2012;134(13):5857–5862. doi: 10.1021/ja211007t. [DOI] [PubMed] [Google Scholar]
- 31.Georgieva ER, Borbat PP, Ginter C, Freed JH, Boudker O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat Struct Mol Biol. 2013;20(2):215–221. doi: 10.1038/nsmb.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hänelt I, Wunnicke D, Bordignon E, Steinhoff HJ, Slotboom DJ. Conformational heterogeneity of the aspartate transporter Glt(Ph) Nat Struct Mol Biol. 2013;20(2):210–214. doi: 10.1038/nsmb.2471. [DOI] [PubMed] [Google Scholar]
- 33.Islam SM, Stein RA, McHaourab HS, Roux B. Structural refinement from restrained-ensemble simulations based on EPR/DEER data: application to T4 lysozyme. J Phys Chem B. 2013;117(17):4740–4754. doi: 10.1021/jp311723a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L, Weinstein H. Conformational rearrangements to the intracellular open states of the LeuT and ApcT transporters are modulated by common mechanisms. Biophys J. 2010;99(12):L103–L105. doi: 10.1016/j.bpj.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudnick G. Cytoplasmic permeation pathway of neurotransmitter transporters. Biochemistry. 2011;50(35):7462–7475. doi: 10.1021/bi200926b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loo DD, Hirayama BA, Karakossian MH, Meinild AK, Wright EM. Conformational dynamics of hSGLT1 during Na+/glucose cotransport. J Gen Physiol. 2006;128(6):701–720. doi: 10.1085/jgp.200609643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raba M, et al. Extracellular loop 4 of the proline transporter PutP controls the periplasmic entrance to ligand binding sites. Structure. 2014;22(5):769–780. doi: 10.1016/j.str.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142(2):331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 39.Jeschke G, et al. DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl Magn Reson. 2006;30(3-4):473–498. [Google Scholar]
- 40.Chiang YW, Borbat PP, Freed JH. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J Magn Reson. 2005;172(2):279–295. doi: 10.1016/j.jmr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 41.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 43.Adelman SADoll JD Generalized Langevin equation approach for atom/solid-surface scattering: General formulation for classical scattering off harmonic solids. J Chem Phys. 1976;64(6):2375–2388. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.